Abstract
Background: Ethionamide and prothionamide are now included in group C of the WHO recommended drugs for the treatment of tuberculosis resistant to rifampicin and multidrug-resistant tuberculosis. The clinical relevance of ethionamide and prothionamide has increased with the wide spread of resistant tuberculosis. Methods: We retrospectively analyzed 349 clinical isolates obtained between 2016 and 2020. The susceptibility to ethionamide was tested using both the BactecTM MGITTM 960 system and the SensititreTM MYCOTB plate. Results: The MIC of ethionamide increases with the total resistance of the isolates in a row from susceptible to XDR strains. A significant part of the isolates have a MIC below the breakpoint: 25%, 36%, and 50% for XDR, pre-XDR, and MDR strains. Sensitivity and specificity of detection of mutations were 96% and 86% using MGIT resistance as a reference. Conclusions: Phenotypic methods for testing ethionamide are imperfectly correlated, and the isolates with MIC of 5 mg/L have the intermediate resistance. A significant proportion of resistant TB cases are susceptible and eligible for ethionamide treatment. Resistance could be explained using only analysis of loci ethA, PfabG1, and inhA for most isolates in the Moscow region. The promoter mutation PfabG1 c(-15)t predicts resistance to ethionamide with high specificity but low sensitivity.
1. Introduction
Ethionamide (2-ethylthioisonicotinamide) was synthesized in 1952 by D. Liberman and from 1955 onward had limited use in the treatment of tuberculosis [1]. In 1956, a structurally similar prothionamide, with slightly lower efficacy but better tolerability, was discovered [2]. The clinical relevance of ethionamide and prothionamide has increased with the wide spread of resistant tuberculosis. With the successful introduction of new and repurposed drugs bedaquiline, linezolid, clofazimine, and delamanid into treatment regimens, ethionamide and prothionamide are now included in group C of the WHO recommended drugs for rifampicin-resistant and multidrug-resistant tuberculosis (MDR) treatment [3]. This group also includes pyrazinamide, ethambutol, amikacin, PAS, imipenem, meropenem, and streptomycin; these drugs should be prescribed if drugs from groups A and B must be excluded due to resistance or side effects. It can be assumed that ethionamide will not be completely eliminated from therapy schemes in the near future, given the high cost of new drugs and the emergence of resistant forms of these drugs as well [3].
Ethionamide is a prodrug that requires bioactivation, which is also characteristic of other antituberculosis drugs: isoniazid, pyrazinamide, PAS, delamanid, and pretomanid. Ethionamide activation is a multistage process, while detailed mechanisms are still not fully revealed (Figure 1) [4]. The main enzyme that provides the conversion of ethionamide to active radicals, which are further converted to a toxic adduct with NADH, is Baeyer-Villiger monooxygenase EthA [5,6,7]. The majority of resistant clinical strains had alterations at the ethA locus; approximately one third of them are loss-of-function frameshift and nonsense mutations [8,9].
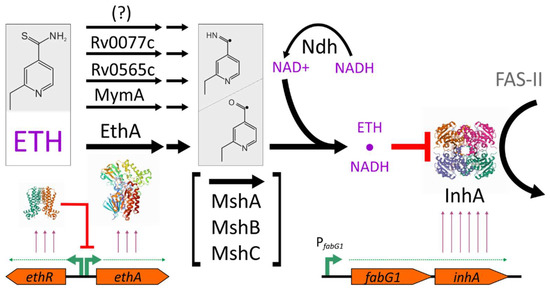
Figure 1.
Model of ethionamide action on M. tuberculosis. Ethionamide is activated in a multistep process, mostly due to EthA monooxygenase activity. Ethionamide-NADH adduct inhibits the enoyl acyl carrier protein reductase InhA, catalyzing one of the steps in FAS-II cycle of mycolic acid biosynthesis. The transcription of the ethA gene is regulated by the EthR repressor. Mycothiol biosynthesis genes (mshA, mshB, mshC) or its product, mycothiol, are also involved in the ethionamide activation, while its particular roles are still not revealed. The protein structures shown are from the RCSB PDB database: EthR—5NJ0 [23], EthA homolog —3UOZ [24], and InhA—4TRN [25].
The resulting adduct of ethionamide and NAD+ inhibits the enoyl acyl carrier protein reductase InhA, an essential FAS-II enzyme of the cell wall mycolic acid synthesis cycle [10,11]. InhA is also a target of isoniazid, which also requires bioactivation to exhibit antituberculosis properties. The activator in the case of isoniazid is catalase/peroxidase KatG. Therefore, partial cross-resistance between ethionamide and isoniazid is observed in clinical isolates [12] due to inhA gene mutations, which affect its binding properties with toxic adducts. Alternatively, promoter mutations of the inhA gene that lead to an increased transcription rate result in an increased number of InhA target proteins in the cell, thus diminishing the toxic action of both drugs [8].
