Metabolism Profile of Mequindox in Sea Cucumbers In Vivo Using LC-HRMS
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of Sample Preparation
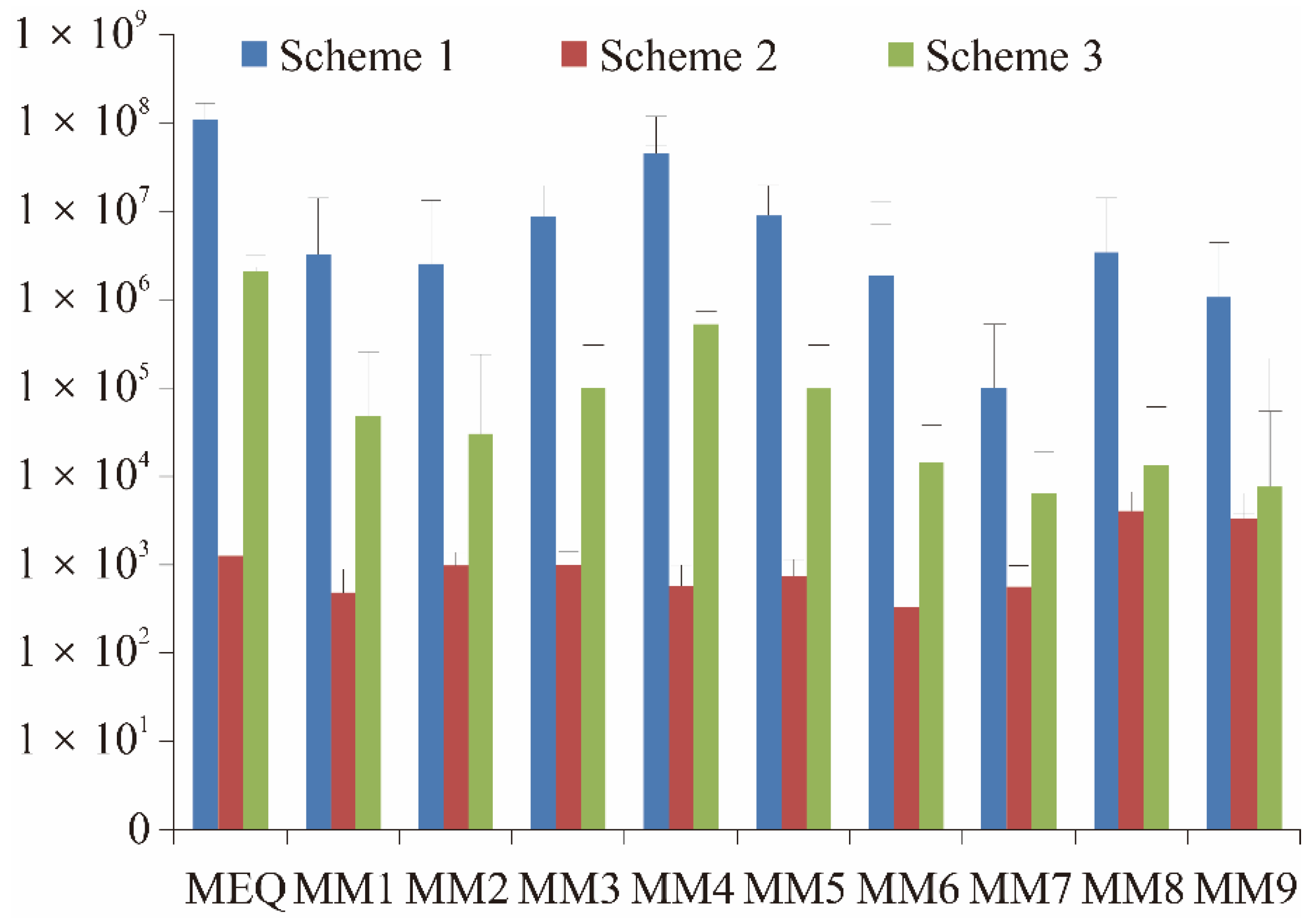
2.2. Identification of Metabolites in Sea Cucumber
2.2.1. Performance of MEQ in Mass Spectrum
2.2.2. Metabolite MM1
2.2.3. Metabolites MM2, MM3, and MM4
2.2.4. Metabolites MM5, MM6, and MM7
2.2.5. Metabolites MM8 and MM9
2.3. Metabolic Pathway of MEQ in Sea Cucumber In Vivo
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Apparatus
3.3. Sample Preparation
3.4. Instrumental Conditions
3.5. Data Processing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suarez-Torres, J.D.; Orozco, C.A.; Ciangherotti, C.E. The numerical probability of carcinogenicity to humans of some antimicrobials: Nitro-monoaromatics (including 5-nitrofurans and 5-nitroimidazoles), quinoxaline-1,4-dioxides (including carbadox), and chloramphenicol. Toxicol. Vitr. 2021, 75, 105172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cheng., G.; Hao, H.; Pan, Y.; Liu, Z.; Dai, M.; Yuan, Z. In vitro antimicrobial activities of animal-used quinoxaline 1, 4-di-N-oxides against mycobacteria, mycoplasma and fungi. BMC Vet. Res. 2016, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, J.; Luo, X.; Ihsan, A.; Liu, X.; Dai, M.; Cheng, G.; Hao, H.; Wang, X.; Yuan, Z. Further investigations into the genotoxicity of quinoxaline-di-N-oxides and their primary metabolites. Food Chem. Toxicol. 2016, 93, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, W.; Chen, Y.; Liu, Y.; Zeng, P.; Xue, F.; Wang, Q. Cytotoxicity of mequindox and its metabolites in HepG2 cells in vitro and murine hepatocytes in vivo. Mutat. Res-Gen. Tox. En. 2016, 797, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Official Journal of the European Union. Commission Regulation No. 2788/98. Off. J. Eur. Union Commun. L347 1998, 34, 3132. [Google Scholar]
- Liu, Q.; Lei, Z.; Gu, C.; Guo, J.; Yu, H.; Fatima, Z.; Zhou, K.; Shabbir, M.A.; Maan, M.K.; Wu, Q. Mequindox induces apoptosis, DNA damage, and carcinogenicity in Wistar rats. Food Chem. Toxicol. 2019, 127, 270–279. [Google Scholar] [CrossRef]
- Rivera, G. Quinoxaline 1, 4-di-N-oxide derivatives: Are They Unselective or Selective Inhibitors? Mini-Rev. Med. Chem. 2022, 22, 15–25. [Google Scholar] [CrossRef]
