Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus
Abstract
1. Antibiotic Resistance and Human Health Risk
2. Emergence of Methicillin-Resistant Staphylococcus aureus (MRSA)
3. Molecular Basis of Non-β-Lactams Resistance
3.1. Target Modification
3.2. Mutation of Target
3.3. Drug Inactivation by Enzymes
3.4. Decreased Antibiotic Uptake or Efflux
4. MRSA Resistance to Non-β-Lactams
4.1. Macrolides (Erythromycin)
4.2. Lincosamide (Clindamycin)
4.3. Aminoglycosides (Gentamicin)
4.4. Glycopeptides (Vancomycin)
4.5. Oxazolidinones (Linezolid)
4.6. Lipopeptides (Daptomycin)
4.7. Fluoroquinolone (Ciprofloxacin)
4.8. Pyrimidines/Sulfonamides (Trimethoprim-Sulfamethoxazole)
4.9. Mupirocin
4.10. Fosfomycin
4.11. Rifampin
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. injluenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance; WHO: Geneva, Switzerlnad, 2021. [Google Scholar]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2016. [Google Scholar]
- Centers for Disease Control and Prevention (U.S.). Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GA, USA, 2019. [Google Scholar]
- Centers for Disease Control and Prevention (U.S.). Methicillin-Resistant Staphylococcus Aureus, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GA, USA, 2019. [Google Scholar]
- Rammelkamp, C.H.; Maxon, T. Resistance of Staphylococcus aureus to the Action of Penicillin. Exp. Biol. Med. 1942, 51, 386–389. [Google Scholar] [CrossRef]
- Kirby, W.M.M. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef]
- Knox, R. A New Penicillin (BRL 1241) Active Against Penicillin-resistant. Staphylococci. BMJ 1960, 2, 690–693. [Google Scholar] [CrossRef]
- Jevons, M.P. “Celbenin”—Resistant. Staphylococci. Br. Med. J. 1961, 1, 124. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; McDougal, L.K.; Goering, R.V.; Killgore, G.; Projan, S.J.; Patel, J.B.; Dunman, P.M. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 2006, 44, 108–118. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Matsuhashi, M.; Song, M.D.; Ishino, F.; Wachi, M.; Doi, M.; Inoue, M.; Ubukata, K.; Yamashita, N.; Konno, M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 1986, 167, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Katayama, Y.; Ito, T.; Hiramatsu, K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 44, 1549–1555. [Google Scholar] [CrossRef]
- Hartman, B.J.; Tomasz, A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984, 158, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Utsui, Y.; Yokota, T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1985, 28, 397–403. [Google Scholar] [CrossRef]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The Basis for Resistance to β-Lactam Antibiotics by Penicillin-binding Protein 2a of Methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef]
- Watkins, R.R.; Holubar, M.; David, M.Z. Antimicrobial resistance in methicillin-resistant Staphylococcus aureus to newer antimicrobial agents. Antimicrob. Agents Chemother. 2019, 63, e01216-19. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Kim, J.-S. Bacterial Targets of Antibiotics in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 398. [Google Scholar] [CrossRef]
- Brown, N.M.; Goodman, A.L.; Horner, C.; Jenkins, A.; Brown, E.M. Treatment of methicillin-resistant Staphylococcus aureus (MRSA): Updated guidelines from the UK. JAC Antimicrob. Resist. 2021, 3, dlaa114. [Google Scholar] [CrossRef]
- Hiramatsu, K. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef]
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef]
- McGehee, R.F.R.; Barre, F.F.; Finland, M. Resistance of Staphylococcus aureus to lincomycin, clinimycin, and erythromycin. Antimicrob. Agents Chemother. 1968, 8, 392–397. [Google Scholar]
- Roberts, M.C.; Sutcliffe, J.; Courvalin, P.; Jensen, L.B.; Rood, J.; Seppala, H. Nomenclature for Macrolide and Macrolide-Lincosamide-Streptogramin B Resistance Determinants. Antimicrob. Agents Chemother. 1999, 43, 2823–2830. [Google Scholar] [CrossRef]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef]
- Schmitz, F.-J.; Jones, M.E.; Hofmann, B.; Hansen, B.; Scheuring, S.; Lückefahr, M.; Fluit, A.; Verhoef, J.; Hadding, U.; Heinz, H.-P.; et al. Characterization of grlA, grlB, gyrA, and gyrB Mutations in 116 Unrelated Isolates of Staphylococcus aureus and Effects of Mutations on Ciprofloxacin MIC. Antimicrob. Agents Chemother. 1998, 42, 1249–1252. [Google Scholar] [CrossRef]
- Ferrero, L.; Cameron, B.; Crouzet, J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 1995, 39, 1554–1558. [Google Scholar] [CrossRef]
- Ng, E.Y.; Trucksis, M.; Hooper, D.C. Quinolone resistance mutations in topoisomerase IV: Relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996, 40, 1881–1888. [Google Scholar] [CrossRef]
- Jaffe, H.W.; Sweeney, H.M.; Weinstein, R.A.; Kabins, S.A.; Nathan, C.; Cohen, S. Structural and phenotypic varieties of gentamicin resistance plasmids in hospital strains of Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob. Agents Chemother. 1982, 21, 773–779. [Google Scholar] [CrossRef]
- Ubukata, K.; Yamashita, N.; Gotoh, A.; Konno, M. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 1984, 25, 754–759. [Google Scholar] [CrossRef]
- Hasanvand, A.; Ghafourian, S.; Taherikalani, M.; Jalilian, F.; Sadeghifard, N.; Pakzad, I. Antiseptic Resistance in Methicillin Sensitive and Methicillin Resistant Staphylococcus aureus Isolates from Some Major Hospitals, Iran. Recent Pat. Antiinfect. Drug Discov. 2015, 10, 105–112. [Google Scholar] [CrossRef]
- Taheri, N.; Ardebili, A.; Amouzandeh-Nobaveh, A.; Ghaznavi-Rad, E. Frequency of Antiseptic Resistance Among Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated From a University Hospital in Central Iran. Oman Med. J. 2016, 31, 426–432. [Google Scholar] [CrossRef]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef]
- Tsai, K.; Stojković, V.; Noda-Garcia, L.; Young, I.D.; Myasnikov, A.G.; Kleinman, J.; Palla, A.; Floor, S.N.; Frost, A.; Fraser, J.S.; et al. Directed evolution of the rRNA methylating enzyme Cfr reveals molecular basis of antibiotic resistance. Elife 2022, 11, e70017. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S.; Jacobsen, L.; Hansen, L.H.; Vester, B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005, 57, 1064–1073. [Google Scholar] [CrossRef]
- Aubry-Damon, H.; Soussy, C.J.; Courvalin, P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1998, 42, 2590–2594. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Roberts, M.C. Tetracycline resistance determinants: Mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 1996, 19, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, L.; Cameron, B.; Manse, B.; Lagneaux, D.; Crouzet, J.; Famechon, A.; Blanche, F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: A primary target of fluoroquinolones. Mol. Microbiol. 1994, 13, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Yoshida, H.; Bogaki-Shonai, M.; Niga, T.; Hattori, H.; Nakamura, S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother. 1994, 38, 2014–2023. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Igarashi, M.; Morimoto, Y.; Baba, T.; Umekita, M.; Akamatsu, Y. Curing bacteria of antibiotic resistance: Reverse antibiotics, a novel class of antibiotics in nature. Int. J. Antimicrob. Agents 2012, 39, 478–485. [Google Scholar] [CrossRef]
- Yun, H.-J.; Min, Y.-H.; Jo, Y.W.; Shim, M.-J.; Choi, E.-C. Increased antibacterial activity of DW286, a novel fluoronaphthyridone antibiotic, against Staphylococcus aureus strains with defined mutations in DNA gyrase and topoisomerase IV. Int. J. Antimicrob. Agents 2005, 25, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Inoue, M.; Endo, Y.; Nakajima, Y. Characteristic expression of three genes, msr (A), mph (C) and erm (Y), that confer resistance to macrolide antibiotics on Staphylococcus aureus. FEMS Microbiol. Lett. 2003, 220, 287–293. [Google Scholar] [CrossRef]
- Thompson, M.K.; Keithly, M.E.; Goodman, M.C.; Hammer, N.D.; Cook, P.D.; Jagessar, K.L.; Harp, J.; Skaar, E.P.; Armstrong, R.N. Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 2014, 53, 755–765. [Google Scholar] [CrossRef]
- Fu, Z.; Liu, Y.; Chen, C.; Guo, Y.; Ma, Y.; Yang, Y.; Hu, F.; Xu, X.; Wang, M. Characterization of Fosfomycin Resistance Gene, fosB, in Methicillin-Resistant Staphylococcus aureus Isolates. PLoS ONE 2016, 11, e0154829. [Google Scholar]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. The remarkably multifunctional fibronectin binding proteins of Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Jang, S. Multidrug efflux pumps in Staphylococcus aureus and their clinical implications. J. Microbiol. 2016, 54, 1–8. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Seo, S.M. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1995, 39, 2650–2655. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 10, 710. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.I.; Lee, Y.-M.; Park, K.-H.; Ryu, B.-H.; Hong, K.-W.; Kim, S.; Bae, I.-G.; Cho, O.-H. Clinical and Molecular Characteristics of qacA- and qacB-Positive Methicillin-Resistant Staphylococcus aureus Causing Bloodstream Infections. Antimicrob. Agents Chemother. 2019, 63, e02157-18. [Google Scholar] [CrossRef]
- Noguchi, N.; Suwa, J.; Narui, K.; Sasatsu, M.; Ito, T.; Hiramatsu, K.; Song, J.H. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J. Med. Microbiol. 2005, 54, 557–565. [Google Scholar] [CrossRef]
- Nakaminami, H.; Takadama, S.; Okita, M.; Sasaki, M.; Noguchi, N. Fast-acting bactericidal activity of olanexidine gluconate against qacA/B-positive methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 2019, 68, 957–960. [Google Scholar] [CrossRef] [PubMed]