The expression of ethA is regulated by the divergently transcribed ethR repressor gene. Therefore, aminoacid substitutions in EthR, providing a more tight binding of the repressor, and thus lowering ethA transcription, could also be selected during treatment [9,13]. Other transcription affecting mutations located in the ethA-ethR intergenic region were associated with a moderate increase in MIC [14,15], and for one of them a lower ethA transcription rate was experimentally demonstrated [16]. More importantly, this mutation was selected independently by many sublineages and impacts the resistance to ethionamide synergistically with mutations in other loci [16].
In addition to EthA, there are five more predicted Baeyer-Villiger monooxygenases in M. tuberculosis: Rv0892, Rv0565c, Rv1393c, Rv3049c, and MymA (Rv3083) [17]. The ability to activate ethionamide has also been demonstrated for MymA and Rv0565c [18]. Mutations in these loci were detected in clinical strains; however, the questions about clinical significance and co-occurrence with ethA mutations are still open [19]. In another report, clinical strains with mymA gene deletion have only a minor increase in MIC far below the clinical breakpoint [16].
A possible role of oxidoreductase Rv0077c in ethionamide activation was discovered during the selection of molecules capable of reversing ethionamide resistance in ethA- strains. The specific inducer SMARt-420 binds to the Rv0078 repressor, leading to the expression of the rv0077c gene and the susceptible phenotype [20].
At least two more metabolic pathways are also involved in ethionamide activation. First, the NAD+:NADH ratio affects the conversion step of ethionamide-derived radicals to ethionamide-NADH adduct [21]. However, in a recent report, ndh mutations were only found in combination with ethA and inhA promoter mutations [9]. The second is a mycothiol biosynthesis pathway: while the exact mechanism of action has not yet been revealed, msh- strains have an EthR phenotype [22]. As with ndh, mshA mutations were identified only in a small proportion of clinical strains, in combination with ethA or inhA promoter mutations [9].
Our aim was to analyze the distribution of minimum inhibitory concentrations of ethionamide (MIC) for clinical isolates of M. tuberculosis circulating in the Moscow region, and to identify molecular determinants of reduced susceptibility of ethionamide. Taking into the account the low frequency of mutations located in ndh, mshA, and ethR genes in ethionamide-resistant clinical strains, the study settings were restricted to the analysis of key resistance determinants in ethA, inhA, ethA-ethR intergenic region and inhA promoter loci. Combining the phenotypic and molecular data, we estimated the proportion of drug-resistant isolates that are eligible for ethionamide treatment.
2. Results
2.1. The MIC of Ethionamide Depends on the Resistance Profile of the Isolate
We retrospectively analyzed 349 clinical isolates, obtained in 2016–2020 from patients who attended the Moscow Research and Clinical Center for Tuberculosis Control. The susceptibility to ethionamide was tested using both the BactecTM MGITTM 960 (MGIT) system (critical concentration of 5 mg/L) and the SensititreTM MYCOTB microtiter plate for MIC determination (critical concentration of 5 mg/L [MYCOTB]). An imperfect correlation of the binary resistant/susceptible (R/S) and MIC data were observed (Table 1). Forty-three percent of resistant isolates had MIC ≤ 5 mg/L and should therefore have been considered susceptible from the Sensititre data. Additionally, 6% of susceptible isolates had MIC ≥ 10 mg/L.

Table 1.
Number of isolates with different resistance profiles.
The isolates studied were divided into groups based on their resistant profiles: susceptible, poly- or monoresistant, MDR, extensively drug-resistant (XDR) tuberculosis, and pre-XDR. Here we used the old definition of XDR tuberculosis, i.e., MDR plus resistance to any fluoroquinolone and to any of the injectable second-line drugs—kanamycin, amikacin, or capreomycin. Pre-XDR are two intermediate categories between MDR and XDR, when additional resistance to fluoroquinolones, or injectable drugs are registered but not both at once. Noticeably, 53 isolates susceptible to all drugs had a more compact MIC distribution compared to 176 isolates susceptible to ethionamide joined from all groups. Its mode of distribution was at a MIC of 2.5 mg/L, while MIC90 was equal to 5 mg/L. Following this finding, the lower breakpoint concentration of 2.5 mg/L for Sensititre assay could be proposed; however, no change of performance compared to MGIT data could be achieved. The MIC of ethionamide increases with the total resistance of isolates in a row from susceptible to XDR strains.
Based on the currently approved critical concentration of 5 mg/L for Sensititre assay, almost 50 % of XDR isolates are eligible for ethionamide treatment. For pre-XDR and MDR isolates, the proportion of presumably susceptible is raised to 63 and 69%, respectively. Even if the lower critical concentration of 2.5 mg/L is used, a significant part of the isolates have a MIC of ethionamide below the breakpoint: 25%, 36% and 50% for XDR, pre-XDR and MDR strains (Table 1).
