Effect of Phorate on the Development of Hyperglycaemia in Mouse and Resistance Genes in Intestinal Microbiota
Abstract
1. Introduction
2. Results
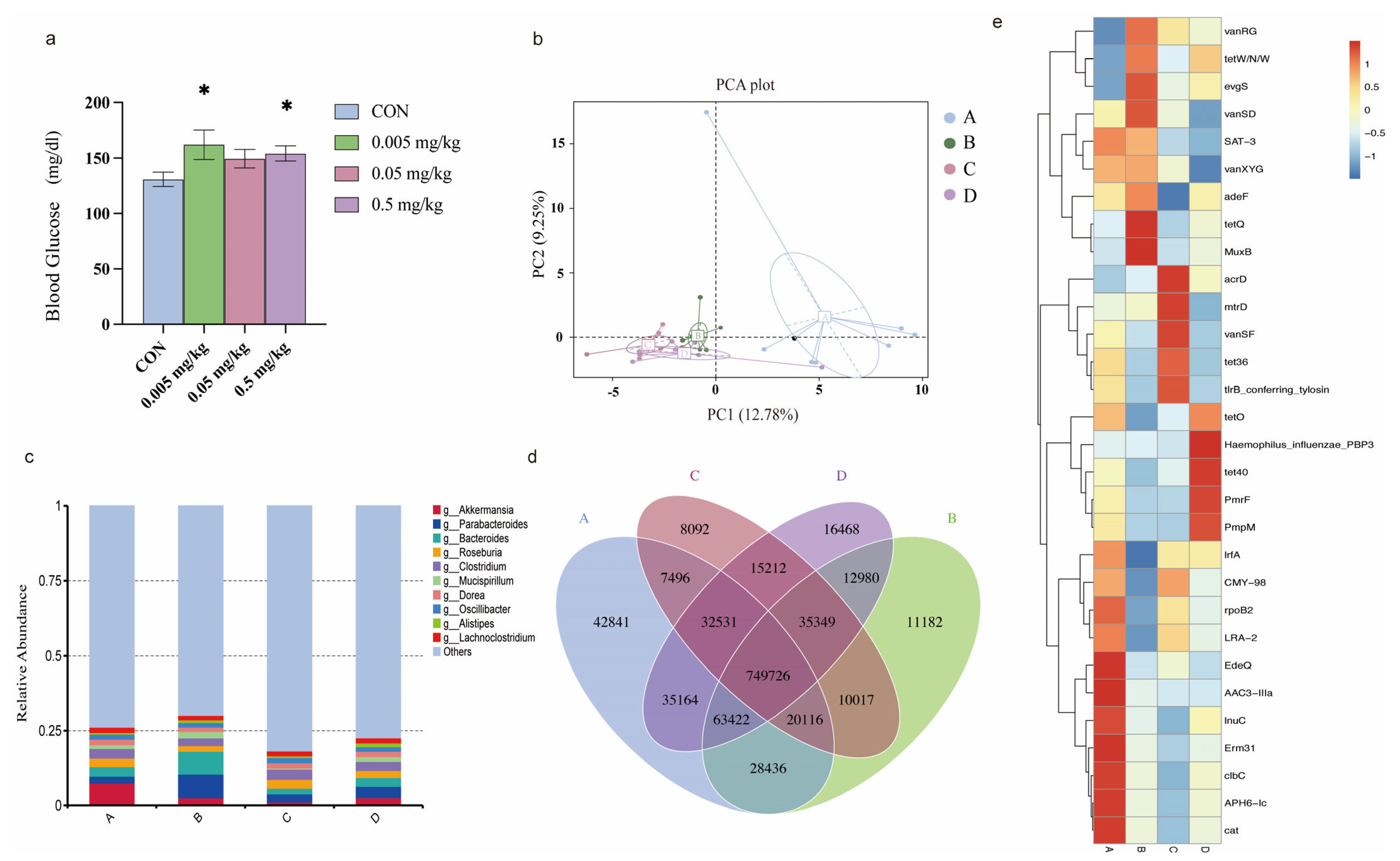
2.1. Phorate Exposure Elevated the Blood Glucose Concentrations in HFD-Fed Mice
2.2. Effects of Phorate on the Beta Diversity of the Gut Microbiota in HFD-Fed Mice
2.3. Relative Abundance of the Intestinal Microbiota in Mice Exposed to Phorate
2.4. Effects of Phorate on Intestinal Microbiota Gene Numbers
2.5. Composition of Intestinal Antibiotic-Resistance Genes after Phorate Exposure
2.6. Relationship between the Resistance Gene Mechanisms and the Intestinal Microbiota Composition
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animals and Treatments
4.3. Blood Glucose Concentrations
4.4. Metagenomic Sequencing
4.5. Annotation of Resistance Genes
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, L.; Li, S.; Qi, L.; Hou, Y.; Zeng, Y.; Xu, W.; Wang, H.; Zhao, X.; Sun, C. Metabonomic analysis of the joint toxic action of long-term low-level exposure to a mixture of four organophosphate pesticides in rat plasma. Mol. Biosyst. 2014, 10, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- De Bleecker, J.L. Organophosphate and carbamate poisoning. Handb. Clin. Neurol. 2008, 91, 401–432. [Google Scholar] [PubMed]
- Saquib, Q.; Attia, S.M.; Siddiqui, M.A.; Aboul-Soud, M.A.; Al-Khedhairy, A.A.; Giesy, J.P.; Musarrat, J. Phorate-induced oxidative stress, DNA damage and transcriptional activation of p53 and caspase genes in male Wistar rats. Toxicol. Appl. Pharmacol. 2012, 259, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, Q.; Bai, R.; Lv, X.; Cao, T.; Shen, S.; Liang, S.; Pang, G. Simultaneous determination and risk assessment of highly toxic pesticides in the market-sold vegetables and fruits in China: A 4-year investigational study. Ecotoxicol. Environ. Saf. 2021, 221, 112428. [Google Scholar] [CrossRef] [PubMed]
- Katsikantami, I.; Colosio, C.; Alegakis, A.; Tzatzarakis, M.N.; Vakonaki, E.; Rizos, A.K.; Sarigiannis, D.A.; Tsatsakis, A.M. Estimation of daily intake and risk assessment of organophosphorus pesticides based on biomonitoring data—The internal exposure approach. Food. Chem. Toxicol. 2019, 123, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Moyer, R.A.; McGarry, K.G., Jr.; Babin, M.C.; Platoff, G.E., Jr.; Jett, D.A.; Yeung, D.T. Kinetic analysis of oxime-assisted reactivation of human, Guinea pig, and rat acetylcholinesterase inhibited by the organophosphorus pesticide metabolite phorate oxon (PHO). Pestic. Biochem. Physiol. 2018, 145, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Bueno, T.S.; Loiko, M.R.; Vidaletti, M.R.; de Oliveira, J.A.; Fetzner, T.; Cerva, C.; de Moraes, L.B.; De Carli, S.; Siqueira, F.M.; Rodrigues, R.O.; et al. Multidrug-resistant Escherichia coli from free-living pigeons (Columba livia): Insights into antibiotic environmental contamination and detection of resistance genes. Zoonoses Public Health 2022, 69, 682–693. [Google Scholar] [CrossRef]
