Prevalence and Antimicrobial Resistance of Escherichia coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the West Coast of Peninsular Malaysia
Abstract
1. Introduction
2. Results
2.1. Farm Demography
2.2. Prevalence of E. coli, Salmonella spp. and Vibrio sp.
2.3. Antibiotic Susceptibility According to Species of Bacteria from Farm-Raised Tilapia
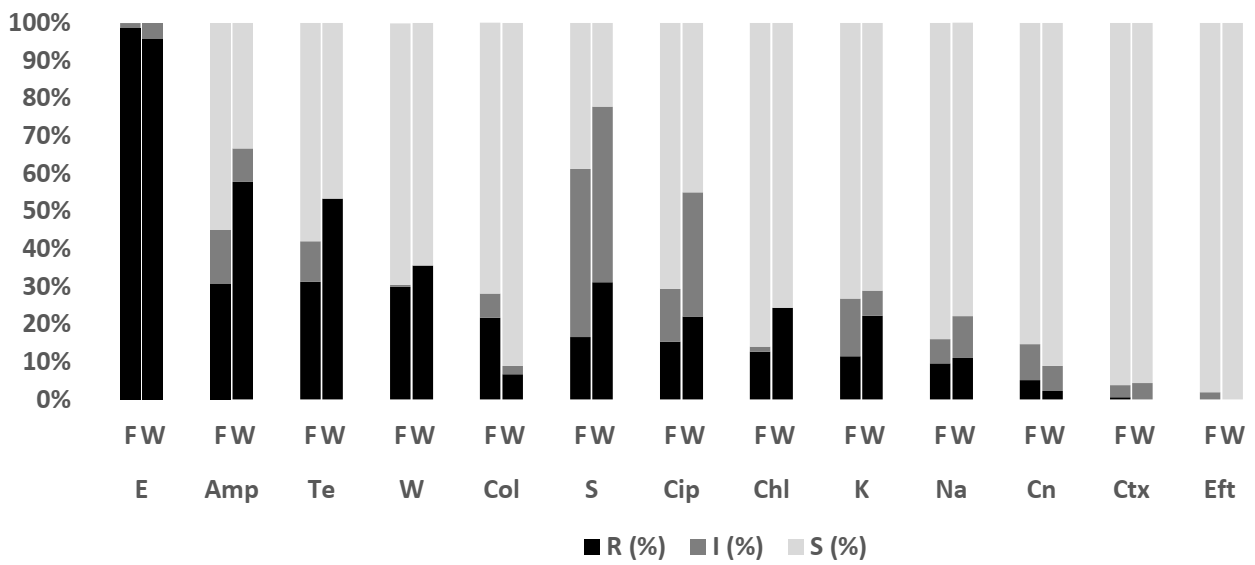
2.3.1. Escherichia coli
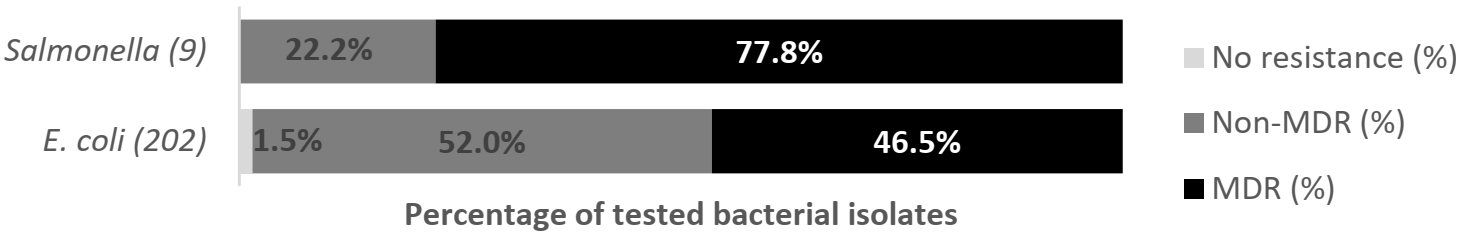
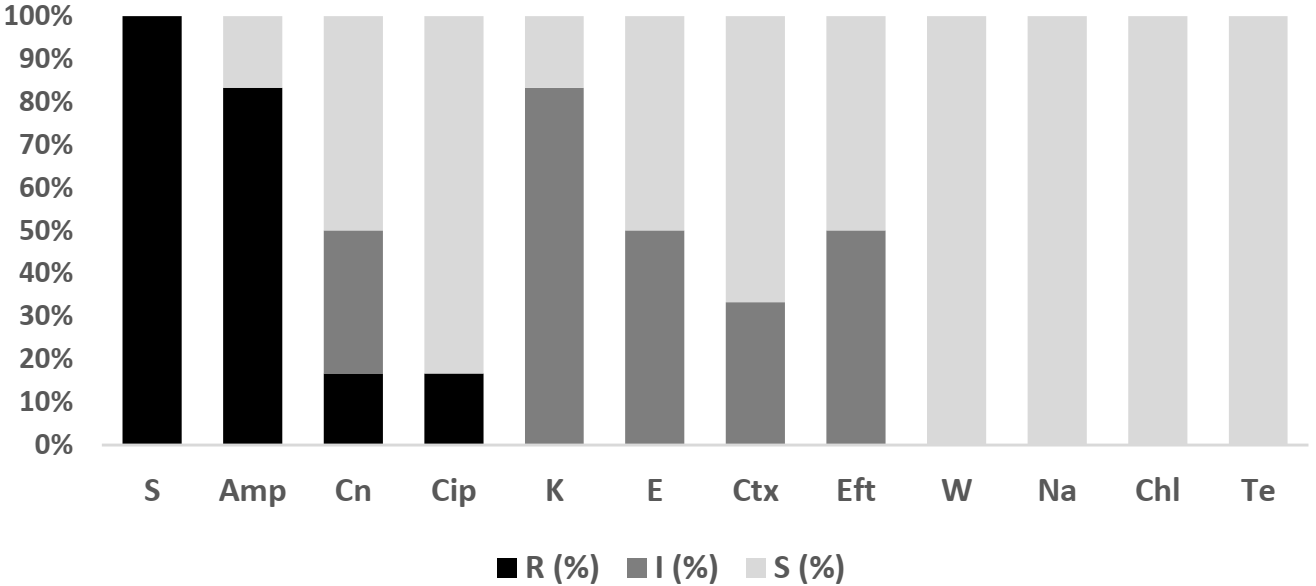
2.3.2. Salmonella spp.
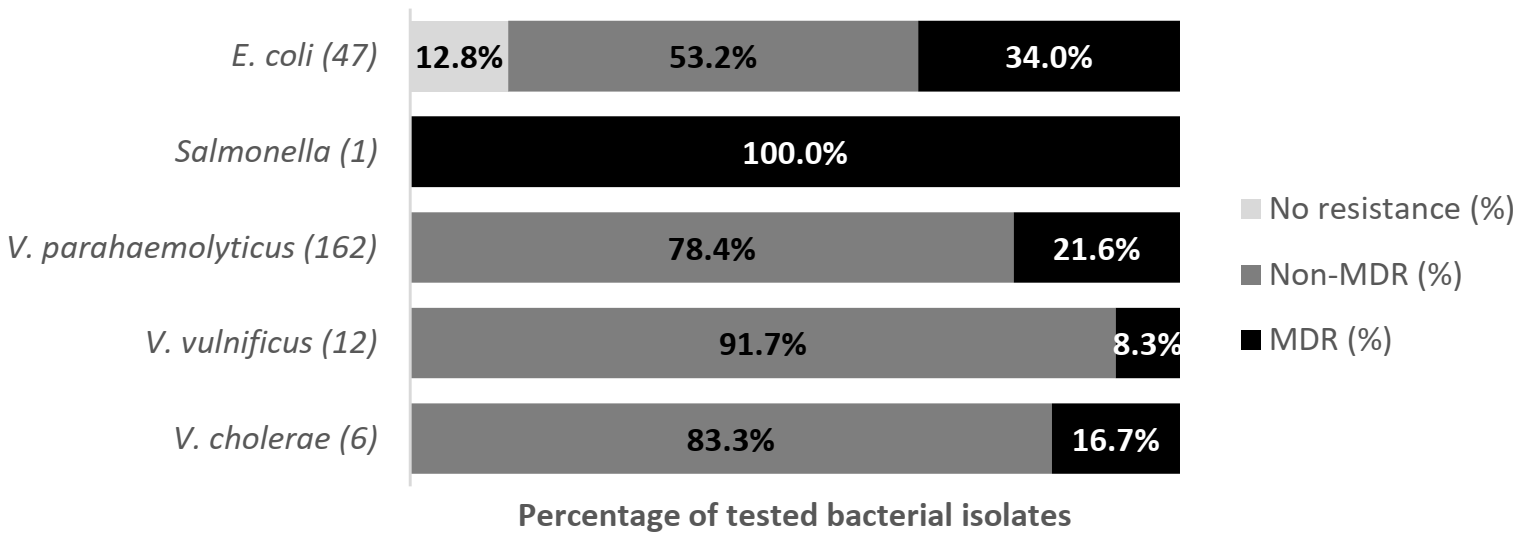
2.4. Antibiotic Susceptibility according to Species of Bacteria from Farm-Raised Asian Seabass
2.4.1. Escherichia coli
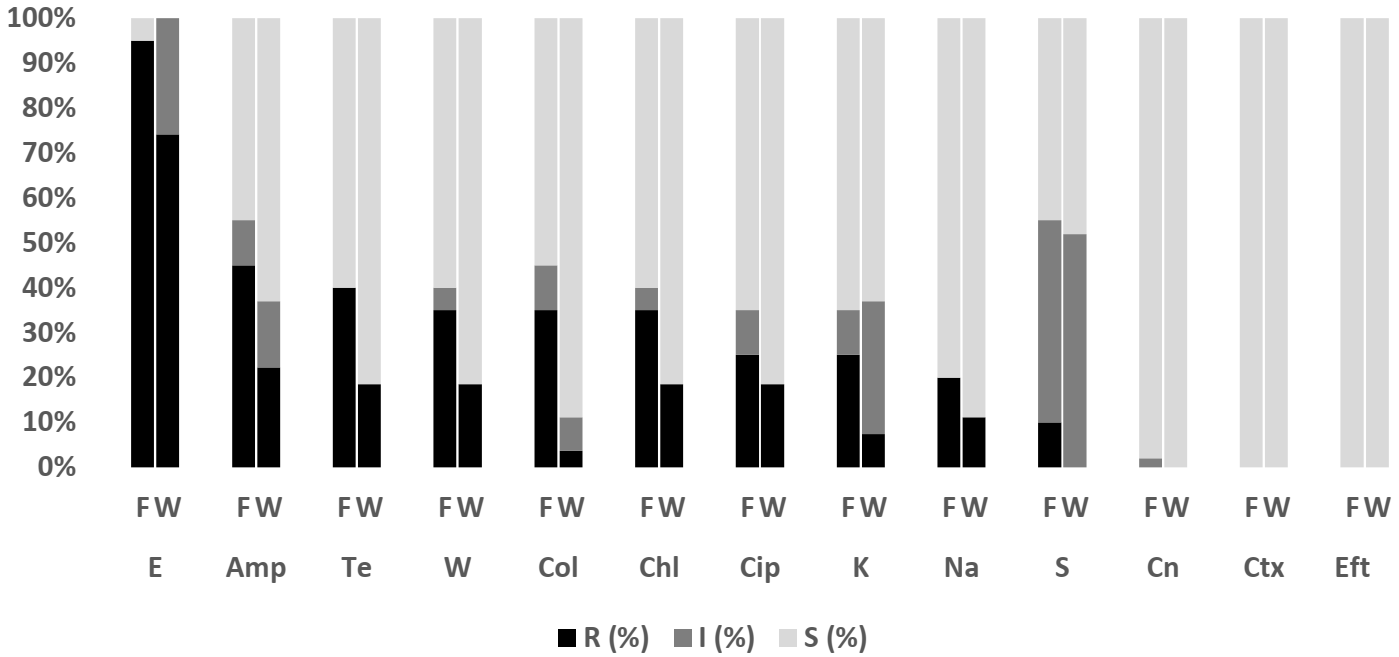
2.4.2. Salmonella spp.
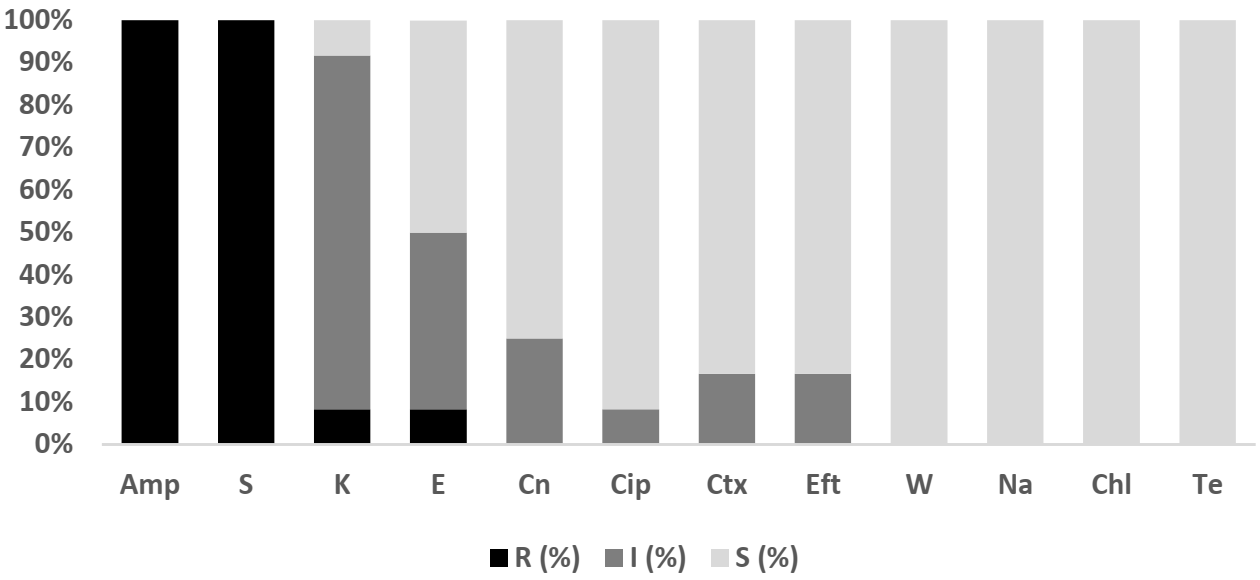
2.4.3. V. parahaemolyticus
2.4.4. V. vulnificus
2.4.5. V. cholerae
2.5. Differences between Resistance Profile of E. coli Isolates from Tilapia and Asian Seabass
2.6. Differences between Resistance Profile of E. coli Isolates from Aquaculture and Livestock
3. Discussion
3.1. E. coli
3.1.1. The Resistance Pattern of E. coli in Tilapia and Asian Seabass
3.1.2. The Comparison of Resistance with Livestock
3.2. Salmonella spp.
3.3. Vibrio sp.
3.4. The Resistance Pattern for E. coli in Tilapia and Asian Seabass
3.5. Resistance to Colistin
4. Materials and Methods
4.1. Study Areas
4.2. Sample Size
4.3. Study Design
4.4. Sample Collection from Farms
4.5. Isolation and Identification of E. coli
4.6. Isolation and Identification of Salmonella
4.7. Isolation and Identification of Vibrio
4.8. Antimicrobial Susceptibility Test