As a reference to the data obtained, the isoniazid resistance of these isolates was also analyzed (Table S1). Isoniazid and ethionamide share common resistance determinants: substitutions in the inhA gene and its promoter mutations. However, the most frequent mutation in isoniazid-resistant isolates is the substitution of KatG S315T, so cross-resistance between these drugs is a rare event. It was confirmed by pairwise comparison of the MIC data (Table 2). Two distinct peaks at 0.06 mg/L and >4 mg/L were observed for the distribution of isoniazid MIC, separating susceptible and resistant isolates. Cross-resistance between two drugs was low, so a significant part of high level isoniazid-resistant strains were ethionamide-susceptible.

Table 2.
Comparison of pairwise MICs of ethionamide and isoniazid.
2.2. Molecular Determinants of Ethionamide Resistance
To study the molecular mechanisms of resistance to ethionamide, we selected only primary culture-positive tuberculosis cases, diagnosed in 2017–2018 (n = 136). We sequenced ethA, inhA, and inhA promoter region upstream the fabG1 gene. Thirty-two isolates had a MIC of 10 mg/L or higher.
Mutations were found in 94% of resistant isolates. However, the detection specificity was approximately 74% (Table 3). Part of the identified mutations were aminoacid substitutions in the EthA protein, which have unknown impact on its activity (Table S2). Its exclusion from prediction increases the specificity to 89%, simultaneously lowering the sensitivity to 84%. Importantly, part of the frameshift and nonsense mutations in ethA did not lead to an increase in MIC (n = 7 from 19 with such type of mutations).

Table 3.
Molecular determinants of ethionamide resistance.
Promoter mutations in the inhA gene, located upstream of fabG1, were previously proposed to be a marker of resistance to ethionamide [26,27]. The most common mutation c(-15)t is usually accompanied by KatG S315T substitution in isoniazid-resistant strains. Taking into account the analyzed set of isolates, the sensitivity and specificity of this mutation in the prediction of ethionamide resistance were 31% and 98%, respectively (Table 3), which is close to previously reported sensitivities of 38% [28] and 43% [29]; however, in one study a sensitivity of 92% was reported [30].
The sensitivity and specificity of detection of any mutation (excluding synonymous) was 97% and 74% for resistance detected with 96-well plate data and 96% and 86% for Bactec MGIT resistance. Exclusion of EthA substitutions leads to decrease of sensitivity with simultaneous increase of specificity: 87% and 86%; 80% and 94%, for Sensititre and MGIT data, respectively.
3. Discussion
Mycolic acid biosynthesis is an attractive target for tuberculosis treatment. One of the first drugs, thiacetazone, developed in 1946 by Gerhard Domagk, inhibits one of the steps in the FAS-II cycle [31]. Although thiacetazone was excluded from treatment schemes due to toxicity and low efficiency, another drug, isoniazid, which inhibits the other step in the FAS-II cycle, remains one of the cornerstones for the treatment of active and latent tuberculosis. Ethionamide shares the target with isoniazid; however, the cross-resistance between them is not frequent. We analyzed the resitance of isoniazid and ethionamide for a large set of clinical isolates from the Moscow region with different resistance profiles.
First, we found a poor correlation of the binary resistance (R/S) of ethionamide obtained with Bactec MGIT 960 and the determination of MIC with Sensititre MYCOTB. MIC ranges for susceptible and resistant isolates intersect significantly, which is in line with previous reports [32]. The cause of such a discrepancy of two methods that use the same liquid 7H9 medium remains unrevealed.
Compared to ethionamide, isoniazid showed a strict difference between susceptible and resistant isolates. Its modes of MIC distribution are separated in six or more dilution steps. Possible explanations for this phenomenon include the wide use of isoniazid as a first-line drug for TB treatment and the high level of transmission of MDR tuberculosis in Russia. We recently compared MDR strains from Russia and Taiwan [33]: while strains from Moscow had narrow spectra of molecular determinants of isoniazid resistance and high MIC, isolates from Taiwan possessed a wider range of MICs and variable spectra of drug-determining mutations reflecting ongoing selection.
Second, a lower critical concentration of ethionamide could be proposed for microdilution plates on the basis of the MIC ranges for susceptible isolates. Currently, the approved concentration is 5 mg/L, while most ethionamide naïve isolates have a MIC < 2.5 mg/L. This value also better fits previously published pharmacokinetic data [34,35,36]. However, the overlap of MIC distributions between resistant and susceptible isolates reflects only the ‘intermediate’ resistance of isolates with MIC = 5 mg/L, and further clinical studies of ethionamide treatment effectiveness in such cases are needed.
Most mutations leading to resistance to ethionamide in clinical strains are found in the ethA and inhA genes. Substitutions in the promoter region of the isoniazid and ethionamide target gene, inhA, leading to increased gene expression should lead to cross-resistance [12]. However, data on the association of the most common PfabG1 c(-15)t mutation and resistance to ethionamide and prothionamide are inconsistent, as resistant and sensitive strains with this mutation have been found [9,37,38,39]. This substitution in the promoter is associated with an increase in the MIC of isoniazid to an intermediate level that cannot be unambiguously interpreted by critical concentration or proportion methods [40]. However, in the case of ethionamide, the variability in the stability level is more pronounced, with a difference of up to two orders of magnitude in the MIC concentration [37]. In our set of isolates, we found high specificity, close to 100%, and low sensitivity of detection of this mutation for the prediction of resistance to ethionamide.