- Guay, G.G.; Khan, S.A.; Rothstein, D.M. The tet(K) Gene of Plasmid pT181 of Staphylococcus aureus Encodes an Efflux Protein That Contains 14 Transmembrane Helices. Plasmid 1993, 30, 163–166. [Google Scholar] [CrossRef]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar]
- McGuire, J.M.; Bunch, R.L.; Anderson, R.C.; Boaz, H.E.; Flynn, E.H.; Powell, H.M.; Smith, J.W. Ilotycin, a new antibiotic. Antibiot. Chemother. 1952, 2, 281–283. [Google Scholar]
- MacCabe, A.F.; Gould, J.C. The Epidemiology of an Erythromycin Resistant Staphylococcus. Scott. Med. J. 1956, 1, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Champney, W.S.; Burdine, R. Macrolide antibiotics inhibit 50S ribosomal subunit assembly in Bacillus subtilis and Staphylococcus aureus. Antimicrob. Agents Chemother. 1995, 39, 2141–2144. [Google Scholar] [CrossRef][Green Version]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M. Mechanisms of Resistance to Macrolide Antibiotics among Staphylococcus aureus. Antibiotics 2021, 10, 1406. [Google Scholar] [CrossRef]
- Schwendener, S.; Donà, V.; Perreten, V. The Novel Macrolide Resistance Genes mef (D), msr (F), and msr (H) Are Present on Resistance Islands in Macrococcus canis, Macrococcus caseolyticus, and Staphylococcus aureus. Antimicrob. Agents Chemother. 2020, 64, e00160-20. [Google Scholar] [CrossRef]
- Ross, J.I.; Eady, E.A.; Cove, J.H.; Cunliffe, W.J.; Baumberg, S.; Wootton, J.C. Inducible erythromycin resistance in staphlyococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 1990, 4, 1207–1214. [Google Scholar] [CrossRef]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile macrolide resistance genes in staphylococci. Plasmid 2018, 99, 2–10. [Google Scholar] [CrossRef]
- Leclercq, R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef]
- Spížek, J.; Řezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 2017, 133, 20–28. [Google Scholar] [CrossRef]
- Wang, H.; Zhuang, H.; Ji, S.; Sun, L.; Zhao, F.; Wu, D.; Shen, P.; Jiang, Y.; Yu, Y.; Chen, Y. Distribution of erm genes among MRSA isolates with resistance to clindamycin in a Chinese teaching hospital. Infect. Genet. Evol. 2021, 96, 105127. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Soussy, C.J.; Bouanchaud, D.H.; Fouace, J.; Dublanchet, A.; Duval, J. A gentamycin resistance plasmid in Staphylococcus aureus. Ann. Microbiol. 1975, 126, 91–94. [Google Scholar]
- Porthouse, A.; Brown, D.F.; Smith, R.G.; Rogers, T. Gentamicin resistance in Staphylococcus aureus. Lancet 1976, 307, 20–21. [Google Scholar] [CrossRef]
- Schluenzen, F.; Tocilj, A.; Zarivach, R.; Harms, J.; Gluehmann, M.; Janell, D.; Bashan, A.; Bartels, H.; Agmon, I.; Franceschi, F.; et al. Structure of Functionally Activated Small Ribosomal Subunit at 3.3 Å Resolution. Cell 2000, 102, 615–623. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Brodersen, D.E.; Clemons, W.M.; Morgan-Warren, R.J.; Carter, A.P.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Magnet, S.; Blanchard, J.S. Molecular Insights into Aminoglycoside Action and Resistance. Chem. Rev. 2005, 105, 477–498. [Google Scholar] [CrossRef]
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on Mechanisms of Action and Resistance and Strategies to Counter Resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef]
- Rouch, D.A.; Byrne, M.E.; Kong, Y.C.; Skurray, R.A. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: Expression and nucleotide sequence analysis. J. Gen. Microbiol. 1987, 133, 3039–3052. [Google Scholar] [CrossRef] [PubMed]
- Ida, T.; Okamoto, R.; Shimauchi, C.; Okubo, T.; Kuga, A.; Inoue, M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: Epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 2001, 39, 3115–3121. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Nonoyama, M.; Okamoto, R.I.T. Antimicrobial activity of arbekacin, a new aminoglycoside antibiotic, against methicillin-resistant Staphylococcus aureus—PubMed. Drugs Exp. Clin. Res. 1994, 20, 233–239. [Google Scholar] [PubMed]
- Tsuchizaki, N.; Ishino, K.; Saito, F.; Ishikawa, J.; Nakajima, M.; Hotta, K. Trends of Arbekacin-resistant MRSA Strains in Japanese Hospitals (1979 to 2000). J. Antibiot. 2006, 59, 229–233. [Google Scholar] [CrossRef][Green Version]
- Tanaka, N.; Matsunaga, K.; Hirata, A.; Matsuhisa, Y.; Nishimura, T. Mechanism of action of habekacin, a novel amino acid-containing aminoglycoside antibiotic. Antimicrob. Agents Chemother. 1983, 24, 797–802. [Google Scholar] [CrossRef]
- Ishino, K.; Ishikawa, J.; Ikeda, Y.; Hotta, K. Characterization of a Bifunctional Aminoglycoside-Modifying Enzyme with Novel Substrate Specificity and Its Gene from a Clinical Isolate of Methicillin-Resistant Staphylococcus aureus with High Arbekacin Resistance. J. Antibiot. 2004, 57, 679–686. [Google Scholar] [CrossRef]
- Ida, T.; Okamoto, R.; Nonoyama, M.; Irinoda, K.; Kurazono, M.; Inoue, M. Antagonism between Aminoglycosides and β-Lactams in a Methicillin-Resistant Staphylococcus aureus Isolate Involves Induction of an Aminoglycoside-Modifying Enzyme. Antimicrob. Agents Chemother. 2002, 46, 1516–1521. [Google Scholar] [CrossRef]
- Kondo, S.; Tamura, A.; Gomi, S.; Ikeda, Y.; Takeuchi, T.; Mitsuhashi, S. Structures of enzymatically modified products of arbekacin by methicillin-resistant Staphylococcus aureus. J. Antibiot. 1993, 46, 310–315. [Google Scholar] [CrossRef][Green Version]
- Matsumoto, T. Arbekacin: Another novel agent for treating infections due to methicillin-resistant Staphylococcus aureus and multidrug-resistant Gram-negative pathogens. Clin. Pharmacol. Adv. Appl. 2014, 6, 139. [Google Scholar] [CrossRef][Green Version]
- Centers for Disease Control and Prevention (CDC). Staphylococcus aureus resistant to vancomycin—United States, 2002. MMWR Morb. Mortal Wkly. Rep. 2002, 51, 565–567. [Google Scholar]
- Reynolds, P.E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Parenti, F. Structure and mechanism of action of teicoplanin. J. Hosp. Infect. 1986, 7, 79–83. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. In The Year in Infection; CRC Press: Boca Raton, FL, USA, 2005; Volume 90, pp. 133–152. [Google Scholar]
- Chang, S.; Sievert, D.M.; Hageman, J.C.; Boulton, M.L.; Tenover, F.C.; Downes, F.P.; Shah, S.; Rudrik, J.T.; Pupp, G.R.; Brown, W.J.; et al. Infection with Vancomycin-Resistant Staphylococcus aureus Containing the vanA Resistance Gene. N. Engl. J. Med. 2003, 348, 1342–1347. [Google Scholar] [CrossRef]
- Cui, L.; Isii, T.; Fukuda, M.; Ochiai, T.; Neoh, H.M.; Da Cunha Camargo, I.L.B.; Watanabe, Y.; Shoji, M.; Hishinuma, T.; Hiramatsu, K. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5222–5233. [Google Scholar] [CrossRef]
- Watanabe, Y.; Cui, L.; Katayama, Y.; Kozue, K.; Hiramatsu, K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 2011, 49, 2680–2684. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Suzuki, S.; Okamura, S.; Miura, Y.; Tsukimori, A.; Nakamura, I.; Ito, N.; Masuya, A.; Shiina, T.; Matsumoto, T. Evolution and single-nucleotide polymorphisms in methicillin-resistant Staphylococcus aureus strains with reduced susceptibility to vancomycin and daptomycin, based on determination of the complete genome. Antimicrob. Agents Chemother. 2015, 59, 3585–3587. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar]
- Arthur, M.; Courvalin, P. Genetics and mechanisms of glycopeptide resistance in Enterococci. Antimicrob. Agents Chemother. 1993, 37, 1563–1571. [Google Scholar] [CrossRef]
- Peng, H.; Hu, Q.; Shang, W.; Yuan, J.; Zhang, X.; Liu, H.; Zheng, Y.; Hu, Z.; Yang, Y.; Tan, L.; et al. WalK(S221P), a naturally occurring mutation, confers vancomycin resistance in VISA strain XN108. J. Antimicrob. Chemother. 2016, 72, dkw518. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, X.; Liu, X.; Chen, C.; Suna, B. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain mw2. Antimicrob. Agents Chemother. 2015, 59, 1352–1355. [Google Scholar] [CrossRef]
- Howden, B.P.; McEvoy, C.R.E.; Allen, D.L.; Chua, K.; Gao, W.; Harrison, P.F.; Bell, J.; Coombs, G.; Bennett-Wood, V.; Porter, J.L.; et al. Evolution of Multidrug Resistance during Staphylococcus aureus Infection Involves Mutation of the Essential Two Component Regulator WalKR. PLoS Pathog. 2011, 7, e1002359. [Google Scholar] [CrossRef]
- Hafer, C.; Lin, Y.; Kornblum, J.; Lowy, F.D.; Uhlemann, A.C. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 5845–5851. [Google Scholar] [CrossRef] [PubMed]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic Pathway in Acquisition and Loss of Vancomycin Resistance in a Methicillin Resistant Staphylococcus aureus (MRSA) Strain of Clonal Type USA300. PLoS Pathog. 2012, 8, e1002505. [Google Scholar] [CrossRef] [PubMed]
- Doddangoudar, V.C.; O’Donoghue, M.M.; Chong, E.Y.C.; Tsang, D.N.C.; Boost, M.V. Role of stop codons in development and loss of vancomycin non-susceptibility in methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2012, 67, 2101–2106. [Google Scholar] [CrossRef]
- Cui, L.; Neoh, H.M.; Shoji, M.; Hiramatsu, K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 1231–1234. [Google Scholar] [CrossRef]
- Neoh, H.M.; Cui, L.; Yuzawa, H.; Takeuchi, F.; Matsuo, M.; Hiramatsu, K. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 2008, 52, 45–53. [Google Scholar] [CrossRef]
- Howden, B.P.; Stinear, T.P.; Allen, D.L.; Johnson, P.D.R.; Ward, P.B.; Davies, J.K. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52, 3755–3762. [Google Scholar] [CrossRef]
- Shoji, M.; Cui, L.; Iizuka, R.; Komoto, A.; Neoh, H.M.; Watanabe, Y.; Hishinuma, T.; Hiramatsu, K. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 3870–3881. [Google Scholar] [CrossRef]
