Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants
Abstract
1. Introduction
2. Terpenoids
2.1. The Antimicrobial Mechanisms of Terpenoids
- Cell membrane destruction: Terpenoids mainly use their lipophilicity to destroy the cell membrane of bacteria. Terpenoids can pass through the phospholipid bilayer of bacteria and diffuse inward, showing antibacterial or bactericidal effects [20]. Since the integrity of the cell membrane is very important for the normal physiological activities of bacteria, the damage of terpenoids to the membrane will affect the bacteria’s basic physiological activities, and the important substances such as proteins and important enzymes in the cell will be lost, finally achieving the antimicrobial effect [21]. It is reported that 1,8-cineole (Table 1), a monoterpene substance extracted from Eucalyptus globulus Labill, showed antibacterial effect against Acinetobacter baumannii, Candida albicans, a methicillin-resistant Staphylococcus aureus (MRSA) strain, and Escherichia coli by destroying the cell membrane [22]. In another study, the researchers exposed Salmonella typhimurium, E. coli O157: H7, Pseudomonas fluorescence, Brochotrix thermophacta, and Staphylococcus aureus cells to cinnamaldehyde (Table 1), carvacrol (Table 1), thymol (Table 1), eugenol (Table 1), and limonene (Table 1), and observed their membrane damage through scanning electron microscopy. These results found that terpenoids can achieve bacteriostatic effects by destroying the membrane structure [23]. The mechanism of action and target sites on microbial cells are graphically illustrated in [20,21].
- Anti-quorum sensing (QS) action: The QS system is an intercellular communication system [20]. It is a communication mode for bacteria to coordinate the interaction between bacteria and other organisms, which is also the main reason for the emergence of antibiotic resistance [19]. The group sensing signal loop of Gram-positive and Gram-negative bacteria has been introduced and illustrated in the literature [24]. Studies have shown that a low concentration of cinnamaldehyde can effectively inhibit the QS effect between bacteria [25]. Low concentrations of carvacrol and thymol can effectively inhibit the self-inducer of bacteria, namely, acyl homoserine lactone (AHL), thus achieving the inhibition of QS [26]. The action mechanism of cinnamaldehyde inhibiting the acyl homoserine lactones and other autoinducers involved in the quorum sensing is illustrated in [27].
- Inhibition of ATP and its enzyme: ATP is the most direct energy source in organisms, and it is also a necessary element for microorganisms to maintain normal operation and work. Terpenoids can act on the cell membrane, resulting in the difference in ATP concentration inside and outside the cell, leading to the disorder of the cell membrane, thus conducting the antibacterial activity [20]. For example, terpenoid eugenol and thymol could target the cell membrane to show fungicidal activity against C. albicans by inhibiting H+-ATPase, which will lead to intracellular acidification and cell death [28]. In another study, the researchers treated the target pathogen with the MIC of carvacrol. The extracellular ATP concentrations of the samples were measured with the help of a luminometer (Biotek). On the basis of absorbance analysis at 260 nm, this study observed that carvacrol disrupted the E. coli membrane, while the release of potassium ions and ATP was also detected [29].
- Inhibition of protein synthesis: The physiological activity of bacteria is inseparable from protein synthesis. Terpenoids, as inhibitors of protein synthesis, can achieve an antibacterial effect by blocking any process of the protein synthesis pathway. Some studies have shown that cinnamaldehyde can reduce the in vitro assembly reaction and the binding reaction of FtsZ (filamenting temperature-sensitive mutant Z)-type protein, a prokaryotic homolog of tubulin that regulates cell division. In addition, cinnamaldehyde can inhibit the hydrolysis of GTP and bind to FtsZ, as well as interfere with the formation of z-loop of cell dynamics, thus showing antibacterial activity against bacteria [30]. In the latest research, the researchers used calculations, biochemistry, and in-vivo-based assays to verify that cinnamaldehyde is a potential inhibitor of S. typhimurium (stFtsZ), and its inhibition rate of stFtsZ GTPase activity and polymerization is up to 70% [31].
- The synergistic effect: For example, the synergistic antibacterial effect of eugenol with carvacrol and thymol is due to the ability of carvacrol and thymol to penetrate the extracellular membrane, thus making it easier for eugenol to enter the cytoplasmic membrane or increasing the number, size, and duration of pores to bind to membrane proteins for better antibacterial activity [32]. The reaction mechanism is shown in the literature [27].
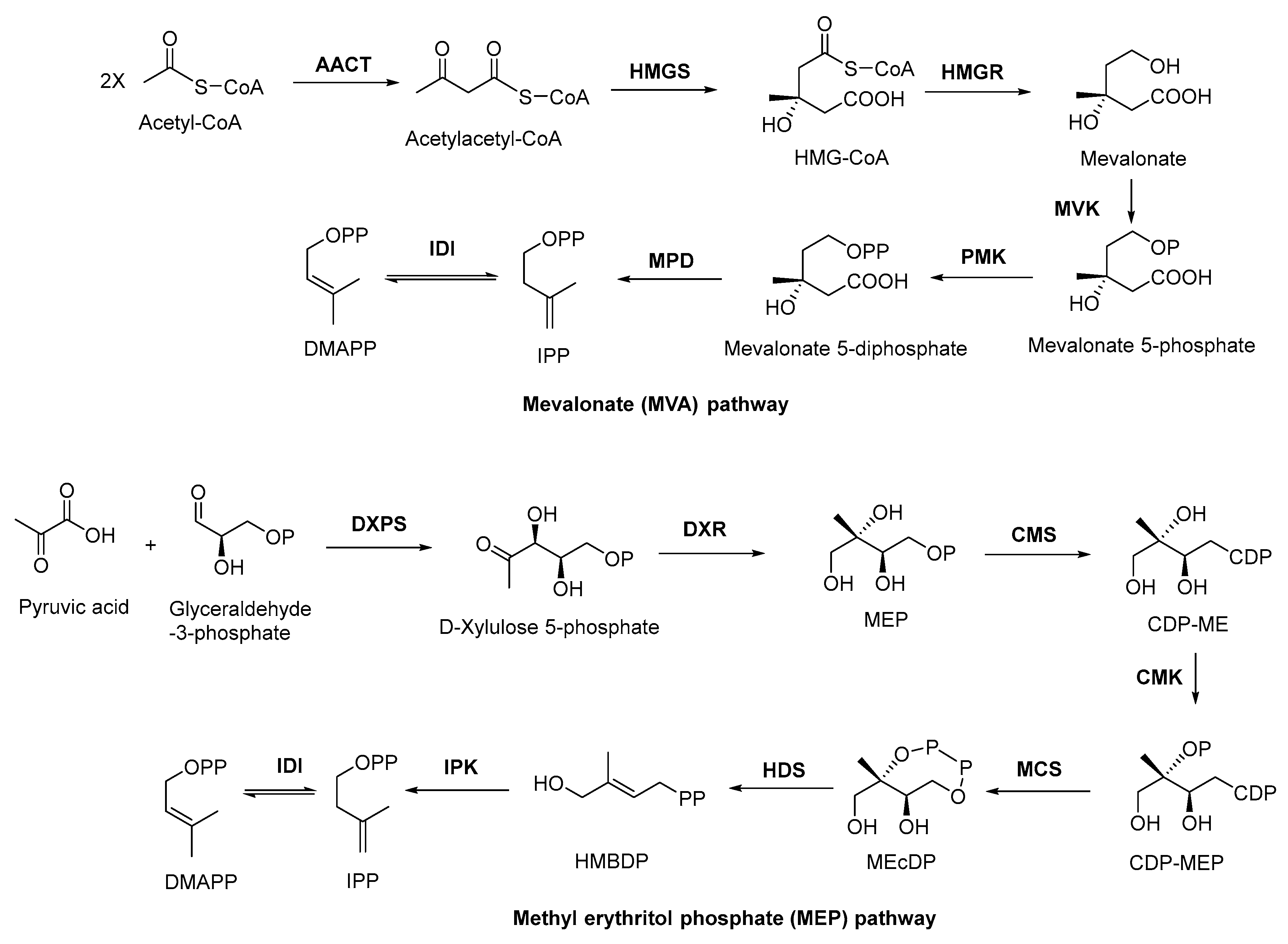
2.2. Biosynthesis of Terpenoid Precursors
2.3. Discovery, Biosynthesis Investigations, and Engineering Strain Construction of the Representative Terpenoid Antimicrobial Agent—Artemisinin
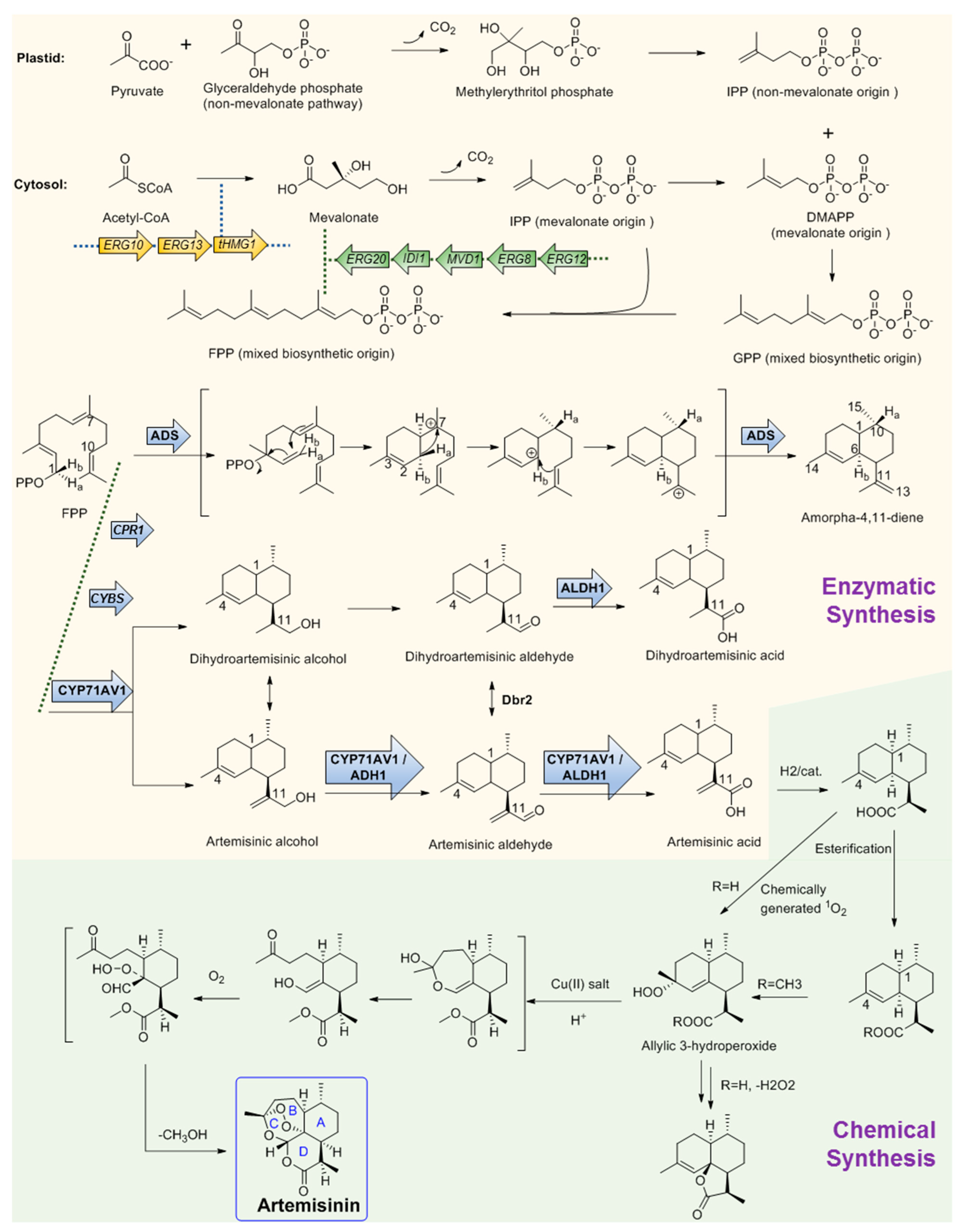
2.3.1. Discovery and Predicted Action Mechanism of Artemisinin
2.3.2. Key Enzymes Involved in The Biosynthesis Pathway of Artemisinin
2.3.3. Microbial Production of Artemisinic Acid
2.3.4. Chemical Conversion to Produce Artemisinin
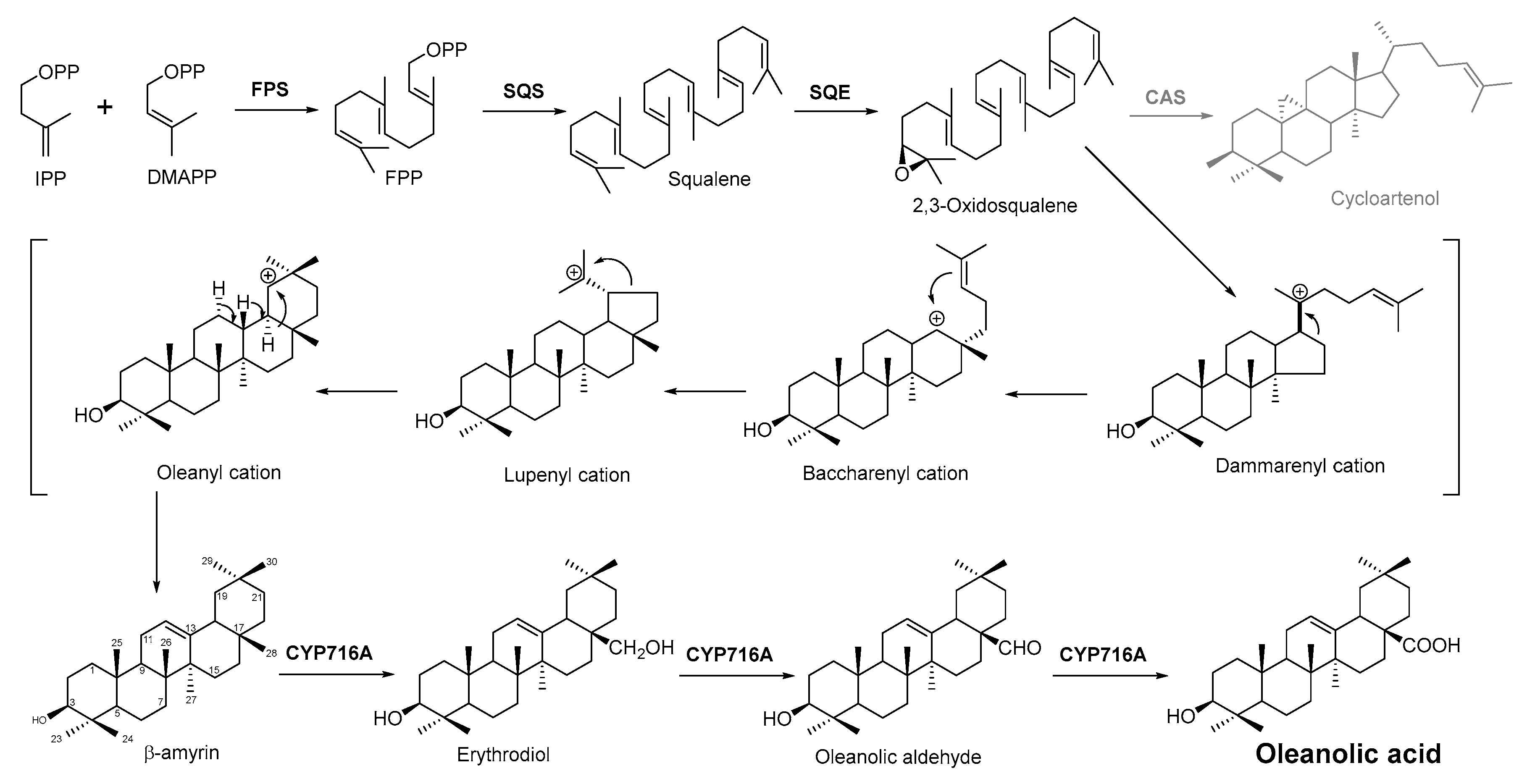
2.4. Biosynthesis Pathway Investigation of the Terpenoid Antimicrobial Agent—Oleanolic Acid
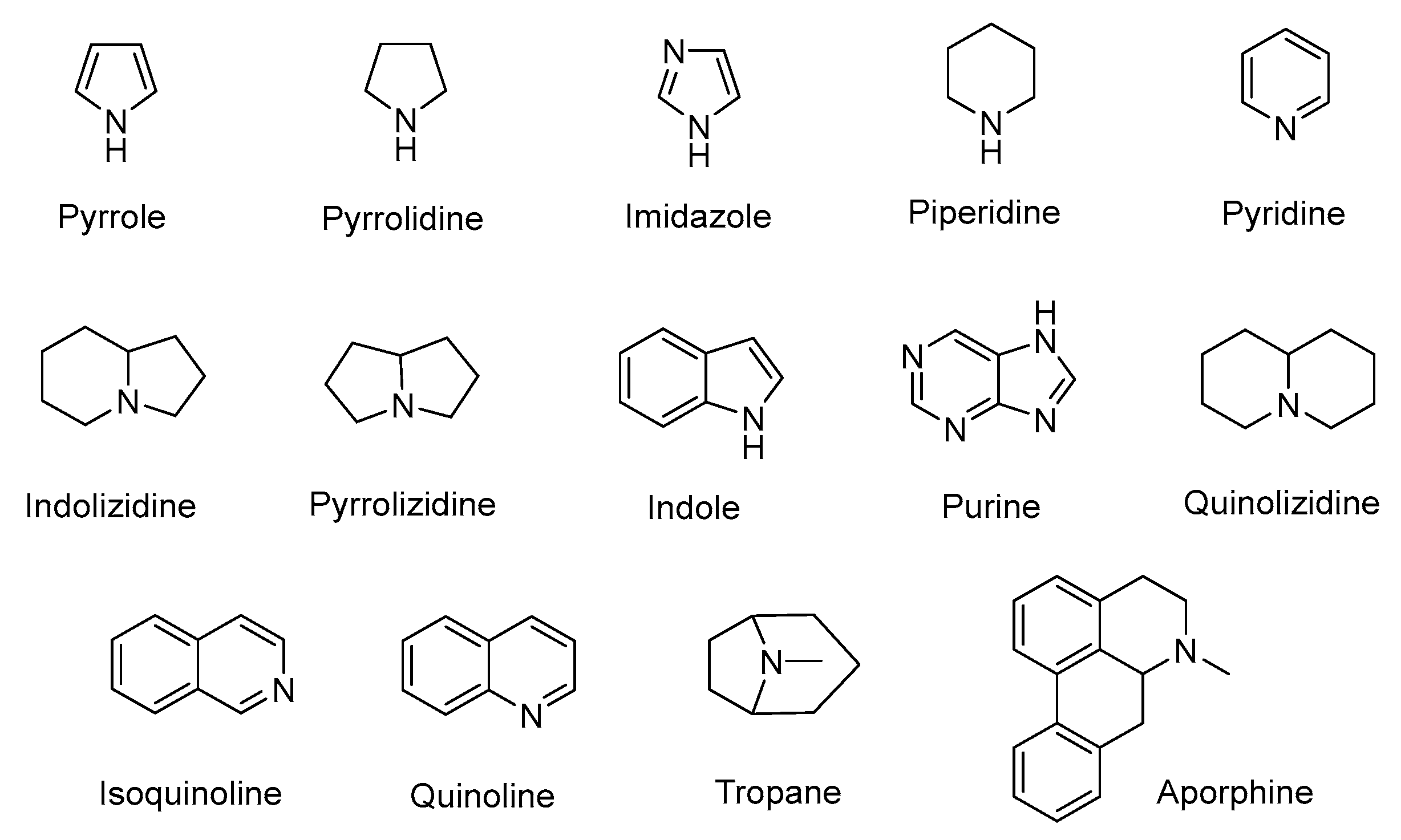
3. Alkaloids
3.1. Plant-Originated Alkaloids with Antimicrobial Bioactivities
3.2. Biosynthesis Investigation of the Representative Antimicrobial Alkaloid Compound—Berberine
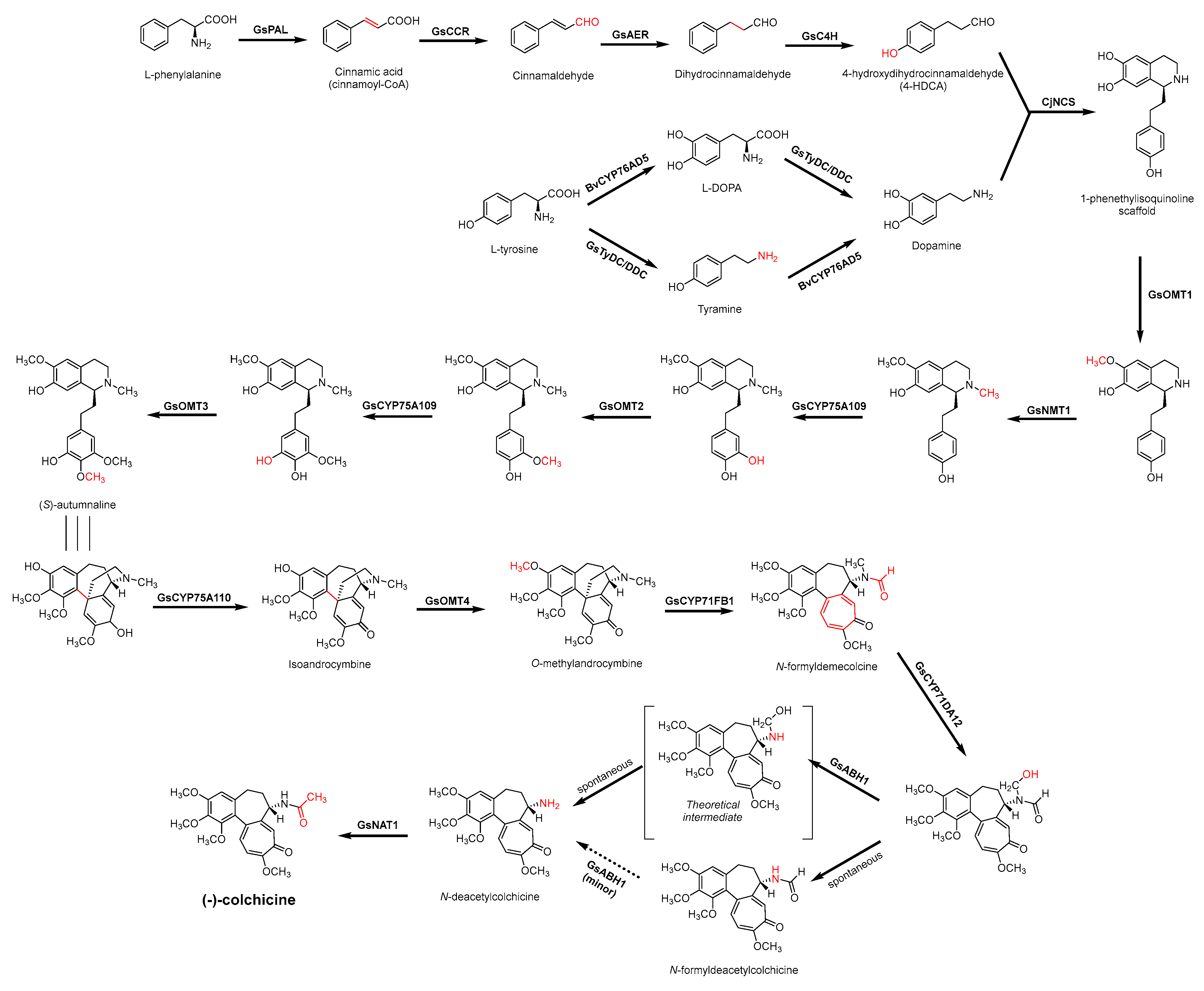
3.3. Biosynthesis Investigations of the Antimicrobial Alkaloid Compound—Colchicine
3.4. De Novo Biosynthetic Production of Colchicine in Nicotiana benthamiana
3.5. Biosynthesis Investigations of Other Antimicrobial Alkaloids
4. Flavonoids
4.1. Structure–Activity Relationship Study on Antimicrobial Activity of Flavonoids
4.2. Antibacterial Effects and Action Mechanisms of Flavonoid Antimicrobial Agents
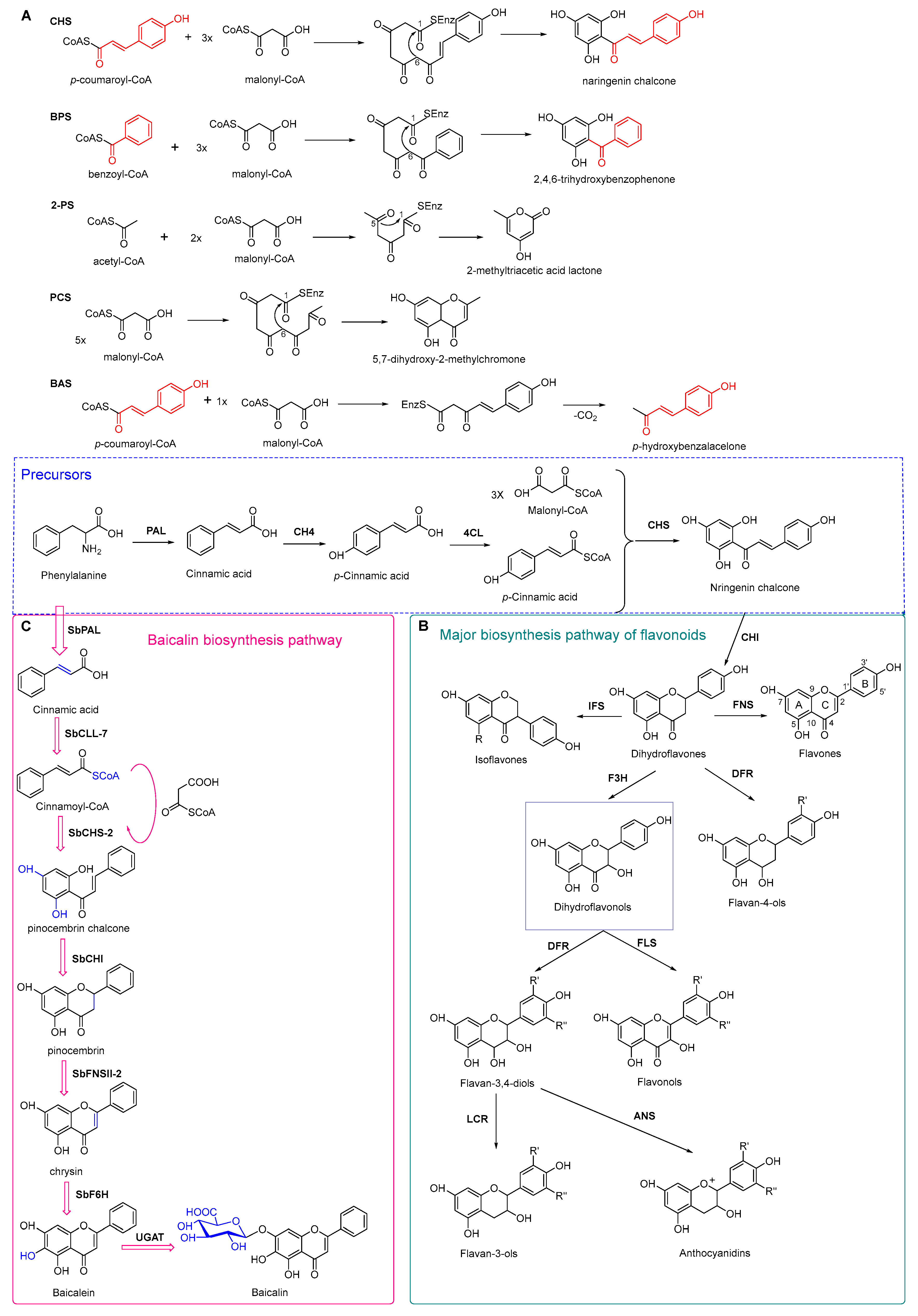
4.3. Plant Type III Polyketide Synthase
4.4. Biosynthesis Investigations of the Antimicrobial Flavonoid Compound—Baicalin
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reinhardt, U.; Cheng, T. The world health report 2000–Health systems: Improving performance. Bull. World Health Organ. 2000, 78, 1064. [Google Scholar]
- Mohr, K.I. History of Antibiotics Research. Curr. Top Microbiol. Immunol. 2016, 398, 237–272. [Google Scholar]
- Ali, S.M.; Siddiqui, R.; Khan, N.A. Antimicrobial discovery from natural and unusual sources. J. Pharm. Pharmacol. 2018, 70, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S. Infectious diseases: Considerations for the 21st century. Clin. Infect. Dis. 2001, 32, 675–685. [Google Scholar] [CrossRef]
- Keita, K.; Darkoh, C.; Okafor, F. Secondary plant metabolites as potent drug candidates against antimicrobial-resistant pathogens. SN Appl. Sci. 2022, 4, 209. [Google Scholar] [CrossRef]