- Shan, Q.; Liu, Y.; He, L.; Ding, H.; Huang, X.; Yang, F.; Li, Y.; Zeng, Z. Metabolism of mequindox and its metabolites identification in chickens using LC-LTQ-Orbitrap mass spectrometry. J. Chromatogr. B 2012, 881, 96–106. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Z. The metabolism of carbadox, olaquindox, mequindox, quinocetone and cyadox: An overview. Med. Chem. 2013, 9, 1017–1027. [Google Scholar] [CrossRef]
- Mu, P.; Zheng, M.; Xu, M.; Zheng, Y.; Tang, X.; Wang, Y.; Wu, K.; Chen, Q.; Wang, L.; Deng, Y. N-oxide reduction of quinoxaline-1, 4-dioxides catalyzed by porcine aldehyde oxidase SsAOX1. Drug Metab. Dispos. 2014, 42, 511–519. [Google Scholar] [CrossRef]
- Li, Y.; Mao, X.; Jiang, L.; Liu, H.; Nie, X.; Liu, X.; Kong, F.; Luo, P.; Li, Y. Reduction and hydroxylation metabolites of mequindox in holothurian analysis by ultra-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. Sci. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ren, C.; Han, D.; Huang, H.; Zou, R.; Zhang, H.; Xu, Y.; Gong, X.; Zhang, X.; Li, Y. UPLC-MS/MS method for simultaneous determination of three major metabolites of mequindox in holothurian. J. Anal. Methods Chem. 2018, 2018, 2768047. [Google Scholar] [CrossRef]
- Hou, R.; Huang, C.; Rao, K.; Xu, Y.; Wang, Z. Characterized in vitro metabolism kinetics of alkyl organophosphate esters in fish liver and intestinal microsomes. Environ. Sci. Technol. 2018, 52, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Koenig, S.; Fernández, P.; Solé, M. Differences in cytochrome P450 enzyme activities between fish and crustacea: Relationship with the bioaccumulation patterns of polychlorobiphenyls (PCBs). Aquat. Toxicol. 2012, 108, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Ru, X.; Zhang, L.; Li, X.; Liu, S.; Yang, H. Development strategies for the sea cucumber industry in China. J. Oceanol. Limnol. 2019, 37, 300–312. [Google Scholar] [CrossRef]
- Yue, H.; Tian, Y.; Feng, X.; Bo, Y.; Leng, Z.; Dong, P.; Xue, C.; Wang, J. Novel peptides from sea cucumber intestinal hydrolysates promote longitudinal bone growth in adolescent mice through accelerating cell cycle progress by regulating glutamine metabolism. Food Funct. 2022, 13, 7730–7739. [Google Scholar] [CrossRef]