- Ito, T.; Okuma, K.; Ma, X.X.; Yuzawa, H.; Hiramatsu, K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: Genomic island SCC. Drug Resist. Update 2003, 6, 41–52. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Lagast, H.; Van der Auwera, P.; Thys, J.P.; Crokaert, F.; Yourassowsky, E.; Meunier-Carpentier, F.; Klastersky, J.; Kains, J.P.; Serruys-Schoutens, E. Clinical evaluation of teicoplanin for therapy of severe infections caused by gram-positive bacteria. Antimicrob. Agents Chemother. 1986, 29, 52–57. [Google Scholar] [CrossRef]
- Manuel, R.J. Detection of teicoplanin resistance in UK EMRSA-17 strains. J. Antimicrob. Chemother. 2002, 50, 1089–1090. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bakthavatchalam, Y.D.; Babu, P.; Munusamy, E.; Dwarakanathan, H.T.; Rupali, P.; Zervos, M.; Victor, P.J.; Veeraraghavan, B. Genomic insights on heterogeneous resistance to vancomycin and teicoplanin in Methicillin-resistant Staphylococcus aureus: A first report from South India. PLoS ONE 2019, 14, e0227009. [Google Scholar] [CrossRef] [PubMed]
- Brandenberger, M.; Tschierske, M.; Giachino, P.; Wada, A.; Berger-Bächi, B. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta Gen. Subj. 2000, 1523, 135–139. [Google Scholar] [CrossRef]
- Maki, H.; McCallum, N.; Bischoff, M.; Wada, A.; Berger-Bächi, B. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 1953–1959. [Google Scholar] [CrossRef]
- Moellering, R.C. Linezolid: The first oxazolidinone antimicrobial. Ann. Intern. Med. 2003, 138, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Tsiodras, S.; Gold, H.S.; Sakoulas, G.; Eliopoulos, G.M.; Wennersten, C.; Venkataraman, L.; Moellering, R.C.; Ferraro, M.J. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 2001, 358, 207–208. [Google Scholar] [CrossRef]
- Swaney, S.M.; Aoki, H.; Ganoza, M.C.; Shinabarger, D.L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 1998, 42, 3251–3255. [Google Scholar] [CrossRef]
- Arias, C.A.; Vallejo, M.; Reyes, J.; Panesso, D.; Moreno, J.; Castañeda, E.; Villegas, M.V.; Murray, B.E.; Quinn, J.P. Clinical and microbiological aspects of linezolid resistance mediated by the cfr gene encoding a 23S rRNA methyltransferase. J. Clin. Microbiol. 2008, 46, 892–896. [Google Scholar] [CrossRef]
- Endimiani, A.; Blackford, M.; Dasenbrook, E.C.; Reed, M.D.; Bajaksouszian, S.; Hujer, A.M.; Rudin, S.D.; Hujer, K.M.; Perreten, V.; Rice, L.B.; et al. Emergence of linezolid-resistant Staphylococcus aureus after prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob. Agents Chemother. 2011, 55, 1684–1692. [Google Scholar] [CrossRef]
- Iguchi, S.; Mizutani, T.; Hiramatsu, K.; Kikuchi, K. Rapid Acquisition of Linezolid Resistance in Methicillin-Resistant Staphylococcus aureus: Role of Hypermutation and Homologous Recombination. PLoS ONE 2016, 11, e0155512. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.-M.; Xiong, L.; Arias, C.A.; Villegas, M.V.; Lolans, K.; Quinn, J.; Mankin, A.S. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 2007, 64, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Meka, V.G.; Pillai, S.K.; Sakoulas, G.; Wennersten, C.; Venkataraman, L.; DeGirolami, P.C.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Gold, H.S. Linezolid Resistance in Sequential Staphylococcus aureus Isolates Associated with a T2500A Mutation in the 23S rRNA Gene and Loss of a Single Copy of rRNA. J. Infect. Dis. 2004, 190, 311–317. [Google Scholar] [CrossRef]
- Locke, J.B.; Hilgers, M.; Shaw, K.J. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 2009, 53, 5265–5274. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Love, R.; Adam, H.; Golden, A.; Zelenitsky, S.; Schweizer, F.; Gorityala, B.; Lagacé-Wiens, P.R.S.; Rubinstein, E.; Walkty, A.; et al. Tedizolid: A Novel Oxazolidinone with Potent Activity Against Multidrug-Resistant Gram-Positive Pathogens. Drugs 2015, 75, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Kisgen, J.J.; Mansour, H.; Unger, N.R.; Childs, L.M. Tedizolid: A new oxazolidinone antimicrobial. Am. J. Heal. Pharm. 2014, 71, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Penewit, K.; Waalkes, A.; Xu, L.; Salipante, S.J.; Nath, A.; Werth, B.J. Identification of a novel tedizolid resistance mutation in rpoB of MRSA after in vitro serial passage. J. Antimicrob. Chemother. 2021, 76, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Dilek, A.R.; Peixe, L.; Novais, C. Dissemination of Staphylococcus epidermidis ST22 With Stable, High-Level Resistance to Linezolid and Tedizolid in the Greek-Turkish Region (2008–2016). Infect. Control Hosp. Epidemiol. 2018, 39, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Contezolid: First Approval. Drugs 2021, 81, 1587–1591. [Google Scholar] [CrossRef]
- Wang, S.; Cai, C.; Shen, Y.; Sun, C.; Shi, Q.; Wu, N.; Zheng, S.; Qian, J.; Zhang, R.; Zhou, H. In vitro Activity of Contezolid Against Methicillin-Resistant Staphylococcus aureus, Vancomycin-Resistant Enterococcus, and Strains With Linezolid Resistance Genes From China. Front. Microbiol. 2021, 12, 2408. [Google Scholar] [CrossRef] [PubMed]
- Mangili, A.; Bica, I.; Snydman, D.R.; Hamer, D.H. Daptomycin-Resistant, Methicillin-Resistant Staphylococcus aureus Bacteremia. Clin. Infect. Dis. 2005, 40, 1058–1060. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.S.; Schneider, T.; Sahl, H.G. Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 2013, 1277, 139–158. [Google Scholar] [CrossRef]
- Müller, A.; Wenzel, M.; Strahl, H.; Grein, F.; Saaki, T.N.V.; Kohl, B.; Siersma, T.; Bandow, J.E.; Sahl, H.G.; Schneider, T.; et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Natl. Acad. Sci. USA 2016, 113, E7077–E7086. [Google Scholar] [CrossRef]
- Pogliano, J.; Pogliano, N.; Silverman, J.A. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 2012, 194, 4494–4504. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Kreiswirth, B.N.; Sakoulas, G.; Yeaman, M.R.; Xiong, Y.Q.; Sawa, A.; Bayer, A.S. Enhanced Expression of dltABCD Is Associated with the Development of Daptomycin Nonsusceptibility in a Clinical Endocarditis Isolate of Staphylococcus aureus. J. Infect. Dis. 2009, 200, 1916–1920. [Google Scholar] [CrossRef]
- Cafiso, V.; Bertuccio, T.; Purrello, S.; Campanile, F.; Mammina, C.; Sartor, A.; Raglio, A.; Stefani, S. dltA overexpression: A strain-independent keystone of daptomycin resistance in methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 2014, 43, 26–31. [Google Scholar] [CrossRef]
- Sulaiman, J.E.; Wu, L.; Lam, H. Mutation in the Two-Component System Regulator YycH Leads to Daptomycin Tolerance in Methicillin-Resistant Staphylococcus aureus upon Evolution with a Population Bottleneck. Microbiol. Spectr. 2022, 10, e01687-22. [Google Scholar] [CrossRef]
- Tran, T.T.; Munita, J.M.; Arias, C.A. Mechanisms of drug resistance: Daptomycin resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 32–53. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Cuirolo, A.X.; Plata, K.B.; Riosa, S.; Silverman, J.A.; Rubio, A.; Rosato, R.R.; Rosato, A.E. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 92–102. [Google Scholar] [CrossRef]
- Miller, W.R.; Bayer, A.S.; Arias, C.A. Mechanism of Action and Resistance to Daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb. Perspect. Med. 2016, 6, a026997. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1-56238-838-X. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Leadbetter, M.R.; Adams, S.M.; Bazzini, B.; Fatheree, P.R.; Karr, D.E.; Krause, K.M.; Lam, B.M.T.; Linsell, M.S.; Nodwell, M.B.; Pace, J.L.; et al. Hydrophobic Vancomycin Derivatives with Improved ADME Properties: Discovery of Telavancin (TD-6424). J. Antibiot. 2004, 57, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Hindy, J.; Haddad, S.F.; Kanj, S.S. New drugs for methicillin-resistant Staphylococcus aureus skin and soft tissue infections. Curr. Opin. Infect. Dis. 2021, 35, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chaftari, A.M.; Hachem, R.; Jordan, M.; Garoge, K.; Al Hamal, Z.; El Zakhem, A.; Viola, G.M.; Granwehr, B.; Mulanovich, V.; Gagel, A.; et al. Case-control study of telavancin as an alternative treatment for gram-positive bloodstream infections in patients with cancer. Antimicrob. Agents Chemother. 2016, 60, 239–244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bassetti, M.; Mikulska, M.; Righi, E.; Nicolini, L.; Viscoli, C. The role of telavancin in the treatment of MRSA infections in hospital. Expert Opin. Investig. Drugs 2009, 18, 521–529. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Nichol, K.; Zhanel, G.G. Telavancin: Mechanisms of Action, In Vitro Activity, and Mechanisms of Resistance. Clin. Infect. Dis. 2015, 61, S58–S68. [Google Scholar] [CrossRef] [PubMed]
- Duggar, B.M. Aureomycin: A product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 1948, 51, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Gale, E.F.; Folkes, J.P. The assimilation of amino-acids by bacteria. 15. Actions of antibiotics on nucleic acid and protein synthesis in Staphylococcus aureus. Biochem. J. 1953, 53, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Epe, B.; Woolley, P. The binding of 6-demethylchlortetracycline to 70S, 50S and 30S ribosomal particles: A quantitative study by fluorescence anisotropy. EMBO J. 1984, 3, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Roberts, M.C.; Werckenthin, C.; Pang, Y.; Lange, C. Tetracycline resistance in Staphylococcus spp. from domestic animals. Vet. Microbiol. 1998, 63, 217–227. [Google Scholar] [CrossRef]
- Stephens, C.R.; Beereboom, J.J.; Rennhard, H.H.; Gordon, P.N.; Murai, K.; Blackwood, R.K.; von Wittenau, M.S. 6-Deoxytetracyclines. IV. 1,2 Preparation, C-6 Stereochemistry, and Reactions. J. Am. Chem. Soc. 1963, 85, 2643–2652. [Google Scholar] [CrossRef]
- Nelson, M.L.; Levy, S.B. The history of the tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, B.S.; Graber, C.J.; Diep, B.A.; Basuino, L.; Perdreau-Remington, F.; Chambers, H.F. doxycycline, not minocycline, induces its own resistance in multidrug-resistant, community-associated methicillin-Resistant Staphylococcus aureus clone usa300. Clin. Infect. Dis. 2009, 48, 1483–1484. [Google Scholar] [CrossRef]