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lulijwa, R.; Rupia, E.J.; Alfaro, A.C. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: A review of the top 15 major producers. Rev. Aqua. 2020, 12, 640–663. [Google Scholar] [CrossRef]
- Noordin, W.N.M.; Misol, G., Jr.; Johari, R.; Science, A.F. Aquaculture Component of National Action Plan on Antimicrobial Resistance in Malaysia. Asian Fish. Sci. 2020, 33, 90–96. [Google Scholar] [CrossRef]
- Ng, W.-K. The current status and future prospects for the aquaculture industry in Malaysia. World Aqua. 2009, 40, 26–30. [Google Scholar]
- Yusoff, A. Status of Resource Management and Aquaculture in Malaysia. In Resource Enhancement and Sustainable Aquaculture Practices in Southeast Asia: Challenges in Responsible Production of Aquatic Species, Proceedings of the International Workshop on Resource Enhancement and Sustainable Aquaculture Practices, South Tigbauan, Iloilo, Philippines, 5–7 March 2014; Aquaculture Department, Southeast Asian Fisheries Development Center: Tigbauan, Philippines, 2015; pp. 52–65. [Google Scholar]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Smith, P. Antimicrobial resistance in aquaculture. Rev. Sci. Technol. Off. Int. Epizoot. 2008, 27, 243–264. [Google Scholar] [CrossRef]
- Feldhusen, F. The role of seafood in bacterial foodborne diseases. Microbes Infect. 2000, 2, 1651–1660. [Google Scholar] [CrossRef]
- Herrera, F.C.; Santos, J.A.; Otero, A.; García-López, M.L. Occurrence of foodborne pathogenic bacteria in retail prepackaged portions of marine fish in Spain. J. Appl. Microbiol. 2006, 100, 527–536. [Google Scholar] [CrossRef]
