Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany
Abstract
1. Introduction
2. Results
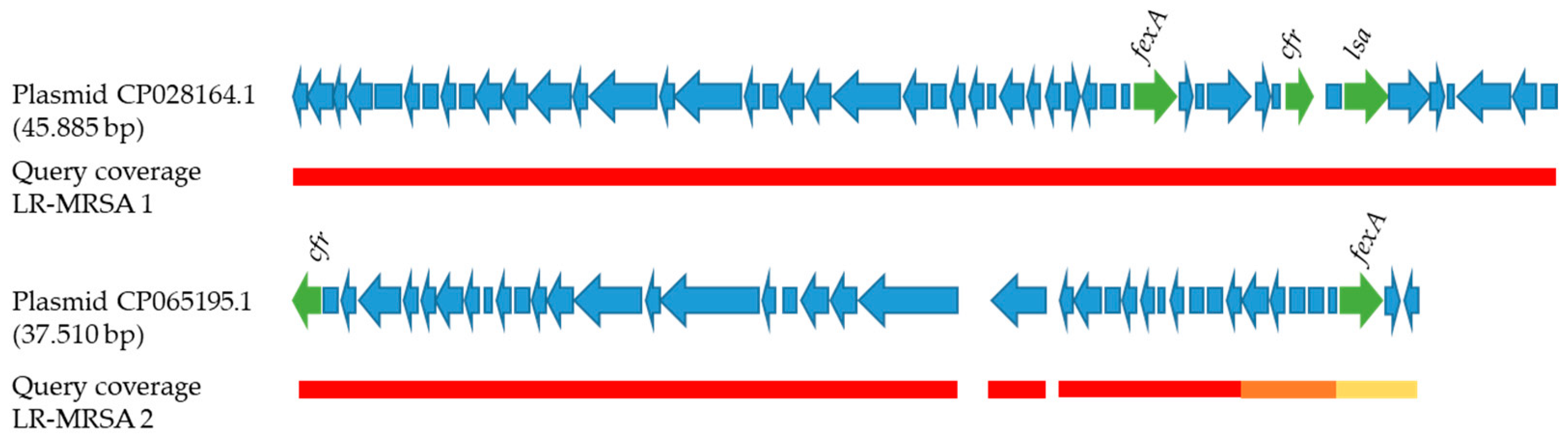
2.1. Genotypic Analyses and AMR
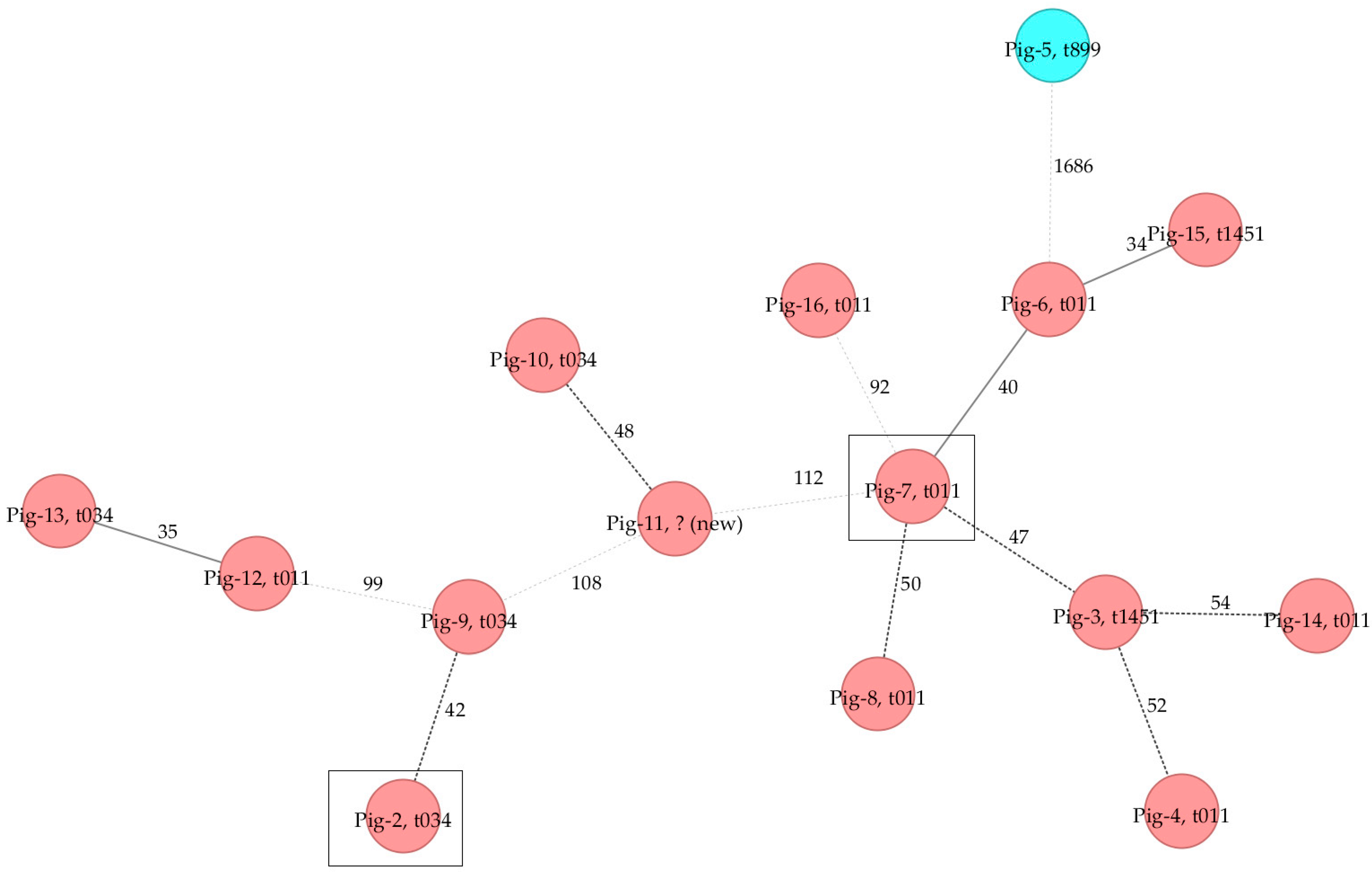
2.2. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Sampling and MRSA Strain Selection
4.2. WGS and Bioinformatics Analyses
4.3. Antimicrobial Susceptibility Testing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aires-de-Sousa, M. Methicillin-resistant Staphylococcus aureus among animals: Current overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef]
- Sergelidis, D.; Angelidis, A.S. Methicillin-resistant Staphylococcus aureus: A controversial food-borne pathogen. Lett. Appl. Microbiol. 2017, 64, 409–418. [Google Scholar] [CrossRef]
- Hashemian, S.M.R.; Farhadi, T.; Ganjparvar, M. Linezolid: A review of its properties, function, and use in critical care. Drug Des. Dev. Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Prystowsky, J.; Siddiqui, F.; Chosay, J.; Shinabarger, D.L.; Millichap, J.; Peterson, L.R.; Noskin, G.A. Resistance to linezolid: Characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 2001, 45, 2154–2156. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 2018, 73, 1763–1769. [Google Scholar] [CrossRef]
- Turner, A.M.; Lee, J.Y.H.; Gorrie, C.L.; Howden, B.P.; Carter, G.P. Genomic Insights into Last-Line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-Resistant Enterococcus. Front. Microbiol. 2021, 12, 637656. [Google Scholar] [CrossRef]
- Schwarz, S.; Zhang, W.; Du, X.D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Lange, D.; Klare, I.; Werner, G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 2018, 52, 819–827. [Google Scholar] [CrossRef]
- Shariati, A.; Dadashi, M.; Chegini, Z.; van Belkum, A.; Mirzaii, M.; Khoramrooz, S.S.; Darban-Sarokhalil, D. The global prevalence of Daptomycin, Tigecycline, Quinupristin/Dalfopristin, and Linezolid-resistant Staphylococcus aureus and coagulase-negative staphylococci strains: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2020, 9, 56. [Google Scholar] [CrossRef]