- Trzcinski, K.; Cooper, B.S.; Hryniewicz, W.; Dowson, C.G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Vaudaux, P.; Fleury, B.; Gjinovci, A.; Huggler, E.; Tangomo-Bento, M.; Lew, D.P. Comparison of tigecycline and vancomycin for treatment of experimental foreign-body infection due to methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 3150–3152. [Google Scholar] [CrossRef]
- Draghi, D.C.; Tench, S.; Dowzicky, M.J.; Sahm, D.F. Baseline in vitro Activity of Tigecycline among Key Bacterial Pathogens Exhibiting Multidrug Resistance. Chemotherapy 2008, 54, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Honeyman, L.; Ismail, M.; Nelson, M.L.; Bhatia, B.; Bowser, T.E.; Chen, J.; Mechiche, R.; Ohemeng, K.; Verma, A.K.; Cannon, E.P.; et al. Structure-activity relationship of the aminomethylcyclines and the discovery of omadacycline. Antimicrob. Agents Chemother. 2015, 59, 7044–7053. [Google Scholar] [CrossRef]
- O’Riordan, W.; Green, S.; Overcash, J.S.; Puljiz, I.; Metallidis, S.; Gardovskis, J.; Garrity-Ryan, L.; Das, A.F.; Tzanis, E.; Eckburg, P.B.; et al. Omadacycline for Acute Bacterial Skin and Skin-Structure Infections. N. Engl. J. Med. 2019, 380, 528–538. [Google Scholar] [CrossRef]
- Tanaka, S.K.; Steenbergen, J.; Villano, S. Discovery, pharmacology, and clinical profile of omadacycline, a novel aminomethylcycline antibiotic. Bioorg. Med. Chem. 2016, 24, 6409–6419. [Google Scholar] [CrossRef]
- Draper, M.P.; Weir, S.; Macone, A.; Donatelli, J.; Trieber, C.A.; Tanaka, S.K.; Levya, S.B. Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob. Agents Chemother. 2014, 58, 1279–1283. [Google Scholar] [CrossRef]
- Macone, A.B.; Caruso, B.K.; Leahy, R.G.; Donatelli, J.; Weir, S.; Draper, M.P.; Tanaka, S.K.; Levy, S.B. In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob. Agents Chemother. 2014, 58, 1127–1135. [Google Scholar] [CrossRef]
- Bodley, J.W.; Zieve, F.J.; Lin, L.; Zieve, S.T. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 1969, 37, 437–443. [Google Scholar] [CrossRef]
- Tanaka, N.; Kinoshita, T.; Masukawa, H. Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem. Biophys. Res. Commun. 1968, 30, 278–283. [Google Scholar] [CrossRef]
- Nagaev, I.; Bjorkman, J.; Andersson, D.I.; Hughes, D. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 2001, 40, 433–439. [Google Scholar] [CrossRef]
- O’Neill, A.J.; Larsen, A.R.; Henriksen, A.S.; Chopra, I. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob. Agents Chemother. 2004, 48, 3594–3597. [Google Scholar] [CrossRef]
- O’Brien, F.G. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 2002, 50, 313–321. [Google Scholar] [CrossRef]
- O’Neill, A.J.; McLaws, F.; Kahlmeter, G.; Henriksen, A.S.; Chopra, I. Genetic basis of resistance to fusidic acid in Staphylococci. Antimicrob. Agents Chemother. 2007, 51, 1737–1740. [Google Scholar] [CrossRef]
- Tanus, T.; Scangarella-Oman, N.E.; Dalessandro, M.; Li, G.; Breton, J.J.; Tomayko, J.F. A Randomized, Double-blind, Comparative Study to Assess the Safety and Efficacy of Topical Retapamulin Ointment 1% Versus Oral Linezolid in the Treatment of Secondarily Infected Traumatic Lesions and Impetigo Due to Methicillin-Resistant Staphylococcus au. Adv. Skin Wound Care 2014, 27, 548–559. [Google Scholar] [CrossRef]
- Hooper, D.C. Mechanisms of fluoroquinolone resistance. Drug Resist. Updates 1999, 2, 38–55. [Google Scholar] [CrossRef]
- LeBel, M. Ciprofloxacin: Chemistry, Mechanism of Action, Resistance, Antimicrobial Spectrum, Pharmacokinetics, Clinical Trials, and Adverse Reactions. Pharmacother. J. Hum. Pharmacol. Drug Ther. 1988, 8, 3–30. [Google Scholar] [CrossRef]
- Hooper, D.C. Mode of Action of Fluoroquinolones. Drugs 1999, 58, 6–10. [Google Scholar] [CrossRef]
- Drlica, K. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 1999, 2, 504–508. [Google Scholar] [CrossRef]
- Papkou, A.; Hedge, J.; Kapel, N.; Young, B.; MacLean, R.C. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat. Commun. 2020, 11, 3970. [Google Scholar] [CrossRef]
- Aeschlimann, J.R.; Dresser, L.D.; Kaatz, G.W.; Rybak, M.J. Effects of NorA inhibitors on in vitro antibacterial activities and postantibiotic effects of levofloxacin, ciprofloxacin, and norfloxacin in genetically related strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 335–340. [Google Scholar] [CrossRef]
- Schmitz, F. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 41, 481–484. [Google Scholar] [CrossRef]
- Nilius, A.M.; Shen, L.L.; Hensey-Rudloff, D.; Almer, L.S.; Beyer, J.M.; Balli, D.J.; Cai, Y.; Flamm, R.K. In vitro antibacterial potency and spectrum of ABT-492, a new fluoroquinolone. Antimicrob. Agents Chemother. 2003, 47, 3260–3269. [Google Scholar] [CrossRef]
- Saravolatz, L.D.; Stein, G.E. Delafloxacin: A New Anti–methicillin-resistant Staphylococcus aureus Fluoroquinolone. Clin. Infect. Dis. 2019, 68, 1058–1062. [Google Scholar] [CrossRef]
- Iregui, A.; Khan, Z.; Malik, S.; Landman, D.; Quale, J. Emergence of Delafloxacin-Resistant Staphylococcus aureus in Brooklyn, New York. Clin. Infect. Dis. 2020, 70, 1758–1760. [Google Scholar] [CrossRef]
- Remy, J.M.; Tow-Keogh, C.A.; McConnell, T.S.; Dalton, J.M.; DeVito, J.A. Activity of delafloxacin against methicillin-resistant Staphylococcus aureus: Resistance selection and characterization. J. Antimicrob. Chemother. 2012, 67, 2814–2820. [Google Scholar] [CrossRef]
- McCurdy, S.; Lawrence, L.; Quintas, M.; Woosley, L.; Flamm, R.; Tseng, C.; Cammarata, S. In vitro activity of delafloxacin and microbiological response against fluoroquinolone-susceptible and nonsusceptible Staphylococcus aureus isolates from two phase 3 studies of acute bacterial skin and skin structure infections. Antimicrob. Agents Chemother. 2017, 61, 2017. [Google Scholar] [CrossRef]
- Kurokawa, I.; Kanayama, S.; Yamasaki, O. Antimicrobial activity of ozenoxacin and other antimicrobials against Staphylococcus aureus strains isolated from clinical skin specimens in Japan in 2019 and 2020. J. Infect. Chemother. 2022, 28, 1693–1696. [Google Scholar] [CrossRef]
- Brogden, R.N.; Carmine, A.A.; Heel, R.C.; Speight, T.M.; Avery, G.S. Trimethoprim: A Review of its Antibacterial Activity, Pharmacokinetics and Therapeutic Use in Urinary Tract Infections. Drugs 1982, 23, 405–430. [Google Scholar] [CrossRef]
- Dale, G.E.; Broger, C.; D’Arcy, A.; Hartman, P.G.; DeHoogt, R.; Jolidon, S.; Kompis, I.; Labhardt, A.M.; Langen, H.; Locher, H.; et al. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance 1 1 Edited by T.Richmond. J. Mol. Biol. 1997, 266, 23–30. [Google Scholar] [CrossRef]
- Nurjadi, D.; Olalekan, A.O.; Layer, F.; Shittu, A.O.; Alabi, A.; Ghebremedhin, B.; Schaumburg, F.; Hofmann-Eifler, J.; Van Genderen, P.J.J.; Caumes, E.; et al. Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J. Antimicrob. Chemother. 2014, 69, 2361–2368. [Google Scholar] [CrossRef]
- Perreten, V.; Kadlec, K.; Schwarz, S.; Gronlund Andersson, U.; Finn, M.; Greko, C.; Moodley, A.; Kania, S.A.; Frank, L.A.; Bemis, D.A.; et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: An international multicentre study. J. Antimicrob. Chemother. 2010, 65, 1145–1154. [Google Scholar] [CrossRef]
- Nurjadi, D.; Schäfer, J.; Friedrich-Jänicke, B.; Mueller, A.; Neumayr, A.; Calvo-Cano, A.; Goorhuis, A.; Molhoek, N.; Lagler, H.; Kantele, A.; et al. Predominance of dfrG as determinant of trimethoprim resistance in imported Staphylococcus aureus. Clin. Microbiol. Infect. 2015, 21, 1095.e5–1095.e9. [Google Scholar] [CrossRef]
- Kadlec, K.; Schwarz, S. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet (L). Antimicrob. Agents Chemother. 2009, 53, 776–778. [Google Scholar] [CrossRef]
- Rouch, D.A.; Messerotti, L.J.; Loo, L.S.L.; Jackson, C.A.; Skurray, R.A. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol. Microbiol. 1989, 3, 161–175. [Google Scholar] [CrossRef]
- Sekiguchi, J.I.; Tharavichitkul, P.; Miyoshi-Akiyama, T.; Chupia, V.; Fujino, T.; Araake, M.; Irie, A.; Morita, K.; Kuratsuji, T.; Kirikae, T. Cloning and characterization of a novel trimethoprim-resistant dihydrofolate reductase from a nosocomial isolate of Staphylococcus aureus CM.S2 (IMCJ1454). Antimicrob. Agents Chemother. 2005, 49, 3948–3951. [Google Scholar] [CrossRef]
- Kadlec, K.; Schwarz, S. Novel ABC Transporter Gene, vga (C), Located on a Multiresistance Plasmid from a Porcine Methicillin-Resistant Staphylococcus aureus ST398 Strain. Antimicrob. Agents Chemother. 2009, 53, 3589–3591. [Google Scholar] [CrossRef]
- Chain, E.B.; Mellows, G. Structure of pseudomonic acid, an antibiotic from Pseudomonas fluorescens. J. Chem. Soc. Chem. Commun. 1974, 20, 847. [Google Scholar] [CrossRef]
- Mehtar, S. New strategies for the use of mupirocin for the prevention of serious infection. J. Hosp. Infect. 1998, 40, S39–S44. [Google Scholar] [CrossRef]
- Rahman, M.; Noble, W.C.; Cookson, B.; Baird, D.; Coia, J. Mupirocin-Resistant Staphylococcus aureus. Lancet 1987, 330, 387–388. [Google Scholar] [CrossRef]
- Kavi, J.; Andrews, J.M.; Wise, R.; Smith, M.D.; Sanghrajka, M.; Lock, S. Mupirocin-Resistant Staphylococcus aureus. Lancet 1987, 330, 1472–1473. [Google Scholar] [CrossRef]
- Hughes, J.; Mellows, G. On the mode of action of pseudomonic acid: Inhibition of protein synthesis in Staphylococcus aureus. J. Antibiot. 1978, 31, 330–335. [Google Scholar] [CrossRef]
- Hurdle, J.G. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J. Antimicrob. Chemother. 2003, 53, 102–104. [Google Scholar] [CrossRef]
- Antonio, M.; McFerran, N.; Pallen, M.J. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 438–442. [Google Scholar] [CrossRef]