- Davies, S.C.; Fowler, T.; Watson, J.; Livermore, D.M.; Walker, D. Annual Report of the Chief Medical Officer: Infection and the rise of antimicrobial resistance. Lancet 2013, 381, 1606–1609. [Google Scholar] [CrossRef]
- Jia, H.; Li, L.; Li, W.; Hou, T.; Ma, H.; Yang, Y.; Wu, A.; Liu, Y.; Wen, J.; Yang, H.; et al. Impact of Healthcare-Associated Infections on Length of Stay: A Study in 68 Hospitals in China. Biomed. Res. Int. 2019, 2019, 2590563. [Google Scholar] [CrossRef]
- Porras, G.; Chassagne, F.; Lyles, J.T.; Marquez, L.; Dettweiler, M.; Salam, A.M.; Samarakoon, T.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. Ethnobotany and the Role of Plant Natural Products in Antibiotic Drug Discovery. Chem. Rev. 2021, 121, 3495–3560. [Google Scholar] [CrossRef]
- Tiwari, P.; Khare, T.; Shriram, V.; Bae, H.; Kumar, V. Plant synthetic biology for producing potent phyto-antimicrobials to combat antimicrobial resistance. Biotechnol. Adv. 2021, 48, 107729. [Google Scholar] [CrossRef] [PubMed]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards Advances in Medicinal Plant Antimicrobial Activity: A Review Study on Challenges and Future Perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef]
- Gonzalez-Lamothe, R.; Mitchell, G.; Gattuso, M.; Diarra, M.S.; Malouin, F.; Bouarab, K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009, 10, 3400–3419. [Google Scholar] [CrossRef] [PubMed]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef]
- Savoia, D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef]
- Shankar, S.R.; Rangarajan, R.; Sarada, D.V.L.; Kumar, C.S. Evaluation of antibacterial activity and phytochemical screening of Wrightia tinctoria L. Pharmacogn. J. 2010, 2, 19–22. [Google Scholar] [CrossRef]
- Li, J.M.; Feng, S.S.; Liu, X.; Jia, X.; Qiao, F.L.; Guo, J.L.; Deng, S.S. Effects of traditional chinese medicine and its active ingredients on drug-resistant bacteria. Front. Pharmacol. 2022, 13, 837907. [Google Scholar] [CrossRef]
- Kannappan, A.; Sivaranjani, M.; Srinivasan, R.; Rathna, J.; Pandian, S.K.; Ravi, A.V. Inhibitory efficacy of geraniol on biofilm formation and development of adaptive resistance in Staphylococcus epidermidis RP62A. J. Med. Microbiol. 2017, 66, 1506–1515. [Google Scholar] [CrossRef]
- Upadhyay, H.C.; Dwivedi, G.R.; Roy, S.; Sharma, A.; Darokar, M.P.; Srivastava, S.K. Phytol derivatives as drug resistance reversal agents. ChemMedChem 2014, 9, 1860–1868. [Google Scholar] [CrossRef]
- Nhiri, M.; Mrid, R.B.; Omari, R.E.; Bouargalne, Y. New insights into the therapeutic effects of phenolic acids from sorghum seeds. J. Rep. Pharm. Sci. 2019, 8, 91–101. [Google Scholar] [CrossRef]
- Sharma, A.; Biharee, A.; Kumar, A.; Jaitak, V. Antimicrobial terpenoids as a potential substitute in overcoming antimicrobial resistance. Curr. Drug. Targets 2020, 21, 1476–1494. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef]
- Burt, S.A.; Reinders, R.D. Antibacterial activity of selected plant essential oils against Escherichia coli O157:H7. Lett. Appl. Microbiol. 2003, 36, 162–167. [Google Scholar] [CrossRef]
- Mulyaningsih, S.; Sporer, F.; Zimmermann, S.; Reichling, J.; Wink, M. Synergistic properties of the terpenoids aromadendrene and 1,8-cineole from the essential oil of Eucalyptus globulus against antibiotic-susceptible and antibiotic-resistant pathogens. Phytomedicine 2010, 17, 1061–1066. [Google Scholar] [CrossRef]
- Di Pasqua, R.; Betts, G.; Hoskins, N.; Edwards, M.; Ercolini, D.; Mauriello, G. Membrane toxicity of antimicrobial compounds from Essential oils. J. Agric. Food Chem. 2007, 55, 4863–4870. [Google Scholar] [CrossRef]
- Mulat, M.; Pandita, A.; Khan, F. Medicinal plant compounds for combating the multi-drug resistant pathogenic bacteria: A Review. Curr. Pharm. Biotechnol. 2019, 20, 183–196. [Google Scholar] [CrossRef]
- Niu, C.; Afre, S.; Gilbert, E.S. Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett. Appl. Microbiol. 2006, 43, 489–494. [Google Scholar] [CrossRef]
- Myszka, K.; Schmidt, M.T.; Majcher, M.; Juzwa, W.; Olkowicz, M.; Czaczyk, K. Inhibition of quorum sensing-related biofilm of Pseudomonas fluorescens KM121 by Thymus vulgare essential oil and its major bioactive compounds. Int. Biodeter. Biodegr. 2016, 114, 252–259. [Google Scholar] [CrossRef]
- Mittal, R.P.; Rana, A.; Jaitak, V. Essential Oils: An impending substitute of synthetic antimicrobial agents to overcome antimicrobial resistance. Curr. Drug Targets 2019, 20, 605–624. [Google Scholar] [CrossRef]
- Ahmad, A.; Khan, A.; Yousuf, S.; Khan, L.A.; Manzoor, N. Proton translocating ATPase mediated fungicidal activity of eugenol and thymol. Fitoterapia 2010, 81, 1157–1162. [Google Scholar] [CrossRef]
- Khan, I.; Bahuguna, A.; Shukla, S.; Aziz, F.; Chauhan, A.K.; Ansari, M.B.; Bajpai, V.K.; Huh, Y.S.; Kang, S.C. Antimicrobial potential of the food-grade additive carvacrol against uropathogenic E. coli based on membrane depolarization, reactive oxygen species generation, and molecular docking analysis. Microb. Pathog. 2020, 142, 104046. [Google Scholar] [CrossRef]
- Domadia, P.; Swarup, S.; Bhunia, A.; Sivaraman, J.; Dasgupta, D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 2007, 74, 831–840. [Google Scholar] [CrossRef]
- Naz, F.; Kumar, M.; Koley, T.; Sharma, P.; Haque, M.A.; Kapil, A.; Kumar, M.; Kaur, P.; Ethayathulla, A.S. Screening of plant-based natural compounds as an inhibitor of FtsZ from Salmonella typhi using the computational, biochemical and in vitro cell-based studies. Int. J. Biol. Macromol. 2022, 219, 428–437. [Google Scholar] [CrossRef]
- Zhou, F.; Ji, B.; Zhang, H.; Jiang, H.; Yang, Z.; Li, J.; Li, J.; Ren, Y.; Yan, W. Synergistic effect of thymol and carvacrol combined with chelators and organic acids against Salmonella Typhimurium. J. Food Prot. 2007, 70, 1704–1709. [Google Scholar] [CrossRef]
- Fontanay, S.; Grare, M.; Mayer, J.; Finance, C.; Duval, R.E. Ursolic, oleanolic and betulinic acids: Antibacterial spectra and selectivity indexes. J. Ethnopharmacol. 2008, 120, 272–276. [Google Scholar] [CrossRef]
- Hikal, D.M. Antibacterial Activity of Piperine and Black Pepper Oil. Biosci. Biotechnol. Res. Asia 2018, 15, 877–880. [Google Scholar] [CrossRef]
- Khan, I.A.; Mirza, Z.M.; Kumar, A.; Verma, V.; Qazi, G.N. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 810–812. [Google Scholar] [CrossRef]
- Mittal, R.P.; Jaitak, V. Plant-derived natural alkaloids as new antimicrobial and adjuvant agents in existing antimicrobial therapy. Curr. Drug Targets 2019, 20, 1409–1433. [Google Scholar] [CrossRef]
- Su, F.; Wang, J. Berberine inhibits the MexXY-OprM efflux pump to reverse imipenem resistance in a clinical carbapenem-resistant Pseudomonas aeruginosa isolate in a planktonic state. Exp. Ther. Med. 2018, 15, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, E.; Cedraro, N.; Mangiaterra, G.; Citterio, B.; Mobbili, G.; Minnelli, C.; Bizzaro, D.; Biavasco, F.; Galeazzi, R. Natural alkaloid berberine activity against Pseudomonas aeruginosa MexXY-mediated aminoglycoside resistance: In Silico and in vitro Studies. J. Nat. Prod. 2019, 82, 1935–1944. [Google Scholar] [CrossRef]