- Lu, Z.; Sun, N.; Dong, L.; Gao, Y.; Lin, S. Production of bioactive peptides from sea cucumber and its potential health benefits: A comprehensive review. J. Agr. Food Chem. 2022, 70, 7607–7625. [Google Scholar] [CrossRef]
- Siddiqui, R.; Boghossian, A.; Khan, N.A. Sea cucumber as a therapeutic aquatic resource for human health. Fish Aquat. Sci. 2022, 25, 251–263. [Google Scholar] [CrossRef]
- Hamed, I.; Özogul, F.; Özogul, Y.; Regenstein, J.M. Marine bioactive compounds and their health benefits: A review. Compr. Rev. Food Sci. F. 2015, 14, 446–465. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, X.H.; Xu, Y.J.; An, H.H.; Zhang, H.W.; Zhou, Q.L.; Zhang, X.Z. Effects of mequindox on non-specific immunity, growth performance and stress resistance of juvenile sea cucumber Apostichopus japonicus. J. Shanghai Ocean Univ. 2014, 23, 848–855. [Google Scholar]
- Liu, Z.Y.; Huang, L.L.; Chen, D.M.; Yuan, Z.H. Metabolism of mequindox in liver microsomes of rats, chicken and pigs. Rapid Commun. Mass. Sp. 2010, 24, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Shen, X.; He, L.; Ding, H.; Tang, Y.; Sun, Y.; Fang, B.; Zeng, Z. Liquid chromatography tandem mass spectrometry for the simultaneous determination of mequindox and its metabolites in porcine tissues. J. Sep. Sci. 2012, 35, 1327–1335. [Google Scholar] [CrossRef]
- Li, Y.; Sun, M.; Mao, X.; Li, J.; Sumarah, M.W.; You, Y.; Wang, Y. Tracing major metabolites of quinoxaline-1,4-dioxides in abalone with high-performance liquid chromatography tandem positive-mode electrospray ionization mass spectrometry. J. Sci. Food Agr. 2019, 99, 5550–5557. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Kavanagh, O.; Gao, H.; Zhang, X.; Deng, S.; Chen, D.; Liu, Z.; Xie, C.; Situ, C.; Yuan, Z. Surface plasmon resonance biosensor for the determination of 3-methyl-quinoxaline-2-carboxylic acid, the marker residue of olaquindox, in swine tissues. Food Chem. 2020, 302, 124623. [Google Scholar] [CrossRef]
- Tan, H.; Pan, Y.; Chen, D.; Tao, Y.; Zhou, K.; Liu, Z.; Yuan, Z.; Huang, L. Discovery of the marker residue of olaquindox in pigs, broilers, and carp. J. Agr. Food Chem. 2019, 67, 6603–6613. [Google Scholar] [CrossRef]
- Li, Y.; Qu, J.; Lin, Y.; Lu, G.; You, Y.; Jiang, G.; Wu, Y. Visible Post-Data analysis protocol for natural mycotoxin production. J. Agr. Food Chem. 2020, 68, 9603–9611. [Google Scholar] [CrossRef]