- Mamfe, L.M.; Akwuobu, C.A.; Ngbede, E.O. Phenotypic detection, antimicrobial susceptibility and virulence profile of staphylococci in the pig production setting, Makurdi, Nigeria. Access Microbiol. 2021, 3, 293. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Bellés-Bellés, A.; Fernández-Fernández, R.; García, M.; Vilaró, A.; Zarazaga, M.; Torres, C. Linezolid-resistant MRSA-CC398 carrying the cfr gene, and MRSA-CC9 isolates from pigs with signs of infection in Spain. J. Appl. Microbiol. 2021, 131, 615–622. [Google Scholar] [CrossRef]
- Timmermans, M.; Bogaerts, B.; Vanneste, K.; De Keersmaecker, S.C.J.; Roosens, N.H.C.; Kowalewicz, C.; Simon, G.; Argudín, M.A.; Deplano, A.; Hallin, M.; et al. Large diversity of linezolid-resistant isolates discovered in food-producing animals through linezolid selective monitoring in Belgium in 2019. J. Antimicrob. Chemother. 2021, 77, 49–57. [Google Scholar] [CrossRef]
- Pletinckx, L.J.; Verhegghe, M.; Crombe, F.; Dewulf, J.; De Bleecker, Y.; Rasschaert, G.; Butaye, P.; Goddeeris, B.M.; De Man, I. Evidence of possible methicillin-resistant Staphylococcus aureus ST398 spread between pigs and other animals and people residing on the same farm. Prev. Vet. Med. 2013, 109, 293–303. [Google Scholar] [CrossRef]
- Cuny, C.; Wieler, L.H.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef]
- Goerge, T.; Lorenz, M.B.; van Alen, S.; Hubner, N.O.; Becker, K.; Kock, R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: Prevalence, preventive options and evidence. Vet. Microbiol. 2017, 200, 6–12. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 2021, 74, 21. [Google Scholar] [CrossRef]
- Sahibzada, S.; Abraham, S.; Coombs, G.W.; Pang, S.; Hernández-Jover, M.; Jordan, D.; Heller, J. Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA ST398 between humans and pigs in Australia. Sci. Rep. 2017, 7, 5273. [Google Scholar] [CrossRef]
- Hansen, J.E.; Ronco, T.; Stegger, M.; Sieber, R.N.; Fertner, M.E.; Martin, H.L.; Farre, M.; Toft, N.; Larsen, A.R.; Pedersen, K. LA-MRSA CC398 in Dairy Cattle and Veal Calf Farms Indicates Spillover from Pig Production. Front. Microbiol. 2019, 10, 2733. [Google Scholar] [CrossRef]