- Mujtaba, M.A.; Akhter, M.H.; Alam, M.S.; Ali, M.D.; Hussain, A. An updated review on therapeutic potential and recent advances in drug delivery of berberine: Current status and future prospect. Curr. Pharm. Biotechnol. 2022, 23, 60–71. [Google Scholar] [CrossRef]
- Wei, W.Y.; Du, H.X.; Shao, C.Y.; Zhou, H.F.; Lu, Y.Y.; Yu, L.; Wan, H.T.; He, Y. Screening of antiviral components of ma huang tang and investigation on the ephedra alkaloids efficacy on influenza virus type A. Front. Pharmacol. 2019, 10, 961. [Google Scholar] [CrossRef]
- He, N.; Wang, P.; Wang, P.; Ma, C.; Kang, W. Antibacterial mechanism of chelerythrine isolated from root of Toddalia asiatica (Linn) Lam. BMC Complement. Altern. Med. 2018, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. 8-Hydroxyquinolines: A review of their metal chelating properties and medicinal applications. Drug Des. Dev. Ther. 2013, 7, 1157–1178. [Google Scholar] [CrossRef]
- Houdkova, M.; Rondevaldova, J.; Doskocil, I.; Kokoska, L. Evaluation of antibacterial potential and toxicity of plant volatile compounds using new broth microdilution volatilization method and modified MTT assay. Fitoterapia 2017, 118, 56–62. [Google Scholar] [CrossRef] [PubMed]
- McMahon, J.B.; Currens, M.J.; Gulakowski, R.J.; Buckheit, R.W.; Lackman-Smith, C.; Hallock, Y.F.; Boyd, M.R. Michellamine B, a novel plant alkaloid, inhibits human immunodeficiency virus-induced cell killing by at least two distinct mechanisms. Antimicrob. Agents Chemother. 1995, 39, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Hamoud, R.; Reichling, J.; Wink, M. Synergistic antibacterial activity of the combination of the alkaloid sanguinarine with EDTA and the antibiotic streptomycin against multidrug resistant bacteria. J. Pharm. Pharmacol. 2015, 67, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Beuria, T.K.; Santra, M.K.; Panda, D. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry 2005, 44, 16584–16593. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.J.; Rao, G.X.; Wang, J.; Luo, L.Y.; He, G.H.; Wang, C.Y.; Ma, C.Y.; Luo, X.X.; Hou, Z.; Xu, G.L. Roemerine improves the survival rate of septicemic BALB/c mice by increasing the cell membrane permeability of Staphylococcus aureus. PLoS ONE 2015, 10, e0143863. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Avci, F.G.; Atas, B.; Aksoy, C.S.; Kurpejovic, E.; Toplan, G.G.; Gurer, C.; Guillerminet, M.; Orelle, C.; Jault, J.M.; Akbulut, B.S. Repurposing bioactive aporphine alkaloids as efflux pump inhibitors. Fitoterapia 2019, 139, 104371. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.S.; Lins, M.O.; Le Hyaric, M.; Barros, T.F.; Velozo, E.S. In vitro antibacterial effects of Zanthoxylum tingoassuiba root bark extracts and two of its alkaloids against multiresistant Staphylococcus aureus. Rev. Bras. Farmacogn. 2017, 27, 195–198. [Google Scholar] [CrossRef]
- Guzman, J.D.; Wube, A.; Evangelopoulos, D.; Gupta, A.; Hufner, A.; Basavannacharya, C.; Rahman, M.M.; Thomaschitz, C.; Bauer, R.; McHugh, T.D.; et al. Interaction of N-methyl-2-alkenyl-4-quinolones with ATP-dependent MurE ligase of Mycobacterium tuberculosis: Antibacterial activity, molecular docking and inhibition kinetics. J. Antimicrob. Chemother. 2011, 66, 1766–1772. [Google Scholar] [CrossRef] [PubMed]
- Hochfellner, C.; Evangelopoulos, D.; Zloh, M.; Wube, A.; Guzman, J.D.; McHugh, T.D.; Kunert, O.; Bhakta, S.; Bucar, F. Antagonistic effects of indoloquinazoline alkaloids on antimycobacterial activity of evocarpine. J. Appl. Microbiol. 2015, 118, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Celiz, G.; Daz, M.; Audisio, M.C. Antibacterial activity of naringin derivatives against pathogenic strains. J. Appl. Microbiol. 2011, 111, 731–738. [Google Scholar] [CrossRef]
- Pyrzynska, K. Hesperidin: A Review on Extraction Methods, Stability and Biological Activities. Nutrients 2022, 14, 2387. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.S.; Babu, T.H.; Srinivas, P.V.; Sastry, B.S.; Kishore, K.H.; Murty, U.S.; Rao, J.M. Synthesis and in vitro study of novel 7-O-acyl derivatives of Oroxylin A as antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 3953–3956. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhao, B.; Wang, D.E.; Yao, T.Y.; Pang, L.; Tu, Q.; Ahmed, S.M.; Liu, J.J.; Wang, J.Y. Nitrogen-containing apigenin analogs: Preparation and biological activity. Molecules 2012, 17, 14748–14764. [Google Scholar] [CrossRef] [PubMed]
- Chinnam, N.; Dadi, P.K.; Sabri, S.A.; Ahmad, M.; Kabir, M.A.; Ahmad, Z. Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. Int. J. Biol. Macromol. 2010, 46, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Martinez, M.D.; Navarro-Peran, E.; Cabezas-Herrera, J.; Ruiz-Gomez, J.; Garcia-Canovas, F.; Rodriguez-Lopez, J.N. Antifolate activity of epigallocatechin gallate against Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2005, 49, 2914–2920. [Google Scholar] [CrossRef] [PubMed]
- Pejin, B.; Ciric, A.; Markovic, J.D.; Glamoclija, J.; Nikolic, M.; Stanimirovic, B.; Sokovic, M. Quercetin potently reduces biofilm formation of the strain Pseudomonas aeruginosa PAO1 in vitro. Curr. Pharm. Biotechnol. 2015, 16, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.; Lamb, A.J. Detection of galangin-induced cytoplasmic membrane damage in Staphylococcus aureus by measuring potassium loss. J. Ethnopharmacol. 2005, 101, 243–248. [Google Scholar] [CrossRef]
- Gradisar, H.; Pristovsek, P.; Plaper, A.; Jerala, R. Green tea catechins inhibit bacterial DNA gyrase by interaction with its ATP binding site. J. Med. Chem. 2007, 50, 264–271. [Google Scholar] [CrossRef]
- Fathima, A.; Rao, J.R. Selective toxicity of Catechin-a natural flavonoid towards bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 6395–6402. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, X.Y.; Martin, C. Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci. Bull. 2016, 61, 1391–1398. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Meng, J.; Qiu, T.; Wang, W.; Wang, R.; Liu, J. Baicalin inhibits biofilm formation by influencing primary adhesion and aggregation phases in Staphylococcus saprophyticus. Vet. Microbiol. 2021, 262, 109242. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, N.; Zhang, S.; Zhang, L.; Liu, Q. Phloretin inhibited the pathogenicity and virulence factors against Candida albicans. Bioengineered 2021, 12, 2420–2431. [Google Scholar] [CrossRef]
- Wang, D.; Xie, K.; Zou, D.; Meng, M.; Xie, M. Inhibitory effects of silybin on the efflux pump of methicillin-resistant Staphylococcus aureus. Mol. Med. Rep. 2018, 18, 827–833. [Google Scholar]
- Yin, Z.; Dickschat, J.S. Engineering fungal terpene biosynthesis. Nat. Prod. Rep. 2022. Advance Article. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically useful plant terpenoids: Biosynthesis, occurrence, and mechanism of action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Rahman, F.I.; Hussain, F.; Rahman, S.M.A.; Rahman, M.M. Antimicrobial diterpenes: Recent development from natural sources. Front. Pharmacol. 2021, 12, 820312. [Google Scholar] [CrossRef]
- Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef]
- Tu, Y. Artemisinin-a gift from traditional Chinese medicine to the world (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 10210–10226. [Google Scholar] [CrossRef]
- O’Neill, P.M.; Barton, V.E.; Ward, S.A. The molecular mechanism of action of artemisinin—The debate continues. Molecules 2010, 15, 1705–1721. [Google Scholar] [CrossRef] [PubMed]
- Meshnick, S.R.; Taylor, T.E.; Kamchonwongpaisan, S. Artemisinin and the antimalarial endoperoxides: From herbal remedy to targeted chemotherapy. Microbiol. Rev. 1996, 60, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Klonis, N.; Crespo-Ortiz, M.P.; Bottova, I.; Abu-Bakar, N.; Kenny, S.; Rosenthal, P.J.; Tilley, L. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. USA 2011, 108, 11405–11410. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Yu, R. Artemisinin biosynthesis and its regulatory enzymes: Progress and perspective. Pharmacogn. Rev. 2011, 5, 189–194. [Google Scholar] [PubMed]
- Schramek, N.; Wang, H.; Römisch-Margl, W.; Keil, B.; Radykewicz, T.; Winzenhörlein, B.; Beerhues, L.; Bacher, A.; Rohdich, F.; Gershenzon, J.; et al. Artemisinin biosynthesis in growing plants of Artemisia annua. A 13CO2 study. Phytochemistry 2010, 71, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, H.J.; Wallaart, T.E.; Janssen, M.H.; van Loo, B.; Jansen, B.J.; Posthumus, M.A.; Schmidt, C.O.; De Kraker, J.W.; König, W.A.; Franssen, M.C. Amorpha-4,11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 1999, 52, 843–854. [Google Scholar] [CrossRef]
- Mercke, P.; Bengtsson, M.; Bouwmeester, H.J.; Posthumus, M.A.; Brodelius, P.E. Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 2000, 381, 173–180. [Google Scholar] [CrossRef]
- Picaud, S.; Mercke, P.; He, X.; Sterner, O.; Brodelius, M.; Cane, D.E.; Brodelius, P.E. Amorpha-4,11-diene synthase: Mechanism and stereochemistry of the enzymatic cyclization of farnesyl diphosphate. Arch. Biochem. Biophys. 2006, 448, 150–155. [Google Scholar] [CrossRef]
- Kim, S.H.; Heo, K.; Chang, Y.J.; Park, S.H.; Rhee, S.K.; Kim, S.U. Cyclization mechanism of amorpha-4,11-diene synthase, a key enzyme in artemisinin biosynthesis. J. Nat. Prod. 2006, 69, 758–762. [Google Scholar] [CrossRef]
- Teoh, K.H.; Polichuk, D.R.; Reed, D.W.; Nowak, G.; Covello, P.S. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 2006, 580, 1411–1416. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Teoh, K.; Polichuk, D.; Reed, D.; Covello, P. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 2009, 87, 635–642. [Google Scholar] [CrossRef]
- Zhang, Y.; Teoh, K.H.; Reed, D.W.; Maes, L.; Goossens, A.; Olson, D.J.; Ross, A.R.; Covello, P.S. The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J. Biol. Chem. 2008, 283, 21501–21508. [Google Scholar] [CrossRef] [PubMed]
- Bertea, C.M.; Freije, J.R.; van der Woude, H.; Verstappen, F.W.; Perk, L.; Marquez, V.; De Kraker, J.W.; Posthumus, M.A.; Jansen, B.J.; de Groot, A.; et al. Identification of intermediates and enzymes involved in the early steps of artemisinin biosynthesis in Artemisia annua. Planta Med. 2005, 71, 40–47. [Google Scholar] [CrossRef]
- Wallaart, T.E.; Pras, N.; Quax, W.J. Isolation and identification of dihydroartemisinic acid hydroperoxide from Artemisia annua: A novel biosynthetic precursor of artemisinin. J. Nat. Prod. 1999, 62, 1160–1162. [Google Scholar] [CrossRef]
- Sy, L.-K.; Brown, G.D. The mechanism of the spontaneous autoxidation of dihydroartemisinic acid. Tetrahedron 2002, 58, 897–908. [Google Scholar] [CrossRef]
- Arsenault, P.R.; Wobbe, K.K.; Weathers, P.J. Recent advances in artemisinin production through heterologous expression. Curr. Med. Chem. 2008, 15, 2886–2896. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Liu, B.Y.; Li, Z.Q.; Ye, H.C.; Wang, H.; Li, G.F.; Han, J.L. Molecular cloning of a classical plant peroxidase from Artemisia annua and its effect on the biosynthesis of artemisinin in vitro. Acta Bot. Sin. 2004, 46, 1338–1346. [Google Scholar]
- Brown, G.D.; Sy, L.-K. In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron 2007, 63, 9548–9566. [Google Scholar] [CrossRef]
- Brown, G.D.; Sy, L.-K. In vivo transformations of dihydroartemisinic acid in Artemisia annua plants. Tetrahedron 2004, 60, 1139–1159. [Google Scholar] [CrossRef]
- Paddon, C.J.; Keasling, J.D. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014, 12, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.J.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.J.; Yoshikuni, Y.; Keasling, J.D. The in vivo synthesis of plant sesquiterpenes by Escherichia coli. Biotechnol. Bioeng. 2001, 75, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.D.; Marshall, J.; Chang, M.; Nowroozi, F.; Paradise, E.; Pitera, D.; Newman, K.L.; Keasling, J.D. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol. Bioeng. 2006, 95, 684–691. [Google Scholar] [CrossRef]
- Chang, M.C.; Keasling, J.D. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2006, 2, 674–681. [Google Scholar] [CrossRef]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Arellanes, A.; Meckes, M.; Torres, J.; Luna-Herrera, J. Antimycobacterial triterpenoids from Lantana hispida (Verbenaceae). J. Ethnopharmacol. 2007, 111, 202–205. [Google Scholar] [CrossRef]
- Kozai, K.; Suzuki, J.; Okada, M.; Nagasaka, N. Effect of oleanolic acid-cyclodextrin inclusion compounds on dental caries by in vitro experiment and rat-caries model. Microbios 1999, 97, 179–188. [Google Scholar]
- Horiuchi, K.; Shiota, S.; Hatano, T.; Yoshida, T.; Kuroda, T.; Tsuchiya, T. Antimicrobial activity of oleanolic acid from Salvia officinalis and related compounds on vancomycin-resistant enterococci (VRE). Biol. Pharm. Bull. 2007, 30, 1147–1149. [Google Scholar] [CrossRef]
- Castellano, J.M.; Ramos-Romero, S.; Perona, J.S. Oleanolic Acid: Extraction, Characterization and Biological Activity. Nutrients 2022, 14, 623. [Google Scholar] [CrossRef]
- Wolska, K.; Grudniak, A.M.; Fiecek, B.; Kraczkiewicz-Dowjat, A.; Kurek, A. Antibacterial activity of oleanolic and ursolic acids and their derivatives. Open Life Sci. 2010, 5, 543–553. [Google Scholar] [CrossRef]
- Pollier, J.; Goossens, A. Oleanolic acid. Phytochemistry 2012, 77, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.R.; Rasbery, J.M.; Bartel, B.; Matsuda, S.P. Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 2006, 9, 305–314. [Google Scholar] [CrossRef]
- Abe, I. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 2007, 24, 1311–1331. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Shibuya, M.; Ebizuka, Y. β-amyrin synthase—Cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. Eur. J. Biochem. 1998, 256, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Abe, I.; Rohmer, M.; Prestwich, G.D. Enzymatic Cyclization of Squalene and Oxidosqualene to Sterols and Triterpenes. Chem. Rev. 1993, 93, 2189–2206. [Google Scholar] [CrossRef]