- Hurdle, J.G.; O’Neill, A.J.; Ingham, E.; Fishwick, C.; Chopra, I. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob. Agents Chemother. 2004, 48, 4366–4376. [Google Scholar] [CrossRef]
- Fujimura, S.; Tokue, Y.; Watanabe, A. Isoleucyl-tRNA Synthetase Mutations in Staphylococcus aureus Clinical Isolates and In Vitro Selection of Low-Level Mupirocin-Resistant Strains. Antimicrob. Agents Chemother. 2003, 47, 3373–3374. [Google Scholar] [CrossRef]
- Udo, E.E.; Jacob, L.E.; Mathew, B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J. Med. Microbiol. 2001, 50, 909–915. [Google Scholar] [CrossRef]
- Seah, C.; Alexander, D.C.; Louie, L.; Simor, A.; Low, D.E.; Longtin, J.; Melano, R.G. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1916–1920. [Google Scholar] [CrossRef]
- Hodgson, J.E.; Curnock, S.P.; Dyke, K.G.H.; Morris, R.; Sylvester, D.R.; Gross, M.S. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 1994, 38, 1205–1208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; ISBN 0956-4624. [Google Scholar]
- Hendlin, D.; Stapley, E.O.; Jackson, M.; Wallick, H.; Miller, A.K.; Wolf, F.J.; Miller, T.W.; Chaiet, L.; Kahan, F.M.; Foltz, E.L.; et al. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science 1969, 166, 122–123. [Google Scholar] [CrossRef]
- Kahan, F.M.; Kahan, J.S.; Cassidy, P.J.; Kropp, H. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 1974, 235, 364–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Fu, Z.; Zhou, Y.; Liu, Y.; Xu, X.; Wang, M. Mutations of the Transporter Proteins GlpT and UhpT Confer Fosfomycin Resistance in Staphylococcus aureus. Front. Microbiol. 2017, 8, 914. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Ma, Y.; Chen, C.; Guo, Y.; Hu, F.; Liu, Y.; Xu, X.; Wang, M. Prevalence of Fosfomycin Resistance and Mutations in murA, glpT, and uhpT in Methicillin-Resistant Staphylococcus aureus Strains Isolated from Blood and Cerebrospinal Fluid Samples. Front. Microbiol. 2016, 6, 1544. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, L.; Liu, Y.; Wang, Y.; Jian, Y.; Zhao, N.; Yang, Z.; Wang, X.; Liu, Q.; Li, M. Mechanisms of high-level fosfomycin resistance in Staphylococcus aureus epidemic lineage ST5. J. Antimicrob. Chemother. 2022, 77, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, T.; Wang, H.; Zeng, W.; Wu, Q.; Yu, K.; Xu, Y.; Zhang, X.; Zhou, T. Molecular Mechanisms and Epidemiology of Fosfomycin Resistance in Staphylococcus aureus Isolated From Patients at a Teaching Hospital in China. Front. Microbiol. 2020, 11, 1290. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Wang, Y.; Hooper, D.C. Tet38 Efflux pump contributes to fosfomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2018, 62, e00927-18. [Google Scholar] [CrossRef]
- Chen, Y.; Ji, S.; Sun, L.; Wang, H.; Zhu, F.; Chen, M.; Zhuang, H.; Wang, Z.; Jiang, S.; Yu, Y.; et al. The novel fosfomycin resistance gene fosY is present on a genomic island in CC1 methicillin-resistant Staphylococcus aureus. Emerg. Microbes Infect. 2022, 11, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Sensi, P. History of the Development of Rifampin. Clin. Infect. Dis. 1983, 5, S402–S406. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.A.; Leon, G.; Miller, N.M.; Reyes, S.P.; Thantrong, C.H.; Thokkadam, A.M.; Lemma, A.S.; Sivaloganathan, D.M.; Wan, X.; Brynildsen, M.P. Rifamycin antibiotics and the mechanisms of their failure. J. Antibiot. 2021, 74, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, G.; Honikel, K.O.; Knüsel, F.; Nüesch, J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1967, 145, 843–844. [Google Scholar] [CrossRef]
- Aiba, Y.; Katayama, Y.; Hishinuma, T.; Murakami-Kuroda, H.; Cui, L.; Hiramatsu, K. Mutation of RNA polymerase β-subunit gene promotes heterogeneous-to-homogeneous conversion of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 4861–4871. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-A. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in Gram-positive cocci isolated in a Korean hospital. J. Antimicrob. Chemother. 2002, 49, 489–495. [Google Scholar] [CrossRef]
- Diep, B.A.; Chambers, H.F.; Graber, C.J.; Szumowski, J.D.; Miller, L.G.; Han, L.L.; Chen, J.H.; Lin, F.; Lin, J.; Phan, T.H.; et al. Emergence of Multidrug-Resistant, Community-Associated, Methicillin-Resistant Staphylococcus aureus Clone USA300 in Men Who Have Sex with Men. Ann. Intern. Med. 2008, 148, 249. [Google Scholar] [CrossRef] [PubMed]
- Martineau, F.; Picard, F.J.; Lansac, N.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000, 44, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, E.; Keynan, Y. Vancomycin Revisited—60 Years Later. Front. Public Health 2014, 2, 217. [Google Scholar] [CrossRef] [PubMed]
- Barna, J.C.J.; Williams, D.H. The Structure and Mode of Action of Glycopeptide Antibiotics of the Vancomycin Group. Annu. Rev. Microbiol. 1984, 38, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.E. Studies on the mode of action of vancomycin. BBA Biochim. Biophys. Acta 1961, 52, 403–405. [Google Scholar] [CrossRef]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [PubMed]
- Dubrac, S.; Bisicchia, P.; Devine, K.M.; Msadek, T. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 2008, 70, 1307–1322. [Google Scholar] [CrossRef]
- Delaune, A.; Poupel, O.; Mallet, A.; Coic, Y.-M.; Msadek, T.; Dubrac, S. Peptidoglycan Crosslinking Relaxation Plays an Important Role in Staphylococcus aureus WalKR-Dependent Cell Viability. PLoS ONE 2011, 6, e17054. [Google Scholar] [CrossRef] [PubMed]
- Herbert, S.; Bera, A.; Nerz, C.; Kraus, D.; Peschel, A.; Goerke, C.; Meehl, M.; Cheung, A.; Götz, F. Molecular Basis of Resistance to Muramidase and Cationic Antimicrobial Peptide Activity of Lysozyme in Staphylococci. PLoS Pathog. 2007, 3, e102. [Google Scholar] [CrossRef]
- Sakoulas, G.; Eliopoulos, G.M.; Moellering, R.C.; Wennersten, C.; Venkataraman, L.; Novick, R.P.; Gold, H.S. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 2002, 46, 1492–1502. [Google Scholar] [CrossRef]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef]
- Münch, D.; Engels, I.; Müller, A.; Reder-Christ, K.; Falkenstein-Paul, H.; Bierbaum, G.; Grein, F.; Bendas, G.; Sahl, H.G.; Schneider, T. Structural variations of the cell wall precursor lipid II and their influence on binding and activity of the lipoglycopeptide antibiotic oritavancin. Antimicrob. Agents Chemother. 2015, 59, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 1993, 175, 117–127. [Google Scholar] [CrossRef]
- Lambert, P. Bacterial resistance to antibiotics: Modified target sites. Adv. Drug Deliv. Rev. 2005, 57, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Sievert, D.M.; Rudrik, J.T.; Patel, J.B.; McDonald, L.C.; Wilkins, M.J.; Hageman, J.C. Vancomycin-Resistant Staphylococcus aureus in the United States, 2002–2006. Clin. Infect. Dis. 2008, 46, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Melo-Cristino, J.; Resina, C.; Manuel, V.; Lito, L.; Ramirez, M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet 2013, 382, 205. [Google Scholar] [CrossRef]
- Birmingham, M.C.; Rayner, C.R.; Meagher, A.K.; Flavin, S.M.; Batts, D.H.; Schentag, J.J. Linezolid for the treatment of multidrug-resistant, gram-positive infections: Experience from a compassionate-use program. Clin. Infect. Dis. 2003, 36, 159–168. [Google Scholar] [CrossRef]
- Barber, K.E.; Smith, J.R.; Raut, A.; Rybak, M.J. Evaluation of tedizolid against Staphylococcus aureus and Enterococci with reduced susceptibility to vancomycin, daptomycin or linezolid. J. Antimicrob. Chemother. 2016, 71, 152–155. [Google Scholar] [CrossRef]
- Perlaza-Jiménez, L.; Tan, K.-S.; Piper, S.J.; Johnson, R.M.; Bamert, R.S.; Stubenrauch, C.J.; Wright, A.; Lupton, D.; Lithgow, T.; Belousoff, M.J. A Structurally Characterized Staphylococcus aureus Evolutionary Escape Route from Treatment with the Antibiotic Linezolid. Microbiol. Spectr. 2022, 10, e00583-22. [Google Scholar] [CrossRef]
- Humphries, R.M.; Pollett, S.; Sakoulas, G. A current perspective on daptomycin for the clinical microbiologist. Clin. Microbiol. Rev. 2013, 26, 759–780. [Google Scholar] [CrossRef]
- Arbeit, R.D.; Maki, D.; Tally, F.P.; Campanaro, E.; Eisenstein, B.I. The Safety and Efficacy of Daptomycin for the Treatment of Complicated Skin and Skin-Structure Infections. Clin. Infect. Dis. 2004, 38, 1673–1681. [Google Scholar] [CrossRef]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef]
- Silverman, J.A.; Oliver, N.; Andrew, T.; Tongchuan, L.I. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 2001, 45, 1799–1802. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; Tinelli, M.; Grundmann, H.; Akkerboom, V.; Monaco, M.; Del Grosso, M.; Errico, G.; Pantosti, A.; Friedrich, A.W. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprF and vraS genes: A new insight into daptomycin resistance. Front. Microbiol. 2018, 9, 2705. [Google Scholar] [CrossRef]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.-J.; Proctor, R.A.; Sahl, H.-G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in Clinical Treatment of Staphylococcus aureus Infection with Daptomycin Are Associated with Alterations in Surface Charge, Membrane Phospholipid Asymmetry, and Drug Binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Van Hal, S.J.; Paterson, D.L.; Gosbell, I.B. Emergence of daptomycin resistance following vancomycin-unresponsive Staphylococcus aureus bacteraemia in a daptomycin-naïve patient—A review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Capone, A.; Cafiso, V.; Campanile, F.; Parisi, G.; Mariani, B.; Petrosillo, N.; Stefani, S. In vivo development of daptomycin resistance in vancomycin-susceptible methicillin-resistant Staphylococcus aureus severe infections previously treated with glycopeptides. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 625–631. [Google Scholar] [CrossRef]