- Budiati, T.; Rusul, G.; Wan-abdullah, W.N.; Mat, Y. Prevalence, antibiotic resistance and plasmid profiling of Salmonella in catfish (Clarias gariepinus) and tilapia (Tilapia mossambica) obtained from wet markets and ponds in Malaysia. Aquaculture 2013, 372, 127–132. [Google Scholar] [CrossRef]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef]
- Le, H.V.; Kawahara, R.; Khong, D.T.; Tran, H.T.; Nguyen, T.N.; Pham, K.N.; Jinnai, M.; Kumeda, Y.; Nakayama, T.; Ueda, S.; et al. Widespread dissemination of extended-spectrum β-lactamase-producing, multidrug-resistant Escherichia coli in livestock and fishery products in Vietnam. Int. J. Food Contam. 2015, 2, 17. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Nguyen, T.A.D.; Le, T.H.; Tran, N.M.D.; Ngo, T.P.; Dang, V.C.; Kawai, T.; Kanki, M.; Kawahara, R.; Jinnai, M.; et al. Dissemination of Extended-Spectrum β -Lactamase- and AmpC β -Lactamase-Producing Escherichia coli within the Food Distribution System of Ho Chi Minh City, Vietnam. Biomed Res. Int. 2016, 2016, 8182096. [Google Scholar] [CrossRef] [PubMed]
- Haulisah, N.A.; Hassan, L.; Bejo, S.K.; Jajere, S.M.; Ahmad, N.I. High Levels of Antibiotic Resistance in Isolates from Diseased Livestock. Front. Vet. Sci. 2021, 8, 300. [Google Scholar] [CrossRef]
- Ramírez-Castillo, F.Y.; Moreno-Flores, A.C.; Avelar-González, F.J.; Márquez-Díaz, F.; Harel, J.; Guerrero-Barrera, A.L. An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: Cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 34. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalization of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- WHO. Global action Plan on Antimicrobial Resistance. 2015. Available online: https://apps.who.int/iris/handle/10665/193736 (accessed on 24 July 2021).