- Seo, S.; Yoshimura, Y.; Uomori, A.; Takeda, K.; Seto, H.; Ebizuka, Y.; Sankawa, U. Biosynthesis of triterpenes, ursolic acid and oleanolic acid in tissue cultures of Rabdosia japonica Hara fed [5-13C2H2] mevalonolactone and [2-13C2H3] acetate. J. Am. Chem. Soc. 1988, 110, 1740–1745. [Google Scholar] [CrossRef]
- Fukushima, E.O.; Seki, H.; Ohyama, K.; Ono, E.; Umemoto, N.; Mizutani, M.; Saito, K.; Muranaka, T. CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 2011, 52, 2050–2061. [Google Scholar] [CrossRef] [PubMed]
- Carelli, M.; Biazzi, E.; Panara, F.; Tava, A.; Scaramelli, L.; Porceddu, A.; Graham, N.; Odoardi, M.; Piano, E.; Arcioni, S.; et al. Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell 2011, 23, 3070–3081. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Sun, Z.; Chen, W. Antimicrobial Susceptibility and Antibacterial Mechanism of Limonene against Listeria monocytogenes. Molecules 2019, 25, 33. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Liu, X.; Jiang, G.; Wu, J.; Zhang, J.L.; Lei, D.; Yuan, Y.J.; Qiao, J.; Zhao, G.R. Orthogonal Engineering of Biosynthetic Pathway for Efficient Production of Limonene in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, A.; Borrego, E.J.; Savka, M.A.; Dobson, R.C.J.; Hudson, A.O. Amino acid-derived defense metabolites from plants: A potential source to facilitate novel antimicrobial development. J. Biol. Chem. 2021, 296, 100438. [Google Scholar] [CrossRef] [PubMed]
- Othman, L.; Sleiman, A.; Abdel-Massih, R.M. Antimicrobial activity of polyphenols and alkaloids in middle eastern plants. Front. Microbiol. 2019, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Cushnie, B.; Lamb, A.J. Alkaloids: An overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents 2014, 44, 377–386. [Google Scholar] [CrossRef]
- Thawabteh, A.; Juma, S.; Bader, M.; Karaman, D.; Scrano, L.; Bufo, S.A.; Karaman, R. The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins 2019, 11, 656. [Google Scholar] [CrossRef]
- Khameneh, B.; Iranshahy, M.; Ghandadi, M.; Atashbeyk, D.G.; Bazzaz, B.S.F.; Iranshahi, M. Investigation of the antibacterial activity and efflux pump inhibitory effect of co-loaded piperine and gentamicin nanoliposomes in methicillin-resistant Staphylococcus aureus. Drug Dev. Ind. Pharm. 2015, 41, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Mabhiza, D.; Chitemerere, T.; Mukanganyama, S. Antibacterial Properties of Alkaloid Extracts from Callistemon citrinus and Vernonia adoensis against Staphylococcus aureus and Pseudomonas aeruginosa. Int. J. Med. Chem. 2016, 2016, 6304163. [Google Scholar] [CrossRef] [PubMed]
- Wink, M.; Ashour, M.L.; El-Readi, M.Z. Secondary metabolites from plants inhibiting ABC transporters and reversing resistance of cancer cells and microbes to cytotoxic and antimicrobial agents. Front. Microbiol. 2012, 3, 130. [Google Scholar] [CrossRef]
- Gibbons, S.; Udo, E.E. The effect of reserpine, a modulator of multidrug efflux pumps, on the in vitro activity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) possessing the tet (K) determinant. Phytother. Res. 2000, 14, 139–140. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Lorenz, P.; Tawara, J.N.; Zenewicz, L.A.; Lewis, K. Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5’-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 2000, 97, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Borsch, C.M.; Neyfakh, A.A.; Schuldiner, S. Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. J. Biol. Chem. 1993, 268, 11086–11089. [Google Scholar] [CrossRef]
- Xie, Q.; Johnson, B.R.; Wenckus, C.S.; Fayad, M.I.; Wu, C.D. Efficacy of berberine, an antimicrobial plant alkaloid, as an endodontic lrrigant against a mixed-culture biofilm in an in vitro tooth model. J. Endodont. 2012, 38, 1114–1117. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.B.; Yan, Y.; Liang, Y.Z.; Bao, Z. Evaluation of the antimicrobial mode of berberine by LC/ESI-MS combined with principal component analysis. J. Pharm. Biomed. Anal. 2007, 44, 301–304. [Google Scholar] [CrossRef]
- Albert, A.; Magrath, D. The choice of a chelating agent for inactivating trace metals: II. Derivatives of oxine (8-hydroxyquinoline). Biochem. J. 1947, 41, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Nakashima, K.; Nishino, K.; Kotani, K.; Tomida, J.; Inoue, M.; Kawamura, Y. Berberine is a novel type efflux inhibitor which attenuates the MexXY-mediated aminoglycoside resistance in Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 1223. [Google Scholar] [CrossRef] [PubMed]
- Tegos, G.; Stermitz, F.R.; Lomovskaya, O.; Lewis, K. Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrob. Agents Chemother. 2002, 46, 3133–3141. [Google Scholar] [CrossRef]
- Das, S.; Kumar, G.S.; Ray, A.; Maiti, M. Spectroscopic and thermodynamic studies on the binding of sanguinarine and berberine to triple and double helical DNA and RNA structures. J. Biomol. Struct. Dyn. 2003, 20, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, K.; Maiti, M.; Kumar, G.S. Berberine-DNA complexation: New insights into the cooperative binding and energetic aspects. Biochim. Biophys. Acta. 2008, 1780, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, K.; Zeller, K.P.; Siehl, H.U.; Berger, S.; Sicker, D. Look, the Yellow is so near: Berberine chloride from berberine bark. Chem. Unserer. Zeit 2017, 51, 344–356. [Google Scholar] [CrossRef]
- He, S.M.; Liang, Y.L.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.M.; Zhang, J.J.; Wang, X.; Dong, Y.; Yang, J.L.; et al. Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in Coptis species. Front. Plant Sci. 2018, 9, 731. [Google Scholar] [CrossRef] [PubMed]
- Samanani, N.; Facchini, P.J. Purification and characterization of norcoclaurine synthase. The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J. Biol. Chem. 2002, 277, 33878–33883. [Google Scholar] [CrossRef]
- Samanani, N.; Liscombe, D.K.; Facchini, P.J. Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 2004, 40, 302–313. [Google Scholar] [CrossRef]
- Minami, H.; Dubouzet, E.; Iwasa, K.; Sato, F. Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 2007, 282, 6274–6282. [Google Scholar] [CrossRef]
- Rueffer, M.; Nagakura, N.; Zenk, M.H. Partial purification and properties of S-adenosylmethionine: (R), (S)-Norlaudanosoline-6-O-methyltransferase from Argemone platyceras cell cultures. Planta Med. 1983, 49, 131–137. [Google Scholar] [CrossRef]
- Sato, F.; Tsujita, T.; Katagiri, Y.; Yoshida, S.; Yamada, Y. Purification and characterization of S-adenosyl-L-methionine: Norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 1994, 225, 125–131. [Google Scholar] [CrossRef]
- Loeffler, S.; Deusneumann, B.; Zenk, M.H. S-adenosyl-L-methionine-(S)-coclaurine-N-methyltransferase from Tinospora-Cordifolia. Phytochemistry 1995, 38, 1387–1395. [Google Scholar] [CrossRef]
- Choi, K.B.; Morishige, T.; Shitan, N.; Yazaki, K.; Sato, F. Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 2002, 277, 830–835. [Google Scholar] [CrossRef]
- Morishige, T.; Tsujita, T.; Yamada, Y.; Sato, F. Molecular characterization of the S-adenosyl-L-methionine: 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 2000, 275, 23398–23405. [Google Scholar] [CrossRef]