- Bæk, K.T.; Thøgersen, L.; Mogenssen, R.G.; Mellergaard, M.; Thomsen, L.E.; Petersen, A.; Skov, S.; Cameron, D.R.; Peleg, A.Y.; Frees, D. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a Compensatory Inactivation of the clpX Gene. Antimicrob. Agents Chemother. 2015, 59, 6983–6991. [Google Scholar] [CrossRef]
- Ernst, C.M.; Slavetinsky, C.J.; Kuhn, S.; Hauser, J.N.; Nega, M.; Mishra, N.N.; Gekeler, C.; Bayer, A.S.; Peschel, A. Gain-of-function mutations in the phospholipid flippase mprf confer specific daptomycin resistance. MBio 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.-J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The Bacterial Defensin Resistance Protein MprF Consists of Separable Domains for Lipid Lysinylation and Antimicrobial Peptide Repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef]
- Bertsche, U.; Yang, S.J.; Kuehner, D.; Wanner, S.; Mishra, N.N.; Roth, T.; Nega, M.; Schneider, A.; Mayer, C.; Grau, T.; et al. Increased Cell Wall Teichoic Acid Production and D-alanylation Are Common Phenotypes among Daptomycin-Resistant Methicillin-Resistant Staphylococcus aureus (MRSA) Clinical Isolates. PLoS ONE 2013, 8, e67398. [Google Scholar] [CrossRef]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2137–2145. [Google Scholar] [CrossRef]
- Mishra, N.N.; Bayer, A.S.; Weidenmaier, C.; Grau, T.; Wanner, S.; Stefani, S.; Cafiso, V.; Bertuccio, T.; Yeaman, M.R.; Nast, C.C.; et al. Phenotypic and Genotypic Characterization of Daptomycin-Resistant Methicillin-Resistant Staphylococcus aureus Strains: Relative Roles of mprF and dlt Operons. PLoS ONE 2014, 9, e107426. [Google Scholar] [CrossRef]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef]
- Meehl, M.; Herbert, S.; Götz, F.; Cheung, A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Pfaller, M.A.; Schmitz, F.J.; Smayevsky, J.; Bell, J.; Jones, R.N.; Beach, M. Survey of Infections Due to Staphylococcus Species: Frequency of Occurrence and Antimicrobial Susceptibility of Isolates Collected in the United States, Canada, Latin America, Europe, and the Western Pacific Region for the SENTRY Antimicrobial Surveillanc. Clin. Infect. Dis. 2001, 32, S114–S132. [Google Scholar] [CrossRef] [PubMed]
- Burgold-Voigt, S.; Monecke, S.; Simbeck, A.; Holzmann, T.; Kieninger, B.; Liebler-Tenorio, E.M.; Braun, S.D.; Collatz, M.; Diezel, C.; Müller, E.; et al. Characterisation and Molecular Analysis of an Unusual Chimeric Methicillin Resistant Staphylococcus Aureus Strain and its Bacteriophages. Front. Genet. 2021, 12, 1823. [Google Scholar] [CrossRef] [PubMed]
- Kaatz, G.W.; Seo, S.M.; Ruble, C.A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1993, 37, 1086–1094. [Google Scholar] [CrossRef]
- Yun, H.J.; Min, Y.H.; Lim, J.A.; Kang, J.W.; Kim, S.Y.; Kim, M.J.; Jeong, J.H.; Choi, Y.J.; Kwon, H.J.; Jung, Y.H.; et al. In vitro and in vivo antibacterial activities of DW286, a new fluoronaphthyridone antibiotic. Antimicrob. Agents Chemother. 2002, 46, 3071–3074. [Google Scholar] [CrossRef][Green Version]
- Schindler, B.D.; Kaatz, G.W. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updates 2016, 27, 1–13. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Ubukata, K.; Itoh-Yamashita, N.; Konno, M. Cloning and expression of the norA gene for fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1989, 33, 1535–1539. [Google Scholar] [CrossRef]
- Neyfakh, A.A.; Borsch, C.M.; Kaatz, G.W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993, 37, 128–129. [Google Scholar] [CrossRef]
- Yoshida, H.; Bogaki, M.; Nakamura, S.; Ubukata, K.; Konno, M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 1990, 172, 6942–6949. [Google Scholar] [CrossRef]
- Paul, M.; Bishara, J.; Yahav, D.; Goldberg, E.; Neuberger, A.; Ghanem-Zoubi, N.; Dickstein, Y.; Nseir, W.; Dan, M.; Leibovici, L. Trimethoprim-sulfamethoxazole versus vancomycin for severe infections caused by meticillin resistant Staphylococcus aureus: Randomised controlled trial. BMJ 2015, 350, h2219. [Google Scholar] [CrossRef] [PubMed]
- Elwell, L.P.; Wilson, H.R.; Knick, V.B.; Keith, B.R. In vitro and in vivo efficacy of the combination trimethoprim-sulfamethoxazole against clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1986, 29, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Frei, C.R.; Miller, M.L.; Lewis, J.S.; Lawson, K.A.; Hunter, J.M.; Oramasionwu, C.U.; Talbert, R.L. Trimethoprim-Sulfamethoxazole or Clindamycin for Community-Associated MRSA (CA-MRSA) Skin Infections. J. Am. Board Fam. Med. 2010, 23, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.D. The Relation of p-Aminobenzoic Acid to the Mechanism of the Action of Sulphanilamide; Wiley-Blackwell: Hoboken, NJ, USA, 1940; Volume 21. [Google Scholar]
- Hitchings, G.H. Mechanism of Action of Trimethoprim-Sulfamethoxazole—I. J. Infect. Dis. 1973, 128, S433–S436. [Google Scholar] [CrossRef]
- Kalkut, G. Sulfonamides and Trimethoprim. Cancer Invest. 1998, 16, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Chernyshev, A.; Fleischmann, T.; Kohen, A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl. Microbiol. Biotechnol. 2007, 74, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A. Role of Folate Antagonists in the Treatment of Methicillin-Resistant Staphylococcus aureus Infection. Clin. Infect. Dis. 2008, 46, 584–593. [Google Scholar]
- Khamash, D.F.; Voskertchian, A.; Tamma, P.D.; Akinboyo, I.C.; Carroll, K.C.; Milstone, A.M. Increasing Clindamycin and Trimethoprim-Sulfamethoxazole Resistance in Pediatric Staphylococcus aureus Infections. J. Pediatric Infect. Dis. Soc. 2019, 8, 351–353. [Google Scholar] [CrossRef]
- Acree, M.E.; Morgan, E.; David, M.Z. S. aureus infections in chicago, 2006–2014: Increase in CA MSSA and decrease in MRSA incidence. Infect. Control Hosp. Epidemiol. 2017, 38, 1226–1234. [Google Scholar] [CrossRef]
- Harris, T.; Bowen, A.; Holt, D.; Sarovich, D.; Stevens, K.; Currie, B.; Howden, B.; Carapetis, J.; Giffard, P.; Tong, S. Investigation of trimethoprim/sulfamethoxazole resistance in an emerging sequence type 5 methicillin-resistant Staphylococcus Aureus clone reveals discrepant resistance reporting. Clin. Microbiol. Infect. 2018, 24, 1027–1029. [Google Scholar] [CrossRef]
- Sato, T.; Ito, R.; Kawamura, M.; Fujimura, S. The Risk of Emerging Resistance to Trimethoprim/Sulfamethoxazole in Staphylococcus aureus. Infect. Drug Resist. 2022, 15, 4779–4784. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.O.; Lyon, B.R. Genetics of antimicrobial resistance in Staphylococcus aureus. Future Microbiol. 2009, 4, 565–582. [Google Scholar] [CrossRef]
- Frey, K.M.; Lombardo, M.N.; Wright, D.L.; Anderson, A.C. Towards the understanding of resistance mechanisms in clinically isolated trimethoprim-resistant, methicillin-resistant Staphylococcus aureus dihydrofolate reductase. J. Struct. Biol. 2010, 170, 93–97. [Google Scholar] [CrossRef]
- Oefner, C.; Bandera, M.; Haldimann, A.; Laue, H.; Schulz, H.; Mukhija, S.; Parisi, S.; Weiss, L.; Lociuro, S.; Dale, G.E. Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity. J. Antimicrob. Chemother. 2009, 63, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.A.; Bradley, S.F.; Kauffman, C.A.; Morton, T.M. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in Staphylococcal isolates. Antimicrob. Agents Chemother. 1996, 40, 2820–2823. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.A.; Bradley, S.F.; Kauffman, C.A.; Morton, T.M.; Patterson, J.E.; Reagan, D.R. Characterization of Mupirocin-Resistant Staphylococcus aureus from Different Geographic Areas. Antimicrob. Agents Chemother. 1998, 42, 1305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bongiorno, D.; Mongelli, G.; Stefani, S.; Campanile, F. Burden of Rifampicin- and Methicillin-Resistant Staphylococcus aureus in Italy. Microb. Drug Resist. 2018, 24, 732–738. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Miyakis, S.; Ward, D.V.; Earl, A.M.; Rubio, A.; Cameron, D.R.; Pillai, S.; Moellering, R.C.; Eliopoulos, G.M. Whole Genome Characterization of the Mechanisms of Daptomycin Resistance in Clinical and Laboratory Derived Isolates of Staphylococcus aureus. PLoS ONE 2012, 7, e28316. [Google Scholar] [CrossRef] [PubMed]
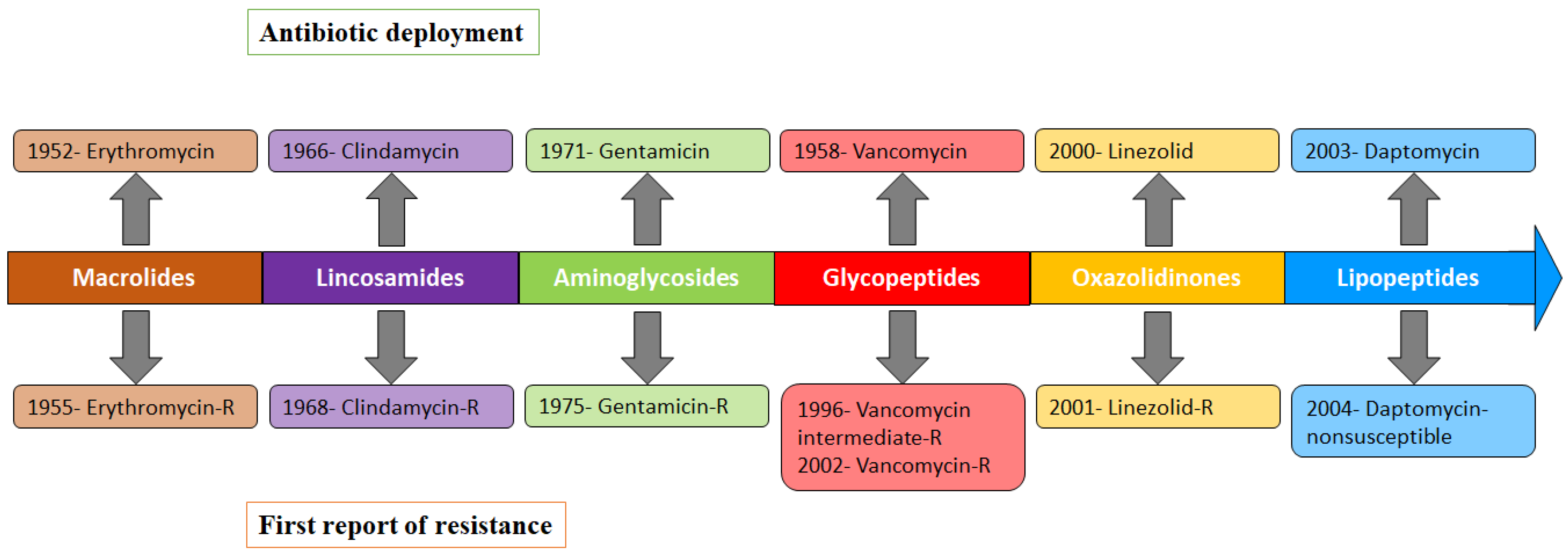
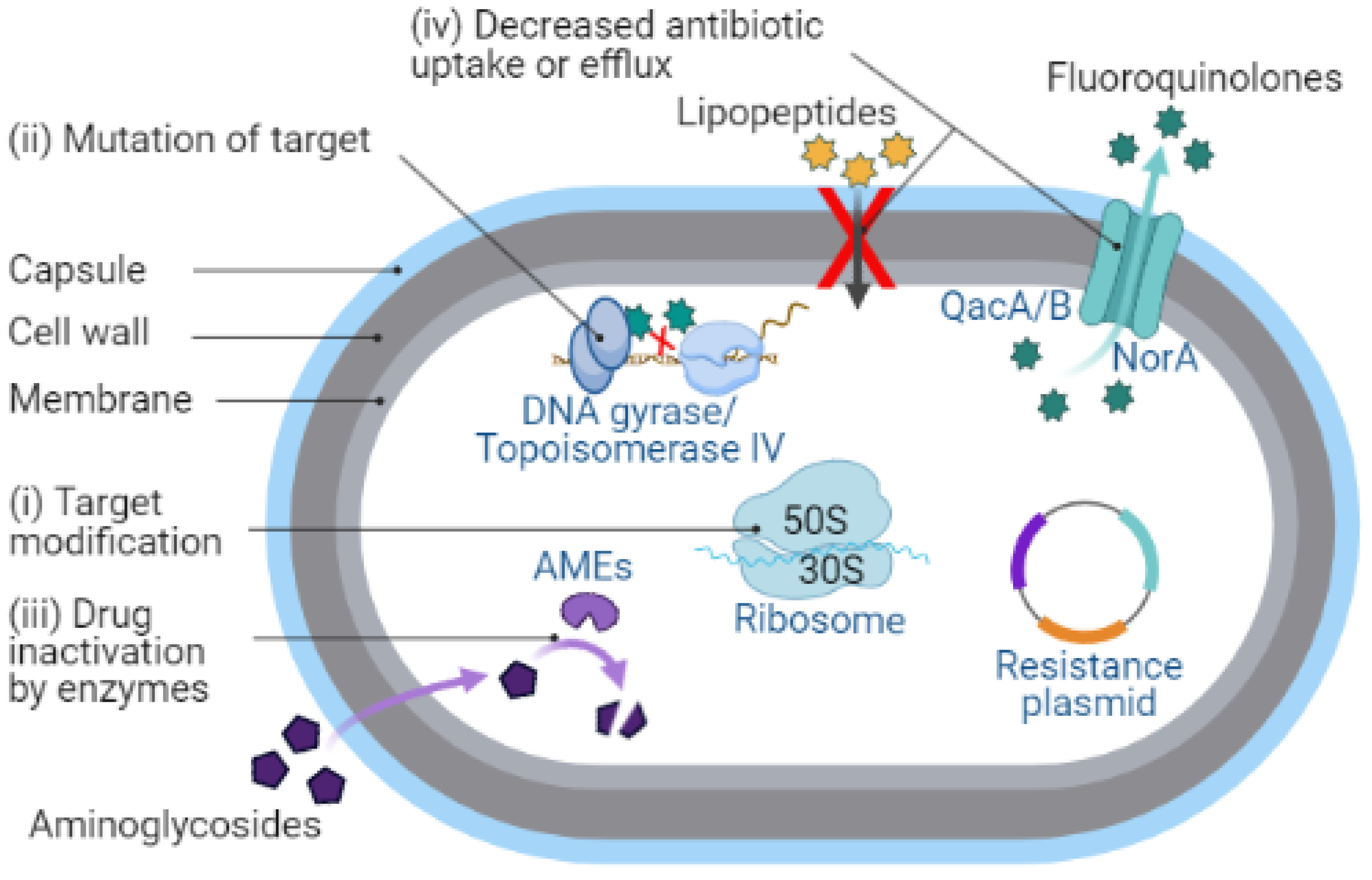
| Antibiotic Class/ Primary Agent | Approve Year and Use | Primary Target and Mechanisms of Action | Resistance Genes | Mechanism(s) of Resistance | Comments |
|---|---|---|---|---|---|
| Macrolides | Protein synthesis | ||||
| Erythromycin | 1952 [66]. SSTI (Resistance 1955) [67] | Erythromycin binds to bacterial 23S rRNA in the 50S ribosomal subunit and stops protein synthesis by inhibiting the transpeptidation/translocation step of protein synthesis and assembly of the 50S ribosomal subunit [68,69]. The target site for macrolides is nucleotides A2058 and A2059 located in the V region of 23S rRNA and, rarely, nucleotide A752 located in domain II [70]. | ermA [31], ermB, ermC [32], ermY [52], msr(F) [71], msrA [72], msrB, ereA, ereB, mphB, mphC [52] | (i) Modification of the bacterial ribosome by 23S rRNA methyltransferase (encoded by erm genes) prevents the binding of erythromycin to ribosomal target [31,32]. (ii) Active efflux of macrolides from cells by ATP-binding-cassette family (ABC-F) transporters (encoded by msrA and msrB genes) protects ribosomes from inhibition [72,73]. (iii) Enzymatic hydrolysis of 14- and 15-membered lactone ring of macrolides by esterase (encoded by ere genes) prevents its binding to the antibiotic target site [74]. (iv) Phosphotransferases (encoded by mph genes) introduce phosphate to the 2′-hydroxyl group of the 14-, 15-, and 16-membered lactone rings of macrolides amino sugar, which interferes with the interaction of the antibiotic with nucleotide A2058 [52]. | Modification of the bacterial ribosome and active efflux from the bacterial cell are important mechanisms of macrolide resistance in S. aureus. |