We also found that frameshift and nonsense mutations in ethA gene do not necessarily lead to the loss of activation and resistance to ethionamide in bacterial cell: 37% of strains had MIC in susceptible range. It is consistent with previous studies, where 35% of the ethA-inactivating mutations of strains were susceptible to prothionamide [14]. It was previously reported that other monooxygenases are also active toward ethionamide. MymA activity levels in wild-type cells are comparable to EthA [18], and it appears that mymA-inactivating mutations can also be selected by ethionamide-based drug therapy [14]. However, it is not clear whether mutations in ethA in ethionamide-susceptible isolates were selected by the drug action, are the result of neutral evolution and genetic drift, or are just a drawback in currently used phenotypic methods. The latter could be explained by the selection of more tolerant, not resistant strains, i.e., a longer survival time at sub-inhibitory concentrations of the drug [41].
The correlation between genotype and ethionamide-resistant phenotype is still an open question because while mutations in the ethA, mshA, ndh and inhA loci were detected in 96.5% of cases [28], in most other studies mutations were not detected in a significant percentage of strains. For example, in a 2011 study, 8 of 47 isolates (17%) were found to have no mutations when analyzing inhA, ethA/R, ndh, mshA [13]. Similarly, a recent study found no mutations in 36 of 178 isolates (20%) when analyzing the same determinants [9]. Furthermore, when a wider range of loci (inhA, ethA/R, ndh, mshA/B/C/D) were analyzed, no mutations were found in 9 of 46 prothionamide resistant strains [42]. It should be noted that 74% of selected in vitro ethionamide-resistant strains had no mutations in ethA/R, katG and inhA also [43]. We analyze the loci ethA, ethA-ethR, inhA, and PfabG1 and achieved 97% sensitivity with 74% specificity if MIC data were used with a critical concentration of 5 mg/L. Using the Bactec MGIT 960 system resistance data as reference, the specificity increased to 86%, while the sensitivity left unchanged. We suppose that the analysis of wider panel of molecular determinants would not lead to the increase in performance due to limitations in phenotypic methods observed in the study—24% overlap in MICs between susceptible and resistant isolates.
In conclusion, determination of ethionamide MIC before treatment is highly desirable, taking into account the inability to increase doses due to drug toxicity. Furthermore, a large individual variation of pK/pD parameters [36] and interaction with other drugs [35,44] require personalized treatment with therapeutic drug monitoring.
4. Materials and Methods
4.1. Mycobacterium Tuberculosis Strains
The M. tuberculosis strains were obtained from clinical specimens collected from TB patients at the Moscow Research and Clinical Center for Tuberculosis Control. In total, 349 clinical isolates, obtained in 2016–2020 from patients who attended the Moscow Research and Clinical Center for Tuberculosis Control were analyzed: 2016—50 isolates, 2017—87 isolates, 2018—66 isolates, 2019—74 isolates, and 202—72 isolates. For molecular analysis only primary isolates from 2017–2018 were used. They were isolated sequentally, one isolate for one patient, before treatment initiation. Drug susceptibility testing for rifampicin, isoniazid, streptomycin, ethambutol, pyrazinamide, ofloxacin, moxifloxacin, kanamycin, capreomycin, and amikacin, PAS, ethionamide was performed using Bactec MGIT 960 as previously described [45,46]. Sensititre MYCOTB MIC determination was performed as described in [47].
The study was approved by the Ethics Committee of the Moscow Government Health Department. The Ethics Committee waived the need for patient consent because the study did not include personal identifiers or clinical data and the samples were analyzed anonymously.
4.2. DNA Isolation and Sequencing
DNA isolation and sequencing of the ethA, inhA and PfabG1 fragments were performed as previously described [48]. The following PCR primers were used: ethAR-F1: 5′-cgacgttgaaatcacgctgg-3′, ethAR-R1: 5′-gtgaccgacaccattgaacg-3′; ethAR-F2: 5′-ttcaaccccgt-tgcggtaat-3′; ethAR-R2: 5′-ctctttctgtgcagcggcta-3′; ethAR-F3: 5′-atgatcggcccgacgaaatc-3′; ethAR-R3: 5′-ccctggcagcttactacgtg-3′; PfabG1-F: 5′-cctcgctgcccagaaaggga-3′; PfabG1-R: 5′-atcc-cccggtttcctccggt-3′-3′; inhA-F2: 5′-gagctatatctccggtgcgg-3′; inhA-R2: 5′-gcgaccgtcatcca-gttgta-3′; inhA-F3: 5′-ccacatctcggcgtattcgt-3′; inhA-R3: 5′-cggtgataccccaccgaaat-3′.