| Compound Name | RT (min) | [M + H]+ (m/z) | Predicted Composition ([M + H]+) | RDB | Error ppm | Major Fragment Ions | Relative Percentage |
|---|---|---|---|---|---|---|---|
| MEQ, M0 | 4.56 | 219.0764 | C11H11O3N2 | 7.5 | 0.143 | 143.0596, 185.0698, 160.0622, 177.0648, 202.0722, 132.0675 | 33.3% |
| 2-iso-MEQ, M1 | 4.34 | 221.0921 | C11H13O3N2 | 6.5 | 0.367 | 169.0760, 187.0866, 203.0815, 160.0632, 177.0659, 143.0604 | 1.9% |
| 1-DMEQ, M2 | 5.36 | 203.0803 | C11H11O2N2 | 7.5 | 0.817 | 203.0803, 186.0777, 161.0701, 144.0674 | 1.1% |
| 4-DMEQ, M3 | 5.69 | 203.0804 | C11H11O2N2 | 7.5 | 0.718 | 186.0778, 144.0674, 158.0829 | 3.4% |
| 2-iso-1-DMEQ, M4 | 4.60 | 205.0960 | C11H13O2N2 | 6.5 | 0.370 | 169.0751, 187.0855, 205.0960, 171.0900, 145.0752 | 17.4% |
| BDMEQ, M5 | 7.32 | 187.0856 | C11H11ON2 | 7.5 | −5.503 | 159.0909, 145.0752 | 5.2% |
| 2-iso-BDMEQ, M6 | 6.52 | 189.1011 | C11H13ON2 | 6.5 | −5.762 | 171.0907, 143.0598 | 36.6% |
| MQCA, M7 | 5.51 | 189.0654 | C10H9O2N2 | 7.5 | −2.507 | 145.0752, 143.0597, 171.0901 | 0.4% |
| deacetyl-MEQ, M8 | 3.42 | 177.0651 | C9H9O2N2 | 6.5 | 2.010 | 160.0622, 143.0606 | 0.5% |
| Deacetyl-1-DMEQ, M9 | 5.35 | 161.0706 | C9H9ON2 | 6.5 | −4.963 | 144.0644 | 0.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, X.; Zhou, X.; He, J.; Liu, G.; Liu, H.; Zhao, H.; Luo, P.; Wu, Y.; Li, Y. Metabolism Profile of Mequindox in Sea Cucumbers In Vivo Using LC-HRMS. Antibiotics 2022, 11, 1599. https://doi.org/10.3390/antibiotics11111599
Mao X, Zhou X, He J, Liu G, Liu H, Zhao H, Luo P, Wu Y, Li Y. Metabolism Profile of Mequindox in Sea Cucumbers In Vivo Using LC-HRMS. Antibiotics. 2022; 11(11):1599. https://doi.org/10.3390/antibiotics11111599
Chicago/Turabian StyleMao, Xin, Xiaozhen Zhou, Jun He, Gongzhen Liu, Huihui Liu, Han Zhao, Pengjie Luo, Yongning Wu, and Yanshen Li. 2022. "Metabolism Profile of Mequindox in Sea Cucumbers In Vivo Using LC-HRMS" Antibiotics 11, no. 11: 1599. https://doi.org/10.3390/antibiotics11111599
APA StyleMao, X., Zhou, X., He, J., Liu, G., Liu, H., Zhao, H., Luo, P., Wu, Y., & Li, Y. (2022). Metabolism Profile of Mequindox in Sea Cucumbers In Vivo Using LC-HRMS. Antibiotics, 11(11), 1599. https://doi.org/10.3390/antibiotics11111599