- Frenzel, T.; Zenk, M.H. S-Adenosyl-L-Methionine—3′-Hydroxy-N-Methyl-(S)-Coclaurine-4′-O-Methyl Transferase, a Regioselective and Stereoselective Enzyme of the (S)-Reticuline Pathway. Phytochemistry 1990, 29, 3505–3511. [Google Scholar] [CrossRef]
- Chapple, C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 311–343. [Google Scholar] [CrossRef]
- Mizutani, M.; Sato, F. Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 2011, 507, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ikezawa, N.; Tanaka, M.; Nagayoshi, M.; Shinkyo, R.; Sakaki, T.; Inouye, K.; Sato, F. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003, 278, 38557–38565. [Google Scholar] [CrossRef] [PubMed]
- Kutchan, T.M.; Dittrich, H. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J. Biol. Chem. 1995, 270, 24475–24481. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Xia, L.Q.; Sun, M.S.; Huang, P.; Zeng, J.G. Effects of codon optimization, N-terminal truncation and gene dose on the heterologous expression of berberine bridge enzyme. World J. Microb. Biot. 2022, 38, 77. [Google Scholar] [CrossRef]
- Muemmler, S.; Rueffer, M.; Nagakura, N.; Zenk, M.H. S-adenosyl-L-methionine: (S)-scoulerine 9-O-methyltransferase, a highly stereo- and regio-specific enzyme in tetrahydroprotoberberine biosynthesis. Plant Cell Rep. 1985, 4, 36–39. [Google Scholar] [CrossRef][Green Version]
- Takeshita, N.; Fujiwara, H.; Mimura, H.; Fitchen, J.H.; Yamada, Y.; Sato, F. Molecular cloning and characterization of S-adenosyl-L-methionine: Scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 1995, 36, 29–36. [Google Scholar]
- Galneder, E.; Rueffer, M.; Wanner, G.; Tabata, M.; Zenk, M.H. Alternative final steps in berberine biosynthesis in Coptis japonica cell cultures. Plant Cell Rep. 1988, 7, 1–4. [Google Scholar] [CrossRef]
- Okada, N.; Koizumi, N.; Tanaka, T.; Ohkubo, H.; Nakanishi, S.; Yamada, Y. Isolation, sequence, and bacterial expression of a cDNA for (S)-tetrahydroberberine oxidase from cultured berberine-producing Coptis japonica cells. Proc. Natl. Acad. Sci. USA 1989, 86, 534–538. [Google Scholar] [CrossRef]
- Payne, J.T.; Valentic, T.R.; Smolke, C.D. Complete biosynthesis of the bisbenzylisoquinoline alkaloids guattegaumerine and berbamunine in yeast. Proc. Natl. Acad. Sci. USA 2021, 118, e2112520118. [Google Scholar] [CrossRef]
- Hawkins, K.M.; Smolke, C.D. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008, 4, 564–573. [Google Scholar] [CrossRef]
- Park, S.U.; Facchini, P.J. Agrobacterium rhizogenes-mediated transformation of opium poppy, Papaver somniferum L., and California poppy, Eschscholzia californica cham., root cultures. J. Exp. Bot. 2000, 51, 1005–1016. [Google Scholar] [CrossRef]
- Sivakumar, G. Upstream biomanufacturing of pharmaceutical colchicine. Crit. Rev. Biotechnol. 2018, 38, 83–92. [Google Scholar] [CrossRef]
- Jana, S.; Shekhawat, G.S. Critical review on medicinally potent plant species: Gloriosa superba. Fitoterapia 2010, 82, 293–301. [Google Scholar] [CrossRef]
- Slobodnick, A.; Shah, B.; Pillinger, M.H.; Krasnokutsky, S. Colchicine: Old and New. Am. J. Med. 2015, 128, 461–470. [Google Scholar] [CrossRef]
- Karamanou, M.; Tsoucalas, G.; Pantos, K.; Androutsos, G. Isolating Colchicine in 19th Century: An old drug revisited. Curr. Pharm. Des. 2018, 24, 654–658. [Google Scholar] [CrossRef]
- Zhang, F.S.; He, Q.Z.; Qin, C.H.; Little, P.J.; Weng, J.P.; Xu, S.W. Therapeutic potential of colchicine in cardiovascular medicine: A pharmacological review. Acta. Pharmacol. Sin. 2022, 43, 2173–2190. [Google Scholar] [CrossRef]
- Bhattacharyya, B.; Panda, D.; Gupta, S.; Banerjee, M. ChemInform Abstract: Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. ChemInform 2008, 28, 155–183. [Google Scholar] [CrossRef]
- Chiu, L.; Lo, C.H.; Shen, M.; Chiu, N.; Aggarwal, R.; Lee, J.; Choi, Y.G.; Lam, H.; Prsic, E.H.; Chow, R.; et al. Colchicine use in patients with COVID-19: A systematic review and meta-analysis. PLoS ONE 2021, e0261358. [Google Scholar] [CrossRef]
- Leete, E.; Nemeth, P.E. The Biogenesis of the Alkaloids of Colchicum. I. The incorporation of phenylalanine into colchicine. J. Am. Chem. Soc. 1960, 82, 6055–6057. [Google Scholar] [CrossRef]
- Herbert, R.B. The biosynthesis of plant alkaloids and nitrogenous microbial metabolites. Nat. Prod. Rep. 2003, 20, 494–508. [Google Scholar] [CrossRef]
- Battersby, A.R.; Herbert, R.B.; McDonald, E.; Ramage, R.; Clements, J.H. Alkaloid biosynthesis. 18. Biosynthesis of colchicine from the 1-phenethylisoquinoline system. J. Chem. Soc. Perkin. 1 1972, 14, 1741–1746. [Google Scholar] [CrossRef]
- Herbert, R.B.; Kattah, A.E.; Knagg, E. The Biosynthesis of the phenethylisoquinoline alkaloid colchicine. Early and intermediate stages. Tetrahedron 1990, 46, 7119–7138. [Google Scholar] [CrossRef]
- Nasreen, A.; Gundlach, H.; Zenk, M.H. Incorporation of phenethylisoquinolines into colchicine in isolated seeds of Colchicum autumnale. Phytochemistry 1997, 46, 107–115. [Google Scholar] [CrossRef]
- Leete, E. Biosynthesis of the tropolone ring of colchicine. Tetrahedron Lett. 1965, 6, 333–336. [Google Scholar] [CrossRef]
- Herbert, R.B.; Knagg, E. The biosynthesis of the phenethylisoquinoline alkaloid, colchicine, from cinnamaldehyde and dihydrocinnamaldehyde. Tetrahedron Lett. 1986, 27, 1099–1102. [Google Scholar] [CrossRef]
- Maier, U.H.; Zenk, M.H. Colchicine is formed by para-para phenol coupling from autumnaline. Tetrahedron Lett. 1997, 38, 7357–7360. [Google Scholar] [CrossRef]
- Rueffer, M.; Zenk, M.H. Microsome-mediated transformation of O-methylandrocymbine to demecolcine and colchicine. FEBS Lett. 1998, 438, 111–113. [Google Scholar] [CrossRef]
- Sheldrake, P.W.; Suckling, K.E.; Woodhouse, R.N.; Murtagh, A.J. Biosynthesis Part 30.1 Colchicine: Studies on the ring expansion step focusing on the fate of the hydrogens at C-4 of autumnaline. J. Chem. Soc. Perkin Trans. 1 1998, 18, 3003–3010. [Google Scholar] [CrossRef]
- Nett, R.S.; Lau, W.; Sattely, E.S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 2020, 584, 148–153. [Google Scholar] [CrossRef]
- Nett, R.S.; Sattely, E.S. Total biosynthesis of the tubulin-binding alkaloid colchicine. J. Am. Chem. Soc. 2021, 143, 19454–19465. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Sunnadeniya, R.; Bean, A.; Brown, M.; Akhavan, N.; Hatlestad, G.; Gonzalez, A.; Symonds, V.V.; Lloyd, A. Tyrosine Hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS ONE 2016, 11, e0149417. [Google Scholar] [CrossRef] [PubMed]
- Luk, L.Y.P.; Bunn, S.; Liscombe, D.K.; Facchini, P.J.; Tanner, M.E. Mechanistic studies on norcoclaurine synthase of benzylisoquinoline alkaloid biosynthesis: An enzymatic Pictet-Spengler reaction. Biochemistry 2007, 46, 10153–10161. [Google Scholar] [CrossRef] [PubMed]
- Ruff, B.M.; Brase, S.; O’Connor, S.E. Biocatalytic production of tetrahydroisoquinolines. Tetrahedron Lett. 2012, 53, 1071–1074. [Google Scholar] [CrossRef]
- Nishihachijo, M.; Hirai, Y.; Kawano, S.; Nishiyama, A.; Minami, H.; Katayama, T.; Yasohara, Y.; Sato, F.; Kumagai, H. Asymmetric synthesis of tetrahydroisoquinolines by enzymatic Pictet-Spengler reaction. Biosci. Biotech. Bioch. 2014, 78, 701–707. [Google Scholar] [CrossRef]