5. Conclusions
Phenotypic methods for testing ethionamide susceptibility using liquid media are poorly correlated. The isolates with MIC = 5 mg/L obtained using the Sensititre microtiter assay should be treated as isolates with intermediate resistance. A significant proportion of MDR and XDR cases are susceptible and eligible for treatment with ethionamide. The promoter mutation PfabG1 c(-15)t predicts resistance to ethionamide with high specificity, but low sensitivity. Resistance could be explained using only the analysis of the loci ethA, PfabG1, and inhA for most isolates, which circulate in the Moscow region.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics11020133/s1, Table S1: Isoniazid MIC for isolates with different resistance profiles, Table S2: Determinants of ethionamide resistance for 136 primary isolates.
Author Contributions
Conceptualization, D.Z. and S.S.; methodology, E.K., Y.M. and A.U.; validation, A.U.; formal analysis, A.U., M.M.; investigation, A.U., Y.M. and E.K.; data curation, S.S.; writing—original draft preparation, review and editing, D.Z.; funding acquisition, D.Z.; supervision, M.M. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (project 20-015-00463).
Institutional Review Board Statement
According to the Ethics Committee of the Moscow Research and Clinical Center for Tuberculosis Control of the Moscow Government Health Department, this research did not require ethics approval. All samples used in this study lacked personal information about the patients, in particular, any name or address, making them anonymous samples.
Informed Consent Statement
According to the Ethics Committee of the Moscow Research and Clinical Center for Tuberculosis Control of the Moscow Government Health Department, this research did not require informed consent statement. All samples used in this study lacked personal information about the patients, in particular, any name or address, making them anonymous samples.
Data Availability Statement
Data are contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Rist, N.; Grumbach, F.; Libermann, D. Experiments on the Antituberculous Activity of Alpha-Ethylthioisonicotinamide. Am. Rev. Tuberc. 1959, 79, 1–5. [Google Scholar] [CrossRef]
- Scardigli, A.; Caminero, J.A.; Sotgiu, G.; Centis, R.; D’Ambrosio, L.; Migliori, G.B. Efficacy and Tolerability of Ethionamide versus Prothionamide: A Systematic Review. Eur. Respir. J. 2016, 48, 946–952. [Google Scholar] [CrossRef]
- Kadura, S.; King, N.; Nakhoul, M.; Zhu, H.; Theron, G.; Köser, C.U.; Farhat, M. Systematic Review of Mutations Associated with Resistance to the New and Repurposed Mycobacterium Tuberculosis Drugs Bedaquiline, Clofazimine, Linezolid, Delamanid and Pretomanid. J. Antimicrob. Chemother. 2020, 75, 2031–2043. [Google Scholar] [CrossRef]
- Laborde, J.; Deraeve, C.; Duhayon, C.; Pratviel, G.; Bernardes-Génisson, V. Ethionamide Biomimetic Activation and an Unprecedented Mechanism for Its Conversion into Active and Non-Active Metabolites. Org. Biomol. Chem. 2016, 14, 8848–8858. [Google Scholar] [CrossRef]
- DeBarber, A.E.; Mdluli, K.; Bosman, M.; Bekker, L.G.; Barry, C.E. Ethionamide Activation and Sensitivity in Multidrug-Resistant Mycobacterium Tuberculosis. Proc. Natl. Acad. Sci. USA 2000, 97, 9677–9682. [Google Scholar] [CrossRef]
- Baulard, A.R.; Betts, J.C.; Engohang-Ndong, J.; Quan, S.; McAdam, R.A.; Brennan, P.J.; Locht, C.; Besra, G.S. Activation of the Pro-Drug Ethionamide Is Regulated in Mycobacteria. J. Biol. Chem. 2000, 275, 28326–28331. [Google Scholar] [CrossRef]
- Vannelli, T.A.; Dykman, A.; Ortiz de Montellano, P.R. The Antituberculosis Drug Ethionamide Is Activated by a Flavoprotein Monooxygenase. J. Biol. Chem. 2002, 277, 12824–12829. [Google Scholar] [CrossRef]
- Vilchèze, C.; Jacobs, W.R. Resistance to Isoniazid and Ethionamide in Mycobacterium Tuberculosis: Genes, Mutations, and Causalities. Microbiol. Spectr. 2014, 2, MGM2-0014-2013. [Google Scholar] [CrossRef]