| Lincosamides | Protein synthesis | ||||
| Clindamycin | Discovered in 1966. SSTI caused by CA-MRSA [29] (Resistance 1968) [30] | Clindamycin binds to bacterial 23S rRNA in the 50S ribosomal subunit and impedes both the assembly of ribosomes and the translation process [75]. | ermA, ermB, ermC [76] cfr [41,42] | (i) The rRNA methylase (encoded by erm genes) methylates an adenosine nucleotide within the peptidyl transferase center, resulting in the C-8 methylation of A2503 (m8A2503) [77]. (ii) The acquired cfr gene encoded rRNA methyltransferase methylates an adenine residue of the 23S rRNA in the 50S ribosomal subunit [41], resulting in altered antibiotic binding sites within the ribosome. | |
| Aminoglycosides | Protein synthesis | ||||
| Gentamicin | U.S. FDA 1971. Bacterial meningitis, sepsis of newborns, septicemia, UTI (Resistance 1975) [78,79] | Gentamicin binds to the A-site on the 16S rRNA helix at the mRNA-tRNA decoding center of bacterial 30S ribosome subunit [80,81], causing the inhibition and inaccurate induction of translation, disrupting protein synthesis [82,83,84]. | aac(6′)/aph(2″) aadD (AG O-adenyltransferase) [85] ant(4′) (AG O-nucleotidyltransferase(4′)) ant(9) (AG O-nucleotidyltransferase(9)) | The bifunctional AMEs inactivate aminoglycosides by acetylating, phosphorylating, or adenylating amino or hydroxyl groups [51,85] Gentamicin, tobramycin and kanamycin resistance is generally mediated by a bifunctional AME AAC(6′)-APH(2″) (encoded by aac(6′)/aph(2″) gene) that specifies 6′-acetyltransferase [AAC(6′)] and/or 2″-phosphotransferase [APH(2″)] aminoglycoside modifying activities [36,37]. | The aac(6′)/aph(2″) gene is most prevalent in aminoglycoside resistant S. aureus [86]. |
| Arbekacin (not used clinically in the U.S.) | Japanese PMDA 1990 [87]. Pneumonia and sepsis due to MRSA. (Resistance 1979) [88] | Arbekacin binds to both 50S and the 30S ribosomal subunits, resulting in codon misreading and inhibition of translation [89]. | aac(6′)-aph(2″) [88,90] | (i) A single base alteration (G1126A) of aac(6′)/aph(2″) gene resulted in one amino acid substitution S376N in AAC(6′)/APH(2″), which leads to arbekacin resistance in MRSA strain PRC104 [90]. (ii) β-lactam-inducible arbekacin resistance was reported in MRSA strain by the integration of Tn4001-IS257 hybrid structure containing aac(6′)/aph(2″) gene cointegrated into a region downstream of blaZ gene [91].(iii) The AAC(6′)/APH(2″) modify arbekacin by 6′-N-acetylation and/or 2″-O-phosphorylation of AGs that contain 6′-NH2 and/or 2″-OH [37,92]. | Arbekacin is not inactivated by AMEs (3′)(APH), (4′)(AAD), or AAD(2″) and has a weak affinity to (6′-IV) (AAC) [93]. |
| Glycopeptides | Cell wall synthesis | ||||
| Vancomycin | 1958. Bacteremia, infective endocarditis, osteomyelitis, meningitis, pneumonia, sepsis, and complicated SSTI due to HA-MRSA and CA-MRSA [29]. (Resistance VISA in 1996 [28] and VRSA in 2002 [94]) | Vancomycin bind to D-Ala-D-Ala termini moieties of Lipid II precursor of peptidoglycan through a series of hydrogen bonds, leading to conformational alteration that prevents incorporation of NAM- and NAG-peptide subunits to the growing peptidoglycan chain and consequent transpeptidation [95,96,97]. This alters membrane integrity and increases permeability, leading to bacterial death. | vanA [97,98] Mutations in walKR, vraSR, graSR, and clpP Mutation in rpoB [99,100] SNPs in capB (E58K) and lytN (I16V) gene [101] | (i) VRSA: The Tn1546-borne vanA gene cluster encodes 9 proteins (D-Ala:D-Lac ligases) that modify D-Ala-D-Ala termini of peptidoglycan chains to D-Ala-D-Lactate, thereby inhibiting target binding by vancomycin [102,103]. (ii) VISA: Mutations in TCSs like essential WalKR [104,105,106,107], VraSR [108,109,110], and GraSR [107,109,110,111,112] affect cell wall biosynthesis, resulting in reduced susceptibility to vancomycin. (iii) Mutation in rpoB (encoding RNA polymerase subunit B) [99,100]. (iv) Mutation in TCS walKR and proteolytic regulatory gene clpP leads to raised vancomycin resistance in laboratory VISA strain N315LR5P1 [113]. (v) SNPs in capB (E58K) gene (encoding tyrosine kinase) and lytN (I16V) gene (encoding N-acetylmuramyl-L-alanine amidase) cause increased S. aureus resistance to vancomycin in the absence of van genes [101]. | VRSA is mediated by the vanA gene cluster, which is transferred from vancomycin-resistant Enterococcus [114]. |
| Teicoplanin (formerly known as teichomycin A2) | 1988. Approved in Europe for SSTI, pneumonia, and sepsis [115]. Never approved for use in the U.S. (Resistance 2000) [116] | Teicoplanin inhibits peptidoglycan polymerization, leading to the inhibition of bacterial cell-wall synthesis. | tcaRAB [117,118], tcaA [119] | (i) The tcaRAB operon may be involved in increased teicoplanin resistance in S. aureus [118]. (ii) Mutation in tcaRAB may influence the transcription of the cell wall biosynthesis gene and may contribute to increasing teicoplanin resistance [117]. (iii) The tcaA gene within tcaRAB plays a relevant role in teicoplanin resistance in S. aureus clinical isolates [119]. | BSAC recommended breakpoint for teicoplanin are susceptible (MIC ≤ 2 mg/L) and resistant (MIC > 2 mg/L). |
| Oxazolidinones | Protein synthesis | ||||
| Linezolid | U.S. FDA 2000. ABSSSI, pneumonia, BJI, catheter- related bacteremia [120] (Resistance 2001) [121] | Linezolid binds to bacterial 23S rRNA in the 50S ribosome subunit, thereby preventing the formation of functional 70S ribosomal initiation complex with 30S subunit, mRNA, initiation factors, and N-formylmethionyl-tRNA (tRNAfMet) [122]. | cfr [123] Mutations in 23S rRNA [121,124] | (i) Acquisition of cfr gene encoding 23S rRNA methyltransferase [125], which modifies adenosine at position 2503 in 23S rRNA in the large ribosomal subunit [126]. (ii) The T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA [127]. (iii) Mutations G2576T, G2576T, G2447T in domain V of 23S rRNA [121,124] and amino acid changes in ribosomal proteins L3 and L4 [128] lead to conformational changes in the ribosome. | |
| Tedizolid | U.S. FDA 2014; E.U. EMA 2015.ABSSSI and pneumonia | Tedizolid binds to 23S rRNA in the 50S ribosome subunit and prevents the formation of 70S ribosomal initial complex, resulting in inhibition of bacterial protein synthesis [129,130]. | cfr rplC, rplD, rplV, rpoB [131] | (i) Mutations in domain V region of 23S rRNA target confer resistance to tedizolid. (ii) Mutations in ribosomal proteins L3, L4, and L22 (encoded by rplC, rplD, and rplV genes, respectively) and the 23S rRNA target [132]. (iii) Mutation in rpoB corresponding to amino acid substitution D449N [131]. | Mutation in L3, L4, and L22 also mediate PhLOPSa (phenicol, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A) resistance. |
| Contezolid | NMPA of China 2021 [133]. Complicated SSTI, ABSSSI (Resistance 2021) [134]. | Contezolid binds to the 23S rRNA region adjacent to the peptidyl transferase center of the 50S ribosomal subunit and prevents the formation of a functional 70S initiation complex, thereby interfering with bacterial protein synthesis. | cfr, optrA | Contezolid exhibited limited activity against strains with linezolid resistance genes cfr and optrA [134]. | Contezolid has reduced hematologic toxicity compared to linezolid |
| Lipopeptides | Cell wall synthesis Cell membrane | ||||
| Daptomycin | U.S. FDA 2003. Bacteremia, ABSSSI (Nonsusceptible 2004) [135] | Daptomycin complexes with Ca2+ to form oligomers that insert into bacterial membranes, resulting in depolarization, permeabilization, leakage of ions, and ultimately bacterial death [136]. Daptomycin disrupts the localization of cell wall synthesis enzymes such as MurG, further interfering with cell wall synthesis [137,138]. | mprF, dltA [139,140], yycH, yycI [141], rpoB [99], walKR, vraSR, graSR [142,143] | (i) Alteration of the surface charge of cells due to mutation in mprF gene (encoding phosphatidylglycerol lysyltransferase) which leads to lysinylation of PG and translocation of lysyl-PG [144]. (ii) Mutation in TCSs walKR, vraSR, and graSR which are involved in cell wall synthesis and permeability are associated with daptomycin susceptibility in S. aureus [142,143]. (iii) Mutation in rpoB gene (encoding RNA polymerase) confers dual heteroresistance to daptomycin and vancomycin [99]. (iv) Mutations in yycH and yycI genes lead to the loss of protein functions essential for cell wall synthesis [141]. (v) dltA gene overexpression leads to electrostatic repulsion and indirectly reduces autolysin, resulting in daptomycin nonsusceptibility [139,140]. | S. aureus strains with MIC ≤ 1 μg/mL are referred as daptomycin-susceptible (DAP-S) [145] and MIC >1 μg/mL as daptomycin-non susceptible [146]. |
| Lipoglycopeptides | Cell wall synthesis | ||||
| Telavancin (derivative of vancomycin. Addition of the hydrophobic side chain and hydrophilic group results in enhanced activity [147]. | U.S. FDA 2009 and 2013 [148]. Complicated SSTI, pneumonia, BJI, ABSSSI, bacteremia [149]. | Telavancin inhibits cell wall biosynthesis by binding to late-stage peptidoglycan synthesis, like vancomycin. Additionally, it depolarizes the bacterial cell membrane and disrupts its functional integrity [150]. | - | The vanA-mediated telavancin resistance is rare in MRSA [151]. | |
| Tetracyclines | Protein synthesis | ||||