- Salyers, A.A.; Gupta, A.; Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef]
- Isles, N.S.; Mu, A.; Kwong, J.C.; Howden, B.P.; Stinear, T.P. Gut microbiome signatures and host colonization with multidrug-resistant bacteria. Trends Microbiol. 2022, 30, 853–865. [Google Scholar] [CrossRef]
- Shuai, M.; Zhang, G.; Zeng, F.F.; Fu, Y.; Liang, X.; Yuan, L.; Xu, F.; Gou, W.; Miao, Z.; Jiang, Z.; et al. Human Gut Antibiotic Resistome and Progression of Diabetes. Adv. Sci. 2022, 9, e2104965. [Google Scholar] [CrossRef]
- Wang, B.; Tsakiridis, E.E.; Zhang, S.; Llanos, A.; Desjardins, E.M.; Yabut, J.M.; Green, A.E.; Day, E.A.; Smith, B.K.; Lally, J.S.V.; et al. The pesticide chlorpyrifos promotes obesity by inhibiting diet-induced thermogenesis in brown adipose tissue. Nat. Commun. 2021, 12, 5163. [Google Scholar] [CrossRef] [PubMed]
- Thayer, K.A.; Heindel, J.J.; Bucher, J.R.; Gallo, M.A. Role of environmental chemicals in diabetes and obesity: A National Toxicology Program workshop review. Environ. Health Perspect. 2012, 120, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Starling, A.P.; Umbach, D.M.; Kamel, F.; Long, S.; Sandler, D.P.; Hoppin, J.A. Pesticide use and incident diabetes among wives of farmers in the Agricultural Health Study. Occup. Environ. Med. 2014, 71, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.A.; Belal, M.H.; Abou-Arab, A.A.K.; Gad, M.F. Monitoring of pesticides and heavy metals in cucumber fruits produced from different farming systems. Chemosphere 2009, 75, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Timoroglu, I.; Yuzbasioglu, D.; Unal, F.; Yilmaz, S.; Aksoy, H.; Celik, M. Assessment of the genotoxic effects of organophosphorus insecticides phorate and trichlorfon in human lymphocytes. Environ. Toxicol. 2014, 29, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Clement, K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat. Rev. Nephrol. 2016, 12, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Geach, T. Gut microbiota: Mucin-munching bacteria modulate glucose metabolism. Nat. Rev. Endocrinol. 2017, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Dao, M.C.; Belda, E.; Prifti, E.; Everard, A.; Kayser, B.D.; Bouillot, J.L.; Chevallier, J.M.; Pons, N.; Le Chatelier, E.; Ehrlich, S.D.; et al. Akkermansia muciniphila abundance is lower in severe obesity, but its increased level after bariatric surgery is not associated with metabolic health improvement. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E446–E459. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Q.; Liu, M.; Zhang, X.; He, F.; Wang, G. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog. Dis. 2018, 76, fty028. [Google Scholar] [CrossRef]