- Islam, M.M.; Tan, Y.; Hameed, H.M.A.; Liu, Z.; Chhotaray, C.; Liu, Y.; Lu, Z.; Cai, X.; Tang, Y.; Gao, Y.; et al. Detection of Novel Mutations Associated with Independent Resistance and Cross-Resistance to Isoniazid and Prothionamide in Mycobacterium Tuberculosis Clinical Isolates. Clin. Microbiol. Infect. 2019, 25, 1041.e1–1041.e7. [Google Scholar] [CrossRef]
- Hanoulle, X.; Wieruszeski, J.-M.; Rousselot-Pailley, P.; Landrieu, I.; Locht, C.; Lippens, G.; Baulard, A.R. Selective Intracellular Accumulation of the Major Metabolite Issued from the Activation of the Prodrug Ethionamide in Mycobacteria. J. Antimicrob. Chemother. 2006, 58, 768–772. [Google Scholar] [CrossRef]
- Dover, L.G.; Alahari, A.; Gratraud, P.; Gomes, J.M.; Bhowruth, V.; Reynolds, R.C.; Besra, G.S.; Kremer, L. EthA, a Common Activator of Thiocarbamide-Containing Drugs Acting on Different Mycobacterial Targets. Antimicrob. Agents Chemother. 2007, 51, 1055–1063. [Google Scholar] [CrossRef]
- Larsen, M.H.; Vilchèze, C.; Kremer, L.; Besra, G.S.; Parsons, L.; Salfinger, M.; Heifets, L.; Hazbon, M.H.; Alland, D.; Sacchettini, J.C.; et al. Overexpression of InhA, but Not KasA, Confers Resistance to Isoniazid and Ethionamide in Mycobacterium Smegmatis M. Bovis BCG and M. Tuberculosis. Mol. Microbiol. 2002, 46, 453–466. [Google Scholar] [CrossRef]
- Brossier, F.; Veziris, N.; Truffot-Pernot, C.; Jarlier, V.; Sougakoff, W. Molecular Investigation of Resistance to the Antituberculous Drug Ethionamide in Multidrug-Resistant Clinical Isolates of Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 355–360. [Google Scholar] [CrossRef]
- Casali, N.; Nikolayevskyy, V.; Balabanova, Y.; Harris, S.R.; Ignatyeva, O.; Kontsevaya, I.; Corander, J.; Bryant, J.; Parkhill, J.; Nejentsev, S.; et al. Evolution and Transmission of Drug-Resistant Tuberculosis in a Russian Population. Nat. Genet. 2014, 46, 279–286. [Google Scholar] [CrossRef]
- Farhat, M.R.; Sultana, R.; Iartchouk, O.; Bozeman, S.; Galagan, J.; Sisk, P.; Stolte, C.; Nebenzahl-Guimaraes, H.; Jacobson, K.; Sloutsky, A.; et al. Genetic Determinants of Drug Resistance in Mycobacterium Tuberculosis and Their Diagnostic Value. Am. J. Respir. Crit. Care Med. 2016, 194, 621–630. [Google Scholar] [CrossRef]
- De Welzen, L.; Eldholm, V.; Maharaj, K.; Manson, A.L.; Earl, A.M.; Pym, A.S. Whole-Transcriptome and -Genome Analysis of Extensively Drug-Resistant Mycobacterium Tuberculosis Clinical Isolates Identifies Downregulation of EthA as a Mechanism of Ethionamide Resistance. Antimicrob. Agents Chemother. 2017, 61, e01461-17. [Google Scholar] [CrossRef] [PubMed]
- Bonsor, D.; Butz, S.F.; Solomons, J.; Grant, S.; Fairlamb, I.J.S.; Fogg, M.J.; Grogan, G. Ligation Independent Cloning (LIC) as a Rapid Route to Families of Recombinant Biocatalysts from Sequenced Prokaryotic Genomes. Org. Biomol. Chem. 2006, 4, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.S.; Wellington, S.; Kawate, T.; Desjardins, C.A.; Silvis, M.R.; Wivagg, C.; Thompson, M.; Gordon, K.; Kazyanskaya, E.; Nietupski, R.; et al. Baeyer-Villiger Monooxygenases EthA and MymA Are Required for Activation of Replicating and Non-Replicating Mycobacterium Tuberculosis Inhibitors. Cell Chem. Biol. 2016, 23, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Hicks, N.D.; Carey, A.F.; Yang, J.; Zhao, Y.; Fortune, S.M. Bacterial Genome-Wide Association Identifies Novel Factors That Contribute to Ethionamide and Prothionamide Susceptibility in Mycobacterium Tuberculosis. mBio 2019, 10, e00616-19. [Google Scholar] [CrossRef] [PubMed]
- Blondiaux, N.; Moune, M.; Desroses, M.; Frita, R.; Flipo, M.; Mathys, V.; Soetaert, K.; Kiass, M.; Delorme, V.; Djaout, K.; et al. Reversion of Antibiotic Resistance in Mycobacterium Tuberculosis by Spiroisoxazoline SMARt-420. Science 2017, 355, 1206–1211. [Google Scholar] [CrossRef] [PubMed]
- Vilchèze, C.; Weisbrod, T.R.; Chen, B.; Kremer, L.; Hazbón, M.H.; Wang, F.; Alland, D.; Sacchettini, J.C.; Jacobs, W.R. Altered NADH/NAD+ Ratio Mediates Coresistance to Isoniazid and Ethionamide in Mycobacteria. Antimicrob. Agents Chemother. 2005, 49, 708–720. [Google Scholar] [CrossRef]