- Lienen, T.; Schnitt, A.; Cuny, C.; Maurischat, S.; Tenhagen, B.-A. Phylogenetic Tracking of LA-MRSA ST398 Intra-Farm Transmission among Animals, Humans and the Environment on German Dairy Farms. Microorganisms 2021, 9, 1119. [Google Scholar] [CrossRef]
- Markwart, R.; Willrich, N.; Eckmanns, T.; Werner, G.; Ayobami, O. Low Proportion of Linezolid and Daptomycin Resistance Among Bloodborne Vancomycin-Resistant Enterococcus faecium and Methicillin-Resistant Staphylococcus aureus Infections in Europe. Front. Microbiol. 2021, 12, 1379. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, S.; Lei, T.; Wu, Q.; Zhang, J.; Huang, J.; Dai, J.; Chen, M.; Ding, Y.; Wang, J.; et al. Presence and characterization of methicillin-resistant Staphylococcus aureus co-carrying the multidrug resistance genes cfr and lsa(E) in retail food in China. Int. J. Food Microbiol. 2022, 363, 109512. [Google Scholar] [CrossRef]
- Cuny, C.; Arnold, P.; Hermes, J.; Eckmanns, T.; Mehraj, J.; Schoenfelder, S.; Ziebuhr, W.; Zhao, Q.; Wang, Y.; Fessler, A.T.; et al. Occurrence of cfr-mediated multiresistance in staphylococci from veal calves and pigs, from humans at the corresponding farms, and from veterinarians and their family members. Vet. Microbiol. 2017, 200, 88–94. [Google Scholar] [CrossRef]
- Kang, H.Y.; Moon, D.C.; Mechesso, A.F.; Choi, J.-H.; Kim, S.-J.; Song, H.-J.; Kim, M.H.; Yoon, S.-S.; Lim, S.-K. Emergence of cfr-Mediated Linezolid Resistance in Staphylococcus aureus Isolated from Pig Carcasses. Antibiotics 2020, 9, 769. [Google Scholar] [CrossRef]
- Flor, M.; Käsbohrer, A.; Kaspar, H.; Tenhagen, B.-A.; Wallmann, J. Beiträge der Arbeitsgruppe Antibiotikaresistenz des Bundesinstituts für Risikobewertung (BfR) und des Bundesamtes für Verbraucherschutz und Lebensmittelsicherheit (BVL) zur Evaluierung der 16. AMG-Novelle. Themenkomplex 1: Entwicklung der Antibiotikaabgabe- und -verbrauchsmengen sowie der Therapiehäufigkeit. Bundesminist. Ernähr. Landwirtsch. BMEL 2019, 9, 913197. [Google Scholar]
- Uelze, L.; Grützke, J.; Borowiak, M.; Hammerl, J.A.; Juraschek, K.; Deneke, C.; Tausch, S.H.; Malorny, B. Typing methods based on whole genome sequencing data. One Health Outlook 2020, 2, 3. [Google Scholar] [CrossRef]