- FAO. The FAO Action Plan on Antimicrobial Resistance 2016–2020. 2016. Available online: https://www.fao.org/3/a-i5996e.pdf (accessed on 24 July 2021).
- Ministry of Health Malaysia; Ministry of Agriculture and Agro-Based Industry Malaysia. Malaysian Action Plan on Antimicrobial Resistance (MyAP-AMR) 2017–2021, 1st ed.; Ministry of Health Malaysia: Putrajaya, Malaysia, 2017; pp. 15–16. [Google Scholar]
- Amalina, N.Z.; Santha, S.; Zulperi, D.; Amal, M.N.A.; Yusof, M.T.; Zamri-Saad, M.; Ina-Salwany, M.Y. Prevalence, antimicrobial susceptibility and plasmid profiling of Vibrio spp. isolated from cultured groupers in Peninsular Malaysia. BMC Microbiol. 2019, 19, 251. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Rukayadi, Y.; Hasan, H.; Thung, T.Y.; Lee, E.; Rollon, W.D.; Hara, H.; Kayali, A.Y.; Nishibuchi, M.; Radu, S. Prevalence and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from different types of seafood in Selangor, Malaysia. Saudi J. Biol. Sci. 2020, 27, 1602–1608. [Google Scholar] [CrossRef] [PubMed]
- Department of Veterinary Services Malaysia. Program Survelan AMR dan Data Analisis 2018–2019. 2020. Available online: http://www.dvs.gov.my/index.php/pages/view/3200 (accessed on 24 March 2021).
- Saharan, V.V.; Verma, P.; Singh, A.P. High prevalence of antimicrobial resistance in Escherichia coli, Salmonella spp. and Staphylococcus aureus isolated from fish samples in India. Aquac. Res. 2020, 51, 1200–1210. [Google Scholar] [CrossRef]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated aquaculture contributes to the transfer of mcr-1 between animals and humans via the aquaculture supply chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef]
- Ellis-Iversen, J.; Seyfarth, A.M.; Korsgaard, H.; Bortolaia, V.; Munck, N.; Dalsgaard, A. Antimicrobial resistant E. coli and Enterococci in pangasius fillets and prawns in Danish retail imported from Asia. Food Control. 2020, 114, 106958. [Google Scholar] [CrossRef]
- Kikomeko, H.; Wamala, S.P.; Mugimba, K.K. Antimicrobial resistance of Escherichia coli found in intestinal tract of Oreochromis niloticus. Uganda J. Agric. Sci. 2016, 17, 157–164. [Google Scholar] [CrossRef]
- Reza, R.H.; Shipa, S.A.; Naser, M.N.; Miah, M.F. Surveillance of Escherichia coli in a fish farm of sylhet, Bangladesh. Bangladesh J. Zool. 2020, 48, 335–346. [Google Scholar] [CrossRef]
- Hon, N.T.N.; Hoa, T.T.T.; Thinh, N.Q.; Hinenoya, A.; Nakayama, H.; Harada, K.; Asayama, M.; Warisaya, M.; Yoshi, K.H.N.T.P.; Yamamot, M. Spread of Antibiotic and Antimicrobial Susceptibility of ESBL-Producing Escherichia coli Isolated from Wild and Captured Fish in the Mekong Delta, Vietnam. In Proceedings of the 9th Symposium on Diseases in Asian Aquaculture, Ho Chi Minh, Vietnam, 24–28 November 2016; pp. 75–82. [Google Scholar]
- Wei, L.S.; Musa, N.; Wee, W. Bacterial flora from a healthy freshwater Asian sea bass (Lates calcarifer) fingerling hatchery with emphasis on their antimicrobial and heavy metal resistance pattern. Vet. Arch. 2010, 80, 411–420. [Google Scholar]
- Ghaderpour, A.; Ho, W.S.; Chew, L.L.; Bong, C.W.; Chong, V.C.; Thong, K.L.; Chai, L.C. Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front. Microbiol. 2015, 6, 977. [Google Scholar] [CrossRef]
- Saif, E.; Mahmoud, A.; Talat, D.; Ibrahim, M. Studies on the Prevalence of E. coli and Salmonella in Mullet Fish from Different Sources. Alexandria J. Vet. Sci. 2017, 55, 162. [Google Scholar] [CrossRef]
- Ng, C.; Chen, H.; Giek, S.; Haller, L.; Wu, Z.; Rathinam, F.; Trottet, A.; Gin, K. Microbial water quality and the detection of multidrug resistant E. coli and antibiotic resistance genes in aquaculture sites of Singapore. Mar. Pollut. 2018, 135, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Lu, Z.; Gao, H.; Zhang, L.; Li, Q.; Li, R.; Yang, F.; Huo, C.; Yao, Z. The effect of environmental factors and migration dynamics on the prevalence of antibiotic-resistant Escherichia coli in estuary environments. Sci. Rep. 2018, 8, 1663. [Google Scholar] [CrossRef]
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tan, L.; Zhang, L.; Tian, W.; Ma, L. A Review of the Distribution of Antibiotics in Water in Different Regions of China and Current Antibiotic Degradation Pathways. Front. Environ. Sci. 2021, 9, 1–24. [Google Scholar] [CrossRef]
- Perez-Rodrigues, F.; Taban, B.M. A State-of-Art Review on Multi-Drug Resistant Pathogens in Foods of Animal Origin: Risk Factors and Mitigation Strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef]
- Ibrahim, S.; Wei Hoong, L.; Lai Siong, Y.; Mustapha, Z.C.W.; Zalati, C.W.S.; Aklilu, E.; Mohamad, M.; Kamaruzzaman, N.F. Prevalence of antimicrobial resistance (AMR) Salmonella spp. and Escherichia coli isolated from broilers in the east coast of Peninsular Malaysia. Antibiotics 2021, 10, 579. [Google Scholar] [CrossRef]
- Roseliza, R.; Khairani-Bejo, J.; Zunita, Z.; Ramlan, M.; Khoo, E.; Rosnah, Y. Antibiotic resistance of Escherichia coli isolated from chicken in Malaysia. Malaysian J. Vet. Res. 2016, 7, 65–76. [Google Scholar]