- Barker, A.C.; Julian, D.R.; Ramage, R.; Woodhouse, R.N.; Hardy, G.; McDonald, E.; Battersby, A.R. Biosynthesis. Part 28.1,2 Colchicine: Definition of intermediates between O-mehylandrocymbine and colchicine and studies on speciosine. J. Chem. Soc. Perk Trans. 1998, 30, 2989–2994. [Google Scholar] [CrossRef]
- McDonald, E.; Ramage, R.; Woodhouse, R.N.; Underhill, E.W.; Wetter, L.R.; Battersby, A.R. Biosynthesis. Part 27.1,2 Colchicine: Studies of the phenolic oxidative coupling and ring-expansion processes based on incorporation of multiply labelled 1-phenethylisoquinolines. J. Chem. Soc. Perk Trans. 1998, 18, 2979–2987. [Google Scholar] [CrossRef]
- Shang, X.F.; Morris-Natschke, S.L.; Liu, Y.Q.; Guo, X.; Xu, X.S.; Goto, M.; Li, J.C.; Yang, G.Z.; Lee, K.H. Biologically active quinoline and quinazoline alkaloids part I. Med. Res. Rev. 2018, 38, 775–828. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Narayanasamy, M.; Feussner, K.D. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech 2017, 7, 172. [Google Scholar] [CrossRef]
- Casciaro, B.; Mangiardi, L.; Cappiello, F.; Romeo, I.; Loffredo, M.R.; Iazzetti, A.; Calcaterra, A.; Goggiamani, A.; Ghirga, F.; Mangoni, M.L.; et al. Naturally-Occurring alkaloids of plant origin as potential antimicrobials against antibiotic-resistant infections. Molecules 2020, 25, 3619. [Google Scholar] [CrossRef]
- Mitchell, G.; Gattuso, M.; Grondin, G.; Marsault, E.; Bouarab, K.; Malouin, F. Tomatidine Inhibits Replication of Staphylococcus aureus Small-Colony Variants in Cystic Fibrosis Airway Epithelial Cells. Antimicrob. Agents Chemother. 2011, 55, 1937–1945. [Google Scholar] [CrossRef]
- Dorsaz, S.; Snaka, T.; Favre-Godal, Q.; Maudens, P.; Boulens, N.; Furrer, P.; Ebrahimi, S.N.; Hamburger, M.; Allemann, E.; Gindro, K.; et al. Identification and mode of action of a plant natural product targeting human fungal pathogens. Antimicrob. Agents Chemother. 2017, 61, e00829-17. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, R.; Lee, J.H.; Nakayasu, M.; Osakabe, K.; Osakabe, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; Sugimoto, Y.; Mizutani, M. Characterization of steroid 5α-reductase involved in α-tomatine biosynthesis in tomatoes. Plant Biotechnol. 2019, 36, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Motaal, F.F.; Nassar, M.S.M.; El-Zayat, S.A.; El-Sayed, M.A.; Ito, S. Responses of fungi to tropane alkaloids produced by a medicinal plant Hyoscyamus muticus (Egyptian Henbane). Folia Microbiol. 2009, 54, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Motaal, F.F.; El-Zayat, S.A.; Kosaka, Y.; El-Sayed, M.A.; Kashima, R.; Maeda, Y.; Nassar, M.S.M.; Ito, S. Antifungal activities of hyoscyamine and scopolamine against two major rice pathogens: Magnaporthe oryzae and Rhizoctonia solani. J. Gen. Plant Pathol. 2010, 76, 102–111. [Google Scholar] [CrossRef]
- Srinivasan, P.; Smolke, C.D. Biosynthesis of medicinal tropane alkaloids in yeast. Nature 2020, 585, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Davison, E.K.; Brimble, M.A. Natural product derived privileged scaffolds in drug discovery. Curr. Opin. Chem. Biol. 2019, 52, 1–8. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.F.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.H.; Hsieh, Y.C.; Chen, Y.H.; Wang, H.Y.; Lu, C.Y.; Chen, C.J.; Li, Y.K. Three important amino acids control the regioselectivity of flavonoid glucosidation in glycosyltransferase-1 from Bacillus cereus. Appl. Microbiol. Biotechnol. 2016, 100, 8411–8424. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Khan, A.; Ahmad, I.; Alghamdi, S.; Rajab, B.S.; Babalghith, A.O.; Alshahrani, M.Y.; Islam, S.; Islam, M.R. Flavonoids a bioactive compound from medicinal plants and its therapeutic applications. Biomed. Res. Int. 2022, 2022, 5445291. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kumar, P.S.; Tan, S.; Huang, C.; Xiang, Z.; Qiu, J.; Tan, X.; Luo, J.; He, M. Anticancer and antibacterial flavonoids from the callus of Ampelopsis grossedentata; a new weapon to mitigate the proliferation of cancer cells and bacteria. RSC Adv. 2022, 12, 24130–24138. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, F.; Iqtadar, S.; Khan, A.; Mumtaz, S.U.; Chaudhry, M.M.; Bertuccioli, A.; Derosa, G.; Maffioli, P.; Togni, S.; Riva, A.; et al. Potential clinical benefits of quercetin in the early stage of COVID-19: Results of a second, pilot, randomized, controlled and open-label clinical trial. Int. J. Gen. Med. 2021, 14, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Biharee, A.; Sharma, A.; Kumar, A.; Jaitak, V. Antimicrobial flavonoids as a potential substitute for overcoming antimicrobial resistance. Fitoterapia 2020, 146, 104720. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.X.; Yang, W.J.; Tang, F.; Chen, X.Q.; Ren, L.C. Antibacterial activities of flavonoids: Structure-activity relationship and mechanism. Curr. Med. Chem. 2015, 22, 132–149. [Google Scholar] [CrossRef] [PubMed]
- Smejkal, K.; Chudík, S.; Kloucek, P.; Marek, R.; Cvacka, J.; Urbanová, M.; Julínek, O.; Kokoska, L.; Slapetová, T.; Holubová, P.; et al. Antibacterial C-geranylflavonoids from Paulownia tomentosa Fruits. J. Nat. Prod. 2008, 71, 706–709. [Google Scholar] [CrossRef]
- Tran, T.D.; Do, T.H.; Tran, N.C.; Ngo, T.D.; Huynh, T.N.; Tran, C.D.; Thai, K.M. Synthesis and anti Methicillin resistant Staphylococcus aureus activity of substituted chalcones alone and in combination with non-beta-lactam antibiotics. Bioorg. Med. Chem. Lett. 2012, 22, 4555–4560. [Google Scholar] [CrossRef] [PubMed]
- Rütschlin, S.; Böttcher, T. Inhibitors of Bacterial Swarming Behavior. Chemistry 2020, 26, 964–979. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Reimold, U.; Kröger, M.; Kreuzaler, F.; Hahlbrock, K. Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. Embo. J. 1983, 2, 1801–1805. [Google Scholar] [CrossRef]
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone synthases (CHSs): The symbolic type III polyketide synthases. Planta 2019, 251, 15. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar] [PubMed]
- Jones, J.A.; Vernacchio, V.R.; Sinkoe, A.L.; Collins, S.M.; Ibrahim, M.H.A.; Lachance, D.M.; Hahn, J.; Koffas, M.A.G. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab. Eng. 2016, 35, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Lyu, Y.; Li, H.; Koffas, M.A.G.; Zhou, J. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol. Bioeng. 2019, 116, 1392–1404. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, Y.; Wang, G.; Hill, L.; Weng, J.K.; Chen, X.Y.; Xue, H.; Martin, C. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2016, 2, e1501780. [Google Scholar] [CrossRef]
- Kitamura, K.; Honda, M.; Yoshizaki, H.; Yamamoto, S.; Nakane, H.; Fukushima, M.; Ono, K.; Tokunaga, T. Baicalin, an inhibitor of HIV-1 production in vitro. Antiviral Res. 1998, 37, 131–140. [Google Scholar] [CrossRef]
- Li, B.Q.; Fu, T.; Dongyan, Y.; Mikovits, J.A.; Ruscetti, F.W.; Wang, J.M. Flavonoid baicalin inhibits HIV-1 infection at the level of viral entry. Biochem. Biophys. Res. Commun. 2000, 276, 534–538. [Google Scholar] [CrossRef]