- Phiri, B.S.J.; Hang’ombe, B.M.; Mulenga, E.; Mubanga, M.; Maurischat, S.; Wichmann-Schauer, H.; Schaarschmidt, S.; Fetsch, A. Prevalence and diversity of Staphylococcus aureus in the Zambian dairy value chain: A public health concern. Int. J. Food Microbiol. 2022, 375, 109737. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2022, 20, e07209. [Google Scholar] [CrossRef]
- Deneke, C.; Brendebach, H.; Uelze, L.; Borowiak, M.; Malorny, B.; Tausch, S.H. Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with AQUAMIS. Genes 2021, 12, 644. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- IWG-SCC. Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- ISO 20776-1:2006; Clinical Laboratory Testing and In Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Reference Method for Testing the In Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. ISO: Geneva, Switzerland, 2006.
- CLSI. M07. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Approved Standard-Ninth Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- EFSA. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin-resistant Staphylococcus aureus in food-producing animals and food. EFSA J. 2012, 10, 2742. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lienen, T.; Grobbel, M.; Tenhagen, B.-A.; Maurischat, S. Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany. Antibiotics 2022, 11, 1802. https://doi.org/10.3390/antibiotics11121802
Lienen T, Grobbel M, Tenhagen B-A, Maurischat S. Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany. Antibiotics. 2022; 11(12):1802. https://doi.org/10.3390/antibiotics11121802
Chicago/Turabian StyleLienen, Tobias, Mirjam Grobbel, Bernd-Alois Tenhagen, and Sven Maurischat. 2022. "Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany" Antibiotics 11, no. 12: 1802. https://doi.org/10.3390/antibiotics11121802
APA StyleLienen, T., Grobbel, M., Tenhagen, B.-A., & Maurischat, S. (2022). Plasmid-Coded Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus from Food and Livestock in Germany. Antibiotics, 11(12), 1802. https://doi.org/10.3390/antibiotics11121802