- Hinthong, W.; Pumipuntu, N.; Santajit, S.; Kulpeanprasit, S.; Buranasinsup, S.; Sookrung, N.; Chaicumpa, W.; Aiumurai, P.; Indrawattana, N. Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. PeerJ 2017, 2017, e3431. [Google Scholar] [CrossRef]
- Khine, N.O.; Lugsomya, K.; Kaewgun, B.; Honhanrob, L.; Pairojrit, P.; Jermprasert, S.; Prapasarakul, N. Multidrug Resistance and Virulence Factors of Escherichia coli Harboring Plasmid-Mediated Colistin Resistance: Mcr-1 and mcr-3 Genes in Contracted Pig Farms in Thailand. Front. Vet. Sci. 2020, 7, 582899. [Google Scholar] [CrossRef] [PubMed]
- Hang, B.P.T.; Wredle, E.; Börjesson, S.; Sjaunja, K.S.; Dicksved, J.; Duse, A. High level of multidrug-resistant Escherichia coli in young dairy calves in southern Vietnam. Trop. Anim. Health Prod. 2019, 51, 1405–1411. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Carrique-Mas, J.J.; Ngo, T.H.; Ho, H.M.; Ha, T.T.; Campbell, J.I.; Nguyen, T.N.; Hoang, N.N.; Pham, V.M.; Wagenaar, J.A.; et al. Prevalence and risk factors for carriage of antimicrobial-resistant Escherichia coli on household and small-scale chicken farms in the Mekong Delta of Vietnam. J. Antimicrob. Chemother. 2015, 70, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Hardiati, A.; Safika, S. Isolation and detection of antibiotics resistance genes of Escherichia coli from broiler farms in Sukabumi, Indonesia. J. Adv. Vet. Anim. Res. 2021, 7710, 84–90. [Google Scholar] [CrossRef]
- Harold, N.; Kallau, G.; Wibawan, I.W.T.; Lukman, D.W.; Sudarwanto, M.B. Detection of multi-drug resistant (MDR) Escherichia coli and tet gene prevalence at a pig farm in Kupang, Indonesia. J. Adv. Veterinary Anim. Res. 2018, 7710, 388–396. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the aquatic environment A review-Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Lai, H.T.; Shiu Mei, L.; Chien, Y. Transformation of Chloramphenicol and Oxytetracycline in Aquaculture Pond Sediments. J. Environ. Sci. Health Part A Environ. Sci. Eng. Toxic. 1995, 30, 1897–1923. [Google Scholar] [CrossRef]
- Sing, C.K.; Khan, M.Z.I.; Mohd Daud, H.H.; Aziz, A.R. Prevalence of Salmonella sp. in African Catfish (Clarias gariepinus) Obtained from Farms and Wet Markets in Kelantan, Malaysia and their antibiotic resistance. Sains Malays. 2016, 45, 1597–1602. [Google Scholar]
- Raufu, I.A.; Lawan, F.A.; Bello, H.S.; Musa, A.S.; Ameh, J.A.; Ambali, A.G. Occurrence and antimicrobial susceptibility profiles of Salmonella serovars from fish in Maiduguri, sub-Saharah, Nigeria. Egypt. J. Aquat. Res. 2014, 40, 59–63. [Google Scholar] [CrossRef]
- Agoba, E.E.; Adu, F.; Agyare, C.; Boamah, V.E. Antibiotic resistance patterns of bacterial isolates from hatcheries and selected fish farms in the Ashanti region of Ghana. J. Microbiol. Antimicrob. 2017, 9, 35–46. [Google Scholar] [CrossRef]
- Nair, D.V.T.; Venkitanarayanan, K.; Johny, A.K. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 2018, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- OIE. OIE List of Antimicrobials of Veterinary Importance. 2007. Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/AMR/A_OIE_List_antimicrobials_May2018.pdf (accessed on 19 August 2021).
- Lopatek, M.; Wieczorek, K.; Oseka, J. Antimicrobial Resistance, Virulence Factors, and Genetic Profiles of Vibrio parahaemolyticus from Seafood. Appl. Environ. Microbiol. 2018, 84, e00537-18. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, J.; Li, H.; Tan, S.; Chen, Y.; Yu, H. Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Front. Microbiol. 2017, 8, 2566. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.M.; Eisa, A.A.; ElBanna, N.I. Characterization of Vibrio parahaemolyticus Infection in Gilthead Seabream (Sparus auratus) Cultured in Egypt. Egypt. J. Aquat. Biol. Fish. 2020, 24, 553–571. [Google Scholar] [CrossRef]
- Igbinosa, E.O. Detection and Antimicrobial Resistance of Vibrio Isolates in Aquaculture Environments: Implications for Public Health. Microb. Drug Resist. 2016, 22, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Sudha, S.; Mridula, C.; Silvester, R.; Hatta, A.A. Prevalence and antibiotic resistance of pathogenic Vibrio in shellfishes from Cochin market. Indian J. Geo-Mar. Sci. 2014, 43, 815–824. [Google Scholar]
- Al-Dulaimi, M.M.K.; Mutalib, S.A.; Ghani, M.A.; Zaini, N.A.M.; Ariffin, A.A. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among vibrio vulnificus isolates. Antibiotics 2019, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Noorlis, A.; Ghazali, F.M.; Cheah, Y.K.; Tuan Zainazor, T.C.; Wong, W.C.; Tunung, R.; Pui, C.F.; Nishibuchi, M.; Nakaguchi, Y.; Son, R. Antibiotic resistance and biosafety of Vibrio cholerae and Vibrio parahaemolyticus from freshwater fish at retail level. Int. Food Res. J. 2011, 18, 1523–1530, ISSN 19854668. [Google Scholar]
- Singh, B.; Tyagi, A.; Billekallu Thammegowda, N.K.; Ansal, M.D. Prevalence and antimicrobial resistance of vibrios of human health significance in inland saline aquaculture areas. Aquac. Res. 2018, 49, 2166–2174. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States. 2013. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 5 June 2021).