- Xu, X.; Vilchèze, C.; Av-Gay, Y.; Gómez-Velasco, A.; Jacobs, W.R. Precise Null Deletion Mutations of the Mycothiol Synthesis Genes Reveal Their Role in Isoniazid and Ethionamide Resistance in Mycobacterium Smegmatis. Antimicrob. Agents Chemother. 2011, 55, 3133–3139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tatum, N.J.; Liebeschuetz, J.W.; Cole, J.C.; Frita, R.; Herledan, A.; Baulard, A.R.; Willand, N.; Pohl, E. New Active Leads for Tuberculosis Booster Drugs by Structure-Based Drug Discovery. Org. Biomol. Chem. 2017, 15, 10245–10255. [Google Scholar] [CrossRef] [PubMed]
- Leisch, H.; Shi, R.; Grosse, S.; Morley, K.; Bergeron, H.; Cygler, M.; Iwaki, H.; Hasegawa, Y.; Lau, P.C.K. Cloning, Baeyer-Villiger Biooxidations, and Structures of the Camphor Pathway 2-Oxo-Δ(3)-4,5,5-Trimethylcyclopentenylacetyl-Coenzyme A Monooxygenase of Pseudomonas Putida ATCC 17453. Appl. Environ. Microbiol. 2012, 78, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Chollet, A.; Mourey, L.; Lherbet, C.; Delbot, A.; Julien, S.; Baltas, M.; Bernadou, J.; Pratviel, G.; Maveyraud, L.; Bernardes-Génisson, V. Crystal Structure of the Enoyl-ACP Reductase of Mycobacterium Tuberculosis (InhA) in the Apo-Form and in Complex with the Active Metabolite of Isoniazid Pre-Formed by a Biomimetic Approach. J. Struct. Biol. 2015, 190, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; de Lisle, G.; Jacobs, W.R. InhA, a Gene Encoding a Target for Isoniazid and Ethionamide in Mycobacterium Tuberculosis. Science 1994, 263, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Heym, B.; Honoré, N.; Truffot-Pernot, C.; Banerjee, A.; Schurra, C.; Jacobs, W.R.; van Embden, J.D.; Grosset, J.H.; Cole, S.T. Implications of Multidrug Resistance for the Future of Short-Course Chemotherapy of Tuberculosis: A Molecular Study. Lancet 1994, 344, 293–298. [Google Scholar] [CrossRef]
- Rueda, J.; Realpe, T.; Mejia, G.I.; Zapata, E.; Rozo, J.C.; Ferro, B.E.; Robledo, J. Genotypic Analysis of Genes Associated with Independent Resistance and Cross-Resistance to Isoniazid and Ethionamide in Mycobacterium Tuberculosis Clinical Isolates. Antimicrob. Agents Chemother. 2015, 59, 7805–7810. [Google Scholar] [CrossRef]
- Farhat, M.R.; Freschi, L.; Calderon, R.; Ioerger, T.; Snyder, M.; Meehan, C.J.; de Jong, B.; Rigouts, L.; Sloutsky, A.; Kaur, D.; et al. GWAS for Quantitative Resistance Phenotypes in Mycobacterium Tuberculosis Reveals Resistance Genes and Regulatory Regions. Nat. Commun. 2019, 10, 2128. [Google Scholar] [CrossRef]
- Sandoval, R.; Monteghirfo, M.; Salazar, O.; Galarza, M. Cross-resistance between isoniazid and ethionamide and its strong association with mutation C-15T in Mycobacterium tuberculosis isolates from Peru. Rev. Argent Microbiol. 2020, 52, 36–42. [Google Scholar] [CrossRef]
- Coxon, G.D.; Craig, D.; Corrales, R.M.; Vialla, E.; Gannoun-Zaki, L.; Kremer, L. Synthesis, Antitubercular Activity and Mechanism of Resistance of Highly Effective Thiacetazone Analogues. PLoS ONE 2013, 8, e53162. [Google Scholar] [CrossRef]
- Heysell, S.K.; Pholwat, S.; Mpagama, S.G.; Pazia, S.J.; Kumburu, H.; Ndusilo, N.; Gratz, J.; Houpt, E.R.; Kibiki, G.S. Sensititre MycoTB Plate Compared to Bactec MGIT 960 for First- and Second-Line Antituberculosis Drug Susceptibility Testing in Tanzania: A Call to Operationalize MICs. Antimicrob. Agents Chemother. 2015, 59, 7104–7108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jou, R.; Lee, W.-T.; Kulagina, E.V.; Weng, J.-Y.; Isakova, A.I.; Lin, W.-H.; Antonova, O.V.; Wu, M.-H.; Arslanbaeva, L.R.; Tasi, H.-Y.; et al. Redefining MDR-TB: Comparison of Mycobacterium Tuberculosis Clinical Isolates from Russia and Taiwan. Infect. Genet. Evol. 2019, 72, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, D.; Pasipanodya, J.G.; Mpagama, S.G.; Srivastava, S.; Bendet, P.; Koeuth, T.; Lee, P.S.; Heysell, S.K.; Gumbo, T. Ethionamide Pharmacokinetics/Pharmacodynamics-Derived Dose, the Role of MICs in Clinical Outcome, and the Resistance Arrow of Time in Multidrug-Resistant Tuberculosis. Clin. Infect. Dis. 2018, 67, S317–S326. [Google Scholar] [CrossRef]