- Liu, W.X.; Feng, Y.; Yu, S.H.; Fan, Z.Q.; Li, X.L.; Li, J.Y.; Yin, H.F. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Pei, T.L.; Yan, M.X.; Li, T.; Li, X.Q.; Yin, Y.J.; Cui, M.Y.; Fang, Y.M.; Liu, J.; Kong, Y.; Xu, P.; et al. Characterization of UDP-glycosyltransferase family members reveals how major flavonoid glycoside accumulates in the roots of Scutellaria baicalensis. BMC Genom. 2022, 23, 169. [Google Scholar] [CrossRef] [PubMed]
- Han, H.D.; Lee, Y.S.; Ahn, J.H. Biological synthesis of baicalein derivatives using Escherichia coli. J. Microbiol. Biotechnol. 2016, 26, 1918–1923. [Google Scholar] [CrossRef]
- Liao, J.J.; Xie, L.; Liu, T.Y.; Mo, C.M.; Cui, S.R.; Jia, X.L.; Huang, X.Y.; Luo, Z.L.; Ma, X.J. Heterologous biosynthesis of health-promoting baicalein in Lycopersicon esculentum. Molecules 2022, 27, 3086. [Google Scholar] [CrossRef]
- Caputi, L.; Franke, J.; Farrow, S.C.; Chung, K.; Payne, R.M.E.; Nguyen, T.D.; Dang, T.T.; Soares Teto Carqueijeiro, I.; Koudounas, K.; Dugé de Bernonville, T.; et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrübbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, 609, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.; Sattely, E.S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 2015, 349, 1224–1228. [Google Scholar] [CrossRef] [PubMed]

| Compounds | Chemical Structures | Target Microorganisms | Antimicrobial Effects | Reference | |
|---|---|---|---|---|---|
| Terpenoids | 1,8-cineole | 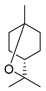 | A. baumannii C. albicans MRSA strain E. coli | Cell membrane destruction | [22] |
| cinnamaldehyde |  | S. typhimurium E. coli O157: H7 P. fluorescence B. thermophacta S. aureus | 1. Cell membrane destruction 2. Anti-quorum sensing action 3. Inhibition of protein synthesis | [23,25,30,31] | |
| carvacrol |  | S. typhimurium E. coli O157: H7 P. fluorescence B. thermophacta S. aureus P. fluorescens KM121 | 1. Cell membrane destruction 2. Anti-quorum sensing action 3. Inhibition of nucleic acid synthesis 4. The synergistic effect 5. Inhibits cell movement and bacterial invasion | [23,26,27,29,32] | |
| thymol | 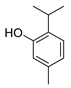 | S. typhimurium E. coli O157: H7 P. fluorescence B. thermophacta S. aureus P. fluorescens KM121 | 1. Cell membrane destruction 2. Anti-quorum sensing action 3. Inhibition of nucleic acid synthesis 4. The synergistic effect | [23,26,27,28,32] | |
| eugenol | 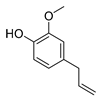 | S. typhimurium E. coli O157: H7 P. fluorescence B. thermophacta S. aureus | 1. Cell membrane destruction 2. Inhibition of nucleic acid synthesis 3. The synergistic effect | [23,27,28,32] | |
| limonene |  | A. baumannii C. albicans MRSA strain E. coli | Cell membrane destruction | [23] | |
| oleanolic acid | 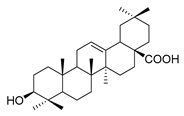 | E. coli S. aureus Enterococcus faecalis P. aeruginosa | Antibacterial | [33] | |
| Alkaloids | piperine | 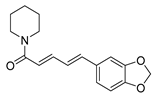 | S. aureus B. subtilis Salmonella sp. E. coli | Efflux pump inhibition | [34,35] |
| reserpine |  | E. coli | Efflux pump inhibition | [36] | |
| berberine |  | E. coli Micrococcus luteus P. aeruginosa Prevotella intermedia Fusobacterium nucleatum MRSA strain | 1. Efflux pump inhibition 2. DNA-intercalating 3. Growth inhibition | [37,38,39] | |
| L-ephedrine | 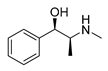 | Influenza A virus | DNA-intercalating | [40] | |
| D-pseudoephedrine | 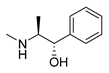 | Influenza A virus | DNA-intercalating | [40] | |
| L-methylephedrine |  | Influenza A virus | DNA-intercalating | [40] | |
| chelerythrine | 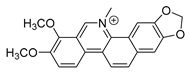 | S. aureus MRSA strain ESBLs-SA | 1. Nucleic acid synthesis and repair inhibition 2. Growth inhibition | [41] | |
| 8-hydroxy quinoline | 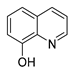 | S. aureus H. influenza S. pneumoniae | Permeability change of membrane | [42,43] | |
| michellamine b | 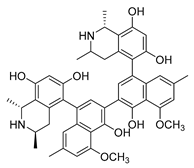 | HIV | Protein activity inhibition | [44] | |
| sanguinarine | 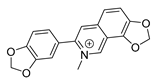 | K. pneumoniae MRSA strain P. aeruginosa Streptococcus pyogenes | 1. DNA-intercalating 2. Growth inhibition | [45,46] | |
| roemerine | 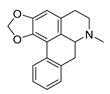 | S. aureus B. subtilis | 1. Efflux pump inhibition 2. Permeability change of membrane | [47,48] | |
| dihydrochelerythrine | 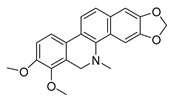 | S. aureus MRSA strain | Growth inhibition | [49] | |
| evodiamine |  | M. tubercolosis | Peptidoglycan biosynthesis inhibitor | [50,51] | |
| Flavonoids | hesperidin |  | S. aureus L. monocytogenes | Inhibit bacterial growth by modulating the expression of virulence factors | [52] [53] |
| oroxylin a |  | B. subtilis S. aureus | / | [54] | |
| apigenin | 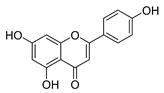 | S. aureus B. subtilis E. coli P. aeruginosa. | 1. Inhibits peptidoglycan synthesis 2. Increases cell membrane permeability | [55] | |
| morin | 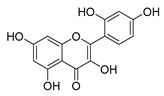 | E. coli | Inhibition of ATP synthetase | [56] | |
| silymarin | 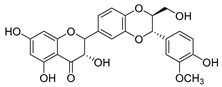 | E. coli | Inhibition of ATP synthetase | [56] | |
| epigallocatechin gallate |  | S. maltophilia | Inhibits dihydrofolate reductase | [57] | |
| quercetin | 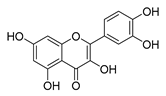 | P. aeruginosa | 1. Inhibits viral polymerase and viral nucleic acid 2. Inhibits the formation of its biofilm | [58] | |
| galangin |  | S. aureus | 1. Destroys the plasma membrane 2. Weakens the cell wall | [59] | |
| catechin | 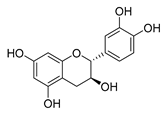 | B. subtilis E. coli | Inhibits the bacterial DNA gyrase | [60] [61] | |
| baicalin | 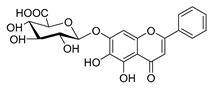 | Salmonella spp. Staphylococcus spp. | Inhibits biofilm formation | [62] [63] | |
| phloretin | 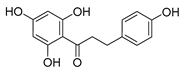 | C. albicans | 1. Inhibits the pathogenicity 2. Inhibits virulence factors | [64] | |
| silybin | 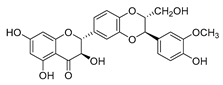 | MRSA strain | Inhibits the efflux pump | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Wang, Y.; Tian, W.; Cui, X.; Tu, P.; Li, J.; Shi, S.; Liu, X. Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants. Antibiotics 2022, 11, 1380. https://doi.org/10.3390/antibiotics11101380
Huang W, Wang Y, Tian W, Cui X, Tu P, Li J, Shi S, Liu X. Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants. Antibiotics. 2022; 11(10):1380. https://doi.org/10.3390/antibiotics11101380
Chicago/Turabian StyleHuang, Wenqian, Yingxia Wang, Weisheng Tian, Xiaoxue Cui, Pengfei Tu, Jun Li, Shepo Shi, and Xiao Liu. 2022. "Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants" Antibiotics 11, no. 10: 1380. https://doi.org/10.3390/antibiotics11101380
APA StyleHuang, W., Wang, Y., Tian, W., Cui, X., Tu, P., Li, J., Shi, S., & Liu, X. (2022). Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants. Antibiotics, 11(10), 1380. https://doi.org/10.3390/antibiotics11101380