- Shaw, K.S.; Rosenberg Goldstein, R.E.; He, X.; Jacobs, J.M.; Crump, B.C.; Sapkota, A.R. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS ONE 2014, 9, e89616. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Prim. 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Elhadi, N.; Nishibuchi, M. Malaysian Journal of Microbiology Characterization of Vibrio parahaemolyticus isolated from coastal water in Eastern. Malays. J. Microbiol. 2011, 14, 1–9. [Google Scholar] [CrossRef]
- Park, K.; Mok, J.S.; Kwon, J.Y.; Ryu, A.R.; Kim, S.H.; Lee, H.J. Food-borne outbreaks, distributions, virulence, and antibiotic resistance profiles of Vibrio parahaemolyticus in Korea from 2003 to 2016: A review. Fish. Aquat. Sci. 2018, 21, 3. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti. Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef]
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in aquaculture. Sci. Rep. 2020, 10, 21878. [Google Scholar] [CrossRef]
- Chiew, I.K.M.; Salter, A.M.; Lim, Y.S. The significance of major viral and bacterial diseases in Malaysian aquaculture industry. Pertanika J. Trop. Agric. Sci. 2019, 42, 1023–1047. [Google Scholar]
- Basri, L.; Nor, R.M.; Salleh, A.; Yasin, I.S.M.; Saad, M.Z.; Rahaman, N.Y.A.; Barkham, T.; Amal, M.N.A. Co-infections of tilapia lake virus, Aeromonas hydrophila and Streptococcus agalactiae in farmed red hybrid tilapia. Animals 2020, 10, 2141. [Google Scholar] [CrossRef]
- Ismail, N.I.A.; Amal, M.N.A.; Shohaimi, S.; Saad, M.Z.; Abdullah, S.Z. Associations of water quality and bacteria presence in cage cultured red hybrid tilapia, Oreochromis niloticus × O. mossambicus. Aquac. Rep. 2016, 4, 57–65. [Google Scholar] [CrossRef]
- Najiah, M.; Aqilah, N.; Lee, K.; Khairulbariyyah, Z.; Mithun, S.; Jalal, K.; Shaharom-Harrison, F.; Nadirah, M. Massive Mortality Associated with Streptococcus agalactiae Infection in Cage-cultured Red Hybrid Tilapia Oreochromis niloticus in Como River, Kenyir Lake, Malaysia. J. Biol. Sci. 2012, 12, 438–442. [Google Scholar] [CrossRef][Green Version]
- Syuhada, R.; Zamri-Saad, M.; Ina-Salwany, M.Y.; Mustafa, M.; Nasruddin, N.N.; Desa, M.N.M.; Nordin, S.A.; Barkham, T.; Amal, M.N.A. Molecular characterization and pathogenicity of Streptococcus agalactiae serotypes Ia ST7 and III ST283 isolated from cultured red hybrid tilapia in Malaysia. Aquaculture 2020, 515, 734543. [Google Scholar] [CrossRef]
- Mohamad, S.N.; Rafi, I.A.A.; Ismail, N.F.; Ridzuan, M.S.M.; Hamzah, A.; Abdul, M.; Nawawi, R.; Othman, M.F.; Noordin, W.N.M.; Jamari, Z. Evaluation of Resistance Effect of Genetically Improved Red Tilapia Hybrid towards Streptococcus agalactiae Infection. Malays. Fish. J. 2019, 18, 42–49. [Google Scholar]
- Musa, N.; Wei, L.S.; Hamdan, R.H.; Leong, L.K.; Wee, W.; Amal, M.N.; Kutty, B.M.; Abdullah, S.Z. Streptococcosis in red hybrid tilapia (Oreochromis niloticus) commercial farms in Malaysia. Aquac. Res. 2009, 40, 630–632. [Google Scholar] [CrossRef]
- Amal, M.N.A.; Zamri-Saad, M. Streptococcosis in Tilapia (Oreochromis niloticus): A review. Pertanika J. Trop. Agric. Sci. 2011, 34, 195–206. [Google Scholar]
- OIE. OIE Standards, Guidelines and Resolution on Antimicrobial Resistance and the Use of Antimicrobial Agents. World Organisation for Animal Health. 2015. Available online: https://web.oie.int/delegateweb/eng/ebook/AF-book-AMR-ANG_FULL.pdf?WAHISPHPSESSID=03152ead00d06990fa9066b7b71fcabc (accessed on 25 February 2021).
- HAIAP. Antibiotic Use and Antibiotic Resistance in Food Animals in Malaysia; A Threat to Human and Animal Health. 2013. Available online: http://www.haiasiapacific.org/wp-content/uploads/2014/06/Memo-on-Antibiotics-in-animal-feeds-the-case-for-Malaysia-21-Nov-2013-V1.pdf (accessed on 3 June 2021).
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Schafhauser, B.H.; Kristofco, L.A.; de Oliveira, C.M.R.; Brooks, B.W. Global review and analysis of erythromycin in the environment: Occurrence, bioaccumulation and antibiotic resistance hazards. Environ. Pollut. 2018, 238, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Dong, H.; Yuan, X.; Wang, W.; Qiang, Z. Occurrence and removal of antibiotics in ecological and conventional wastewater treatment processes: A field study. J. Environ. Manag. 2016, 178, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, T. Biodegradation and Adsorption of Antibiotics in the Activated Sludge Process. Environ. Sci. Technol. 2010, 44, 3468–3473. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.M.; Ullman, J.L.; Teel, A.L.; Watts, R.J. Chemosphere Hydrolysis of amphenicol and macrolide antibiotics: Chloramphenicol, florfenicol, spiramycin, and tylosin. Chemosphere 2015, 134, 504–511. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment-occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, H.; Xiong, P.; Zhu, Q.; Liao, C.; Jiang, G. Science of the Total Environment Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: A review. Sci. Total Environ. 2021, 753, 141975. [Google Scholar] [CrossRef]
- JECFA. Chloramphenicol. 2004. Available online: https://apps.who.int/food-additives-contaminants-jecfa-database/chemical.aspx?chemID=551 (accessed on 28 July 2021).