- Chirehwa, M.T.; Court, R.; de Kock, M.; Wiesner, L.; de Vries, N.; Harding, J.; Gumbo, T.; Maartens, G.; Warren, R.; Denti, P.; et al. Effect of Isoniazid Intake on Ethionamide Pharmacokinetics and Target Attainment in Multidrug-Resistant Tuberculosis Patients. Antimicrob. Agents Chemother. 2021, 65, e00278-21. [Google Scholar] [CrossRef] [PubMed]
- Mugabo, P.; Mulubwa, M. Ethionamide Population Pharmacokinetics/Pharmacodynamics and Therapeutic Implications in South African Adult Patients with Drug-Resistant Tuberculosis. Br. J. Clin. Pharmacol. 2021, 87, 3863–3870. [Google Scholar] [CrossRef]
- Morlock, G.P.; Metchock, B.; Sikes, D.; Crawford, J.T.; Cooksey, R.C. EthA, InhA, and KatG Loci of Ethionamide-Resistant Clinical Mycobacterium Tuberculosis Isolates. Antimicrob. Agents Chemother. 2003, 47, 3799–3805. [Google Scholar] [CrossRef] [PubMed]
- Abe, C.; Kobayashi, I.; Mitarai, S.; Wada, M.; Kawabe, Y.; Takashima, T.; Suzuki, K.; Sng, L.-H.; Wang, S.; Htay, H.H.; et al. Biological and Molecular Characteristics of Mycobacterium Tuberculosis Clinical Isolates with Low-Level Resistance to Isoniazid in Japan. J. Clin. Microbiol. 2008, 46, 2263–2268. [Google Scholar] [CrossRef]
- Imperiale, B.R.; Di Giulio, Á.B.; Adrián Cataldi, A.; Morcillo, N.S. Evaluation of Mycobacterium Tuberculosis Cross-Resistance to Isoniazid, Rifampicin and Levofloxacin with Their Respective Structural Analogs. J. Antibiot. 2014, 67, 749–754. [Google Scholar] [CrossRef]
- Ghodousi, A.; Tagliani, E.; Karunaratne, E.; Niemann, S.; Perera, J.; Köser, C.U.; Cirillo, D.M. Isoniazid Resistance in Mycobacterium Tuberculosis Is a Heterogeneous Phenotype Composed of Overlapping MIC Distributions with Different Underlying Resistance Mechanisms. Antimicrob. Agents Chemother. 2019, 63, e00092-19. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of Lag Time Underlies Antibiotic Tolerance in Evolved Bacterial Populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef]
- Tan, Y.; Su, B.; Zheng, H.; Song, Y.; Wang, Y.; Pang, Y. Molecular Characterization of Prothionamide-Resistant Mycobacterium Tuberculosis Isolates in Southern China. Front. Microbiol. 2017, 8, 2358. [Google Scholar] [CrossRef] [PubMed]
- Brossier, F.; Sola, C.; Bernard, C.; Jarlier, V.; Veziris, N.; Sougakoff, W. Characterization of a Clone of Mycobacterium Tuberculosis Clinical Isolates with Mutations in KatG (A110V), EthA (Q269STOP), and the InhA Promoter (-15C→T). J. Clin. Microbiol. 2015, 53, 3104. [Google Scholar] [CrossRef]
- Ranjan, R.; Srivastava, A.; Bharti, R.; Roy, T.; Verma, S.; Ray, L.; Misra, A. Preclinical Development of Inhalable D-Cycloserine and Ethionamide To Overcome Pharmacokinetic Interaction and Enhance Efficacy against Mycobacterium Tuberculosis. Antimicrob. Agents Chemother. 2019, 63, e00099-19. [Google Scholar] [CrossRef]
- WHO. Companion Handbook to the WHO Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis; WHO Guidelines Approved by the Guidelines Review Committee; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-154880-9.
- WHO. Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant Tuberculosis; World Health Organization: Geneva, Switzerland, 2018; 106p.
- Nosova, E.Y.; Zimenkov, D.V.; Khakhalina, A.A.; Isakova, A.I.; Krylova, L.Y.; Makarova, M.V.; Galkina, K.Y.; Krasnova, M.A.; Safonova, S.G.; Litvinov, V.I.; et al. A Comparison of the Sensititre MycoTB Plate, the Bactec MGIT 960, and a Microarray-Based Molecular Assay for the Detection of Drug Resistance in Clinical Mycobacterium Tuberculosis Isolates in Moscow, Russia. PLoS ONE 2016, 11, e0167093. [Google Scholar] [CrossRef] [PubMed]
- Zimenkov, D.V.; Nosova, E.Y.; Kulagina, E.V.; Antonova, O.V.; Arslanbaeva, L.R.; Isakova, A.I.; Krylova, L.Y.; Peretokina, I.V.; Makarova, M.V.; Safonova, S.G.; et al. Examination of Bedaquiline- and Linezolid-Resistant Mycobacterium Tuberculosis Isolates from the Moscow Region. J. Antimicrob. Chemother. 2017, 72, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).