| Tetracycline | 1948 [152] SSTI (Resistance 1953) [44] | Tetracycline binds to bacterial 30S ribosomal subunit and prevents the aminoacyl tRNA from binding to A site of the rRNA, resulting in inhibition of translation. To some extent, it also binds to the bacterial 50S ribosomal subunit [44,153,154]. | tetM, tetO, tetK [155], tetS/M, tetA | (i) Ribosomal protection: the tetM and tetO genes encode enzymes that destabilize the interaction between tetracyclines and their cellular target ribosome [44,45]. (ii) Active efflux: the tetK gene encodes efflux protein that couples the tetracycline with proton motive force to pump it out from the cell against the concentration gradient [44,155]. | The tetK gene is normally found in S. aureus. |
| Doxycycline | U.S. FDA 1967 [156,157]. UTI, SSTI [27] | Doxycycline inhibits bacterial protein synthesis by preventing the association of aminoacyl tRNA with the ribosome, an MoA similar to tetracycline. | tetK [158,159] | Active efflux by tetK encoded efflux [158,159]. | |
| Tigecycline | U.S. FDA 2005.ABSSSI, pneumonia | Tigecycline inhibits protein synthesis, an MoA similar to tetracycline but with enhanced binding. | tetM, tetO, tetX | The oxygen-dependent destruction of tigecycline is catalyzed by the enzyme TetX [160,161,162]. | Tigecycline retains activity against both tetM and tetO. |
| Omadacycline (derived from tetracycline) [163] | U.S. FDA 2018.ABSSSI, SSTI [164], pneumonia (CA-associated) | Omadacycline binds to bacterial 30S ribosomal subunit and inhibits protein synthesis, an MoA similar to tetracycline with enhanced binding like tigecycline [165]. | - | Resistance mechanism not reported. | Unaffected by the presence of tetK active efflux gene and ribosomal protection tetM or tetO gene [166,167]. |
| Fusidane | Protein synthesis | ||||
| Fusidic acid | 1962. ABSSSI | Fusidic acid binds to elongation factor G (EF-G) on the ribosome, thereby preventing the release of EF-G-guanosine diphosphate complex and delaying bacterial protein synthesis by inhibiting the next stage in translation [168,169]. | fusA [170], fusB [171,172], fusc, fusD | (i) Mutations in chromosomal fusA (encoding ribosomal translocase and translation elongation factor EF-G) [170] or fusE genes confer high-level resistance to fusidic acid. (ii) Mutation in acquired genes fusB (encoding an inducible protein that protects an in vitro translation) [171,172] and fusD genes mediate low-level resistance. These mutations affect the elongation factor EF-6. | The fusc and fusD are homologs of fusB [173]. |
| Pleuromutilin | Protein synthesis | ||||
| Retapamulin | U.S. FDA 2007. Impetigo [174] | Retapamulin binds to domain V of 23S rRNA on the 50S ribosome subunit, thereby blocking peptide formation directly by interfering with substrate binding. | 23S rRNA | Resistance to retapamulin occurs due to mutations in the genes encoding 23S rRNA methyltransferase. | Retapamulin is a semisynthetic derivative of pleuromutilin |
| Fluoroquinolones | DNA replication | [46,175] | |||
| Ciprofloxacin (2nd-generation fluoroquinolone) | U.S. FDA 1987.UTI | Ciprofloxacin target bacterial DNA topoisomerase IV and DNA gyrase, thus preventing it from supercoiling the bacterial DNA [176], which leads to inhibition of DNA replication [177,178]. | gyrA [33], grlA [33], flqA (formerly ofx/cfx) [35], norA [58,179] | (i) Mutation in the genes grlA (encoding DNA topoisomerase IV subunit A) [33,34,35,46], gyrA (encoding DNA gyrase subunit A) [33,34,35], and flqA (linked to DNA topoisomerase IV) [35]. (ii) Mutations in the gene norA (encoding a membrane-associated active efflux pump NorA) [58,180]. | Elevated norA expression potentiates evolution by increasing the fitness benefit provided by a mutation in DNA topoisomerase [179]. |
| Levofloxacin | U.S. FDA 1996. RTI, UTI, SSTI | Levofloxacin inhibits bacterial DNA replication, an MoA similar to ciprofloxacin. | gyrA, grlA | (i) Mutation in the genes grlA and gyrA [181]. (ii) Mutations in the gene norA [180]. | |
| Delafloxacin (previously referred to as ABT-492) [182] | U.S. FDA 2017 [183]; E.U. EMA 2019. SSTI, ABSSSI (Resistance 2017) [184] | Delafloxacin inhibits bacterial DNA replication by blocking both DNA topoisomerase IV and DNA gyrase, an MoA similar to ciprofloxacin [182]. | grlA | Point mutations in the grlA [185,186]. | Delafloxacin is not active substrate for S. aureus efflux pumps [185]. |
| Quinolones | DNA replication | ||||
| Ozenoxacin (topical quinolone without fluorine at C6-position) | U.S. FDA 2017.Japanese PMDA 2016 [187]. SSTI (impetigo) caused by MRSA | Ozenoxacin inhibits bacterial DNA replication by dual-targeting activity against DNA topoisomerase IV and DNA gyrase [35]. | grlA, grlB | Mutations in QRDR regions of grlA and gyrA are the primary cause of decreased susceptibility to ozenoxacin [35]. | Low MIC of ozenoxacin was observed for MSSA and MRSA strains with reduced susceptibility to nadifloxacin [187]. |
| Pyrimidine/ Sulfonamide | Folate synthesis (DNA synthesis and protein synthesis) | ||||
| Trimethoprim–Sulfamethoxazole (TMP-SMX) | UTI, SSTI, and BJI due to CA-MRSA [29] | TMP binds and inhibits the dihydrofolate reductase, thereby preventing the conversion of dihydrofolic acid (DHF) to tetrahydrofolic acid (THF) [188]. THF is an essential precursor of the thymidine synthesis pathway and interference with this pathway results in inhibition of bacterial DNA synthesis. SMX inhibits bacterial dihydropteroate synthase, an enzyme involved upstream in the thymidine synthesis pathway, resulting in the inhibition of folic acid biosynthesis [188]. | dfrA, dfrB [189], dfrD [189], dfrG [190], dfrK, dfrS1 [191,192] | (i) The acquisition of dfrA gene (encoding DHFR) and mutation of the chromosomal dfrB gene (encoding SaDHFR) are considered key determinants of TMP-SMX resistance [189,193,194,195]. (ii) Point mutation in the dfrB gene resulted in a single amino acid substitution Phe98Tyr of SaDHFR, which was associated with TMP-SMX resistance in S. aureus [189]. (iii) Transposon-located dfrA gene mediates TMP resistance [194,196]. (iv) The dfrG gene (encoding DHFR) mainly mediates the TMP resistance in S. aureus clinical isolates [190,195]. | |
| Other classes | |||||
| Mupirocin (previously pseudomonic acid) | Discovered in 1971 [197] while marketed for clinical use in the UK in 1985 and US in 1988 [198]. SSTI, nasal carriage of S. aureus (Resistance 1987) [199,200]. | Mupirocin binds to bacterial isoleucyl transfer RNA (tRNA) synthetase, leading to depletion of isoleucyl–tRNA and accumulation of the corresponding uncharged tRNA. This results in the inhibition of protein and RNA synthesis [201]. | ileS [202,203,204], mupA [205,206], and mupB [207] | (i) Mutations in the chromosomal ileS gene (encoding native isoleucyl t-RNA synthetase) result in V588F or V631F alterations [202,203,204], which lead to low-level mupirocin resistance [205]. (ii) Acquisition of the plasmid-encoded mupA gene (encoding eukaryotic-like isoleucyl–tRNA synthetase variant) [208] confers high-level resistance to mupirocin [205,206]. (iii) Acquisition of the plasmid-encoded mupB gene (encoding eukaryotic-like isoleucyl–tRNA synthetase variant) confers high-level resistance to mupirocin [207]. | Low-level mupirocin resistance (MIC 8–256 μg/mL) and high-level resistance (MIC ≥ 512 μg/mL) [209]. |
| Fosfomycin | Discovered in 1969 [210]. UTI | Fosfomycin deactivates the enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and catalyzes the addition of phosphoenolpyruvate to UDP-N-acetylglucosamine (UDP-GlcNAc) to form UDP-N-acetylmuramic acid (UDP-MurNAc), thereby inhibiting bacterial cell-wall synthesis [211]. | fosB [54], glpT and uhpT [212,213,214], murA [213,215] tet38 [216], fosY [217] | (i) Thiol-S-transferase (encoded by fosB gene) catalyzes the inactivation of fosfomycin [53,54]. (ii) Mutations in fosfomycin uptake transporter proteins GlpT (Trp137/Arg) (encoded by glpT gene) [213] and UhpT (encoded by uhpT genes) [214] reduce the permeability and subsequently prevent fosfomycin from invading the bacterium [212,213]. (iii) Mutation in target enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (encoded by murA gene) reduces its affinity for fosfomycin [215]. (iv) The major facilitator superfamily efflux transporter Tet38 (encoded by tet38 gene) contributes to fosfomycin resistance [216]. (v) FosY protein, a putative bacillithiol transferase enzyme (encoded by fosY gene) confers resistance to fosfomycin in CC1 S. aureus [217]. | |
| Rifampin | Discovered in 1965, introduced for therapy in Italy in 1968, and approved in the United States in 1971 [218]. Endocarditis; BJI [27]. | Rifampin inhibits transcription (RNA synthesis) by binding to the β-subunit of the bacterial DNA-dependent RNA polymerase [219,220]. | rpoB [43,221] | (i) Mutations in the RRDR region of rpoB gene (encoding RNA polymerase) resulted in amino acid substitutions of Gln468/Arg, His481/Tyr, and Arg484/His and are associated with high-level resistance to rifampicin [43]. (ii) Mutation in the rpoB (N967I) gene causes the substitution Asn967/Ile in the β-subunit of RNA polymerase [221]. | CLSI breakpoint of rifampicin susceptibility is ≤1 μg/mL [146]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lade, H.; Joo, H.-S.; Kim, J.-S. Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus. Antibiotics 2022, 11, 1378. https://doi.org/10.3390/antibiotics11101378
Lade H, Joo H-S, Kim J-S. Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus. Antibiotics. 2022; 11(10):1378. https://doi.org/10.3390/antibiotics11101378
Chicago/Turabian StyleLade, Harshad, Hwang-Soo Joo, and Jae-Seok Kim. 2022. "Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus" Antibiotics 11, no. 10: 1378. https://doi.org/10.3390/antibiotics11101378
APA StyleLade, H., Joo, H.-S., & Kim, J.-S. (2022). Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus. Antibiotics, 11(10), 1378. https://doi.org/10.3390/antibiotics11101378