- Schlüsener, M.P.; Bester, K. Persistence of antibiotics such as macrolides, tiamulin and salinomycin in soil. Environ. Pollut. 2006, 143, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Pak, K.; Pothuluri, J.V.; Cerniglia, C.E. Mineralization of erythromycin A in aquaculture sediments. FEMS Microbiol. Lett. 2004, 234, 169–175. [Google Scholar] [CrossRef][Green Version]
- Richardson, M.L.; Bowron, J.M. The fate of pharmaceutical chemicals in the aquatic environment. J. Pharm. Pharmacol. 1985, 37, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Stanton, I.C.; Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Evolution of antibiotic resistance at low antibiotic concentrations including selection below the minimal selective concentration. Commun. Biol. 2020, 3, 1–11. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf. Health 2020, 2, 71–78. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Yu, C.Y.; Ang, G.Y.; Chin, P.S.; Ngeow, Y.F.; Yin, W.F.; Chan, K.G. Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int. J. Antimicrob. Agents. 2016, 47, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, M.; Angers-Loustau, A.; Kreysa, J. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect. Dis. 2016, 16, 280. [Google Scholar] [CrossRef]
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Kassem, I.I. The mobile colistin resistance gene, mcr-1.1, is carried on incx4 plasmids in multidrug resistant E. coli isolated from rainbow trout aquaculture. Microorganisms 2020, 8, 1636. [Google Scholar] [CrossRef]
- Hoa, T.T.T.; Nakayama, T.; Huyen, H.M.; Harada, K.; Hinenoya, A.; Phuong, N.T.; Yamamoto, Y. Extended-spectrum beta-lactamase-producing Escherichia coli harbouring sul and mcr-1 genes isolates from fish gut contents in the Mekong Delta, Vietnam. Lett. Appl. Microbiol. 2020, 71, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Leon, A.; Garcia-Omil, C.; Dalama, J.; Rodriguez-Souto, R.; Martinez-Urtaza, J.; Gonzalez-Escanola, N. Detection of colistin resistance mcr-1 gene in Salmonella enterica serovar Rissen isolated from mussels, Spain, 2012 to 2016. Eurosurveillance 2019, 24, 1900200. [Google Scholar] [CrossRef]
- Lv, L.; Cao, Y.; Yu, P.; Huang, R.; Wang, J.; Wen, Q.; Zhi, C.; Zhang, Q.; Liu, J.-H. Detection of mcr-1 Gene among Escherichia coli Isolates from Farm Fish and Characterization of mcr-1 Bearing IncP Plasmid. Antimicrob. Agents Chemother. 2018, 62, e02378-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P. Comment on: Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2017, 72, 636–637. [Google Scholar] [CrossRef][Green Version]
- FAO. National Aquaculture Sector Overview Malaysia, National Aquaculture Sector Overview Fact Sheets. 2008. Available online: http://www.fao.org/fishery/countrysector/naso_malaysia/en (accessed on 4 August 2020).
- Thursfield, M. Veterinary Epidemiology, 4th ed.; Willey Blackwell: Oxford, UK, 2018; pp. 230–242. [Google Scholar]
- Abdullah, A.; Ramli, R.; Ridzuan, M.S.M.; Murni, M.; Hashim, S.; Sudirwan, F.; Abdullah, S.Z.; Mansor, N.N.; Amira, S.; Saad, M.Z.; et al. The presence of Vibrionaceae, Betanodavirus and Iridovirus in marine cage-cultured fish: Role of fish size, water physicochemical parameters and relationships among the pathogens. Aquac. Rep. 2017, 7, 57–65. [Google Scholar] [CrossRef]
- Jang, S.; Biberstein, E.; Hirsh, D. A Diagnostic: Manual of Veterinary Clinical Bacteriology and Mycology; University of California: Davis County, UT, USA, 2008. [Google Scholar]
- Ryu, S.H.; Park, S.G.; Choi, S.M.; Hwang, Y.O.; Ham, H.J.; Kim, S.U.; Lee, Y.K.; Kim, M.S.; Park, G.Y.; Kim, K.S.; et al. Antimicrobial resistance and resistance genes in Escherichia coli strains isolated from commercial fish and seafood. Int. J. Food Microbiol. 2012, 152, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Momtaz, H.; Dehkordi, F.S.; Rahimi, E.; Asgarifar, A. Detection of Escherichia coli, Salmonella species, and Vibrio cholerae in tap water and bottled drinking water in Isfahan, Iran. BMC Public Health 2013, 13, 556. [Google Scholar] [CrossRef] [PubMed]
- Mikoleit, M.L. Laboratory Protocol: “Biochemical Identification of Salmonella and Shigella Using an Abbreviated Panel of Tests”, WHO Global Foodborne Infections Network. 2015. Available online: https://antimicrobialresistance.dk/CustomerData/Files/Folders/2-newsletter-pdf/20_07-gfn-biochem-v002-final-16oct2015.pdf (accessed on 15 January 2019).
- Stegniy, B.; Gerilovych, A.; Arefyev, V.; Glebova, K.; Potkonjak, A. A Method for Detecting and Typing of Salmonella by Multiplex PCR. Arch. Vet. Med. 2015, 7, 47–56. [Google Scholar] [CrossRef]
- Huq, A.; Haley, B.J.; Taviani, E.; Chen, A.; Hassan, N.A.; Cowell, R.R. Detection, Isolation, and Identification of Vibrio cholerae from the Environment. Curr. Protoc. Microbiol. 2013, 178, 1–58. [Google Scholar] [CrossRef] [PubMed]
- CDC. Chapter 5: Examination of Food and Environmental Sample; Lab. Methods Diagnosis Vibrio Cholerae. 2014. Available online: https://www.cdc.gov/cholera/pdf/laboratory-methods-for-the-diagnosis-of-vibrio-cholerae-chapter-5.pdf (accessed on 2 February 2018).
- Neogi, S.B.; Chowdhury, N.; Asakura, M.; Hinenoya, A.; Haldar, S.; Saidi, S.M.; Kogure, K.; Lara, R.J.; Yamasaki, S. A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 2010, 51, 293–300. [Google Scholar] [CrossRef]
- WHO. WHO List of Critically Important Antimicrobials (CIA). 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed on 2 February 2018).
- CLSI Standard VET01; Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals. 5th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 19–50. Available online: https://standards.globalspec.com/std/13407119/CLSI%20VET01 (accessed on 2 February 2018).
- CLSI Guideline VET03; Methods for Antimicrobial Broth Dilution and Disk Diffusion Susceptibility Testing of Bacteria Isolated from Aquatic Animals. 2nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; pp. 41–60.
- CLSI Guideline M45; Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 3rd ed. Clinical and laboratory Standards Institute: Wayne, PA, USA, 2016; pp. 56–60.
- CLSI Document M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 9th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; pp. 10–19.
- Bardet, L.; Rolain, J.M. Development of new tools to detect colistin-resistance among Enterobacteriaceae strains. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 3095249. [Google Scholar] [CrossRef]
- WHO. WHONET Tutorial Data Analysis 1 for the Surveillance of Antimicrobial resistance. 2006. Available online: http://www.whonet.org/Docs/WHONET%206.Expert%20system.doc (accessed on 16 September 2020).
- WHO. WHONET Tutorial Data Analysis 2 for the Surveillance of Antimicrobial resistance. 2006. Available online: https://ddgqe0f1ahilg.cloudfront.net/Docs/WHONET%205.Data%20analysis%202.doc (accessed on 20 September 2020).