- Yoon, H.S.; Cho, C.H.; Yun, M.S.; Jang, S.J.; You, H.J.; Kim, J.H.; Han, D.; Cha, K.H.; Moon, S.H.; Lee, K.; et al. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 2021, 6, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Girdhar, K.; Huang, Q.; Chow, I.T.; Vatanen, T.; Brady, C.; Raisingani, A.; Autissier, P.; Atkinson, M.A.; Kwok, W.W.; Kahn, C.R.; et al. A gut microbial peptide and molecular mimicry in the pathogenesis of type 1 diabetes. Proc. Natl. Acad. Sci. USA 2022, 119, e2120028119. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Wang, F.; Yuan, J.; Li, J.; Jiang, D.; Zhang, J.; Li, H.; Wang, R.; Tang, J.; Huang, T.; et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: A 6-month randomised controlled-feeding trial. Gut 2019, 68, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, T.; Sun, J.; Huang, Y.; Liu, T.; Ye, Z.; Hu, J.; Zhang, G.; Chen, H.; Ye, Z.; et al. Calorie restriction ameliorates hyperglycemia, modulates the disordered gut microbiota, and mitigates metabolic endotoxemia and inflammation in type 2 diabetic rats. J. Endocrinol. Investig. 2022. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, W.; Liu, Z.; Yu, J.; Wang, W.; Shang, F.; Xue, T. Role of McbR in the regulation of antibiotic susceptibility in avian pathogenic Escherichia coli. Poult Sci. 2020, 99, 6390–6401. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczak, K.A.; Scott, K.P.; Kelly, D.; Aminov, R.I. Tetracycline resistome of the organic pig gut. Appl. Environ. Microbiol. 2009, 75, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Ruuskanen, M.O.; Erawijantari, P.P.; Havulinna, A.S.; Liu, Y.; Meric, G.; Tuomilehto, J.; Inouye, M.; Jousilahti, P.; Salomaa, V.; Jain, M.; et al. Gut Microbiome Composition Is Predictive of Incident Type 2 Diabetes in a Population Cohort of 5572 Finnish Adults. Diabetes Care 2022, 45, 811–818. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, T.; Guo, Y.; Wang, D.; Liu, Z.; Huang, S.; Peng, C.; Wang, S.; Wang, Y.; Lu, Q.; Xiao, F.; et al. Effect of Phorate on the Development of Hyperglycaemia in Mouse and Resistance Genes in Intestinal Microbiota. Antibiotics 2022, 11, 1584. https://doi.org/10.3390/antibiotics11111584
Cao T, Guo Y, Wang D, Liu Z, Huang S, Peng C, Wang S, Wang Y, Lu Q, Xiao F, et al. Effect of Phorate on the Development of Hyperglycaemia in Mouse and Resistance Genes in Intestinal Microbiota. Antibiotics. 2022; 11(11):1584. https://doi.org/10.3390/antibiotics11111584
Chicago/Turabian StyleCao, Tingting, Yajie Guo, Dan Wang, Zhiyang Liu, Suli Huang, Changfeng Peng, Shaolin Wang, Yang Wang, Qi Lu, Fan Xiao, and et al. 2022. "Effect of Phorate on the Development of Hyperglycaemia in Mouse and Resistance Genes in Intestinal Microbiota" Antibiotics 11, no. 11: 1584. https://doi.org/10.3390/antibiotics11111584
APA StyleCao, T., Guo, Y., Wang, D., Liu, Z., Huang, S., Peng, C., Wang, S., Wang, Y., Lu, Q., Xiao, F., Liang, Z., Zheng, S., Shen, J., Wu, Y., Lv, Z., & Ke, Y. (2022). Effect of Phorate on the Development of Hyperglycaemia in Mouse and Resistance Genes in Intestinal Microbiota. Antibiotics, 11(11), 1584. https://doi.org/10.3390/antibiotics11111584