- CLSI Supplement M100S; Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 30–38.
- CLSI Supplement VET 01S; Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals. 5th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; pp. 25–42. Available online: https://clsi.org/standards/products/veterinary-medicine/documents/vet01s/ (accessed on 20 September 2020).


| Sample Types | Total | Positive (95% CI) | |
|---|---|---|---|
| E. coli | Salmonella sp. | ||
| Tilapia | 312 | 139 (44.5; 39.4–50.4) | 2 (0.6; 0.2–1.5) |
| Asian seabass | 265 | 14 (5.3; 2.6–7.9) | 1 (0.4; 0.3–1.09) |
| Tilapia pond water 1 | 19 | 19 (100; 82.4–100) | 3 (15.7; 3.4–39.6) |
| Asian seabass pond water 1 | 13 | 8 (61.5; 31.6–86.1) | 0 (0) |
| Overall | 609 | 181 (29.7; 26.1–33.5) | 6 (0.9; 0.4–2.1) |
| Sample Type | Total | Positive (95% CI) | ||
|---|---|---|---|---|
| V. parahaemolyticus | V. vulnificus | V. cholera | ||
| Tilapia | 312 | NA 2 | NA 2 | 0 (0) |
| Asian Seabass | 265 | 94 (35.5; 29.7–41.3) | 6 (2.3; 0.8–4.8) | 5 (1.9; 0.61–4.3) |
| Tilapia water 1 | 19 | NA 2 | NA 2 | 0(0) |
| Asian seabass water 1 | 13 | 6 (46.2; 19.2–74.8) | 1 (7.7; 0.2–36) | 1 (7.7; 0.2–3.6) |
| Antimicrobial Agent | Resistance% (95% CI) | |
|---|---|---|
| Tilapia | Asian Seabass | |
| Ampicillin | 36.6 (30.0–43.7) | 31.9 (19.5–47.2) |
| Chloramphenicol | 15.3 (10.8–21.2) | 25.5 (14.4–40.6) |
| Ciprofloxacin | 16.8 (12.1–22.8) | 21.3 (11.2–36.1) |
| Colistin | 18.3 (13.4–24.5) | 17 (8.1–31.3) |
| Cefotaxime | 0.5 (0–3.2) | 0 (0.0–9.4) |
| Erythromycin | 98 (94.6–99.4) | 83 (68.7–91.9) * |
| Gentamycin | 4.5 (2.2–8.6) | 0 (0.0–9.4) |
| Kanamycin | 13.9 (9.6–19.6) | 14.9 (6.7–28.9) |
| Nalidixic Acid | 9.9 (6.3–15.1) | 14.9 (6.7–28.9) |
| Streptomycin | 19.8 (14.7–26.1) | 4.3 (0.8–15.8) * |
| Tetracycline | 36.1 (29.6–43.2) | 27.7 (16.1–42.9) |
| Ceftiofur | 0 (0.0–2.3) | 0 (0.0–9.4) |
| Trimethoprim | 31.2 (25.0–38.1) | 25.5 (14.4–40.6) |
| Antimicrobial Agent | Resistance% (95% CI) | |||
|---|---|---|---|---|
| Fish 1 | Layer 2 | Broiler 2 | Pig 2 | |
| Erythromycin | 95.6 (92–97.7) | NA | NA | NA |
| Ampicillin | 35.7 (29.8–42) | 61 (46.6–78.4) | 92 (74.2–102.8) | 84 (67–104) |
| Tetracycline | 34.5 (28.7–40.8) | 78 (61.6–97.3) | 94 (75.9–115) | 84 (67–104) |
| Trimethoprim | 30.1 (24.6–36.3) | NA | NA | NA |
| Colistin | 18.1 (13.6–23.6) | NA | NA | NA |
| Chloramphenicol | 17.3 (12.9–22.7) | 32 (21.9–45.2) | 80 (63.4–99.5) | 76 (59.9–95.1) |
| Ciprofloxacin | 17.7 (13.3–23.1) | 22 (13.8–33.3) | 48 (35.4–63.4) | 16 (9.1–25.9) |
| Streptomycin | 16.9(12.6–22.3) | 24 (15.4–35.7) | 56 (42.3–72.2) | 60 (45.8–77.2) |
| Kanamycin | 14.1 (10.1–19.2) | NA | NA | NA |
| Nalidixic Acid | 10.4 (7–15) | NA | NA | NA |
| Gentamycin | 3.6 (1.8–7.0) | 4 (1.1–10.4) | 31 (21.1–44) | 16 (9.1–25.9) |
| Cefotaxime | 0.4 (0.0–2.6) | 9 (4.1–17.1) | 15 (8.4–24.7) | 7 (2.8–14.4) |
| Ceftiofur | 0 (0.0–1.9) | 4 (1.1–10.4) | 8 (3.5–15.8) | 7 (2.8–14.4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dewi, R.R.; Hassan, L.; Daud, H.M.; Matori, M.F.; Nordin, F.; Ahmad, N.I.; Zakaria, Z. Prevalence and Antimicrobial Resistance of Escherichia coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the West Coast of Peninsular Malaysia. Antibiotics 2022, 11, 136. https://doi.org/10.3390/antibiotics11020136
Dewi RR, Hassan L, Daud HM, Matori MF, Nordin F, Ahmad NI, Zakaria Z. Prevalence and Antimicrobial Resistance of Escherichia coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the West Coast of Peninsular Malaysia. Antibiotics. 2022; 11(2):136. https://doi.org/10.3390/antibiotics11020136
Chicago/Turabian StyleDewi, Rita Rosmala, Latiffah Hassan, Hassan Mohammad Daud, Mohd. Fuad Matori, Fauziah Nordin, Nur Indah Ahmad, and Zunita Zakaria. 2022. "Prevalence and Antimicrobial Resistance of Escherichia coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the West Coast of Peninsular Malaysia" Antibiotics 11, no. 2: 136. https://doi.org/10.3390/antibiotics11020136
APA StyleDewi, R. R., Hassan, L., Daud, H. M., Matori, M. F., Nordin, F., Ahmad, N. I., & Zakaria, Z. (2022). Prevalence and Antimicrobial Resistance of Escherichia coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the West Coast of Peninsular Malaysia. Antibiotics, 11(2), 136. https://doi.org/10.3390/antibiotics11020136






