Abstract
The purpose of this study was the identification of genetic lineages and antimicrobial resistance (AMR) and virulence genes in Klebsiella pneumoniae isolates associated with severe infections in the neuro-ICU. Susceptibility to antimicrobials was determined using the Vitek-2 instrument. AMR and virulence genes, sequence types (STs), and capsular types were identified by PCR. Whole-genome sequencing was conducted on the Illumina MiSeq platform. It was shown that K. pneumoniae isolates of ST14K2, ST23K57, ST39K23, ST76K23, ST86K2, ST218K57, ST219KL125/114, ST268K20, and ST2674K47 caused severe systemic infections, including ST14K2, ST39K23, and ST268K20 that were associated with fatal incomes. Moreover, eight isolates of ST395K2 and ST307KL102/149/155 were associated with manifestations of vasculitis and microcirculation disorders. Another 12 K. pneumoniae isolates of ST395K2,KL39, ST307KL102/149/155, and ST147K14/64 were collected from patients without severe systemic infections. Major isolates (n = 38) were XDR and MDR. Beta-lactamase genes were identified: blaSHV (n = 41), blaCTX-M (n = 28), blaTEM (n = 21), blaOXA-48 (n = 21), blaNDM (n = 1), and blaKPC (n = 1). The prevalent virulence genes were wabG (n = 41), fimH (n = 41), allS (n = 41), and uge (n = 34), and rarer, detected only in the genomes of the isolates causing severe systemic infections—rmpA (n = 8), kfu (n = 6), iroN (n = 5), and iroD (n = 5) indicating high potential of the isolates for hypervirulence.
1. Introduction
Healthcare-associated infections (HAI) have posed a huge medical burden to public health worldwide. Klebsiella pneumoniae is one of the clinically significant nosocomial pathogens causing broad spectra of diseases and showing increasingly frequent acquisition of resistance to antibiotics including in intensive care units (ICU) [1]. Today, according to the BIGSDB Institute Pasteur database (https://bigsdb.pasteur.fr/ access date 8 August 2021), 5797 K. pneumoniae sequence types and 711 capsular types have been discovered. K. pneumoniae causes different infections in ICU patients associated with high mortality rates, including bloodstream infection, pulmonary infection, and healthcare-associated ventriculitis, meningitis, and urinary tract infection [2,3]. It was reported from China, Spain, and Taiwan that K. pneumoniae was associated with leukocytoclastic vasculitis and microcirculation disorders, but there is no data on the relationship of such a clinical manifestation with any K. pneumoniae genetic lineages [4,5,6].
Current data suggest that genetic determinants associated with antimicrobial resistance (AMR) and virulence are specific for distinct genetic lineages of K. pneumoniae [7]. Classical K. pneumoniae (cKP) are the common healthcare-associated pathogens causing nosocomial infections in immunocompromised patients and characterized as a great possibility to accumulate resistance mechanisms to antimicrobials—multidrug-resistant (MDR), extensive drug-resistant (XDR), and pan drug-resistant (PDR) strains have been described [8]. Carbapenems are the last antibiotics available to control K. pneumoniae infection. However, to date, many mechanisms involved in carbapenem resistance in K. pneumoniae have already been described, including carbapenemase production. Strains producing carbapenemases of functional class A (KPC), class B (NDM), and class D (OXA-48), and co-producing more than one type of carbapenemase are increasingly reported [9].
For hypervirulent K. pneumoniae (hvKP), many critical virulence factors have been discovered, among them rmpA (regulator of mucoid phenotype), aerobactin (an iron siderophore), kfu (an iron uptake system), allS (associated with allantoin metabolism), and K1/K2 capsules [10]. The global spread of ‘convergent’ K. pneumoniae strains that combines the pathogenic potentials of cKP and hvKP has occurred since 2010. Three types of the mechanism for the emergence of ‘convergent’ clones have been identified: (i) cKP acquiring a hypervirulence plasmid; (ii) hvKP acquiring a carbapenem-resistant plasmid; and (iii) K. pneumoniae acquiring both a carbapenem resistance and hypervirulence hybrid plasmid. These strains are at high risk to disseminate and have the potential to cause severe infections [9].
This study aimed to examine the phenotypes, genotypes, and genetic relatedness of K. pneumoniae isolates collected from the patients of neuro-ICU and evaluated the sequence types associated with severe infections between 2017 and 2019.
2. Results
2.1. Patients and Bacterial Strains
During the period from Oct. 2017 to Jan. 2019, the following incidence of infections was detected in neurosurgery ICU: 8.4 infections of the central nervous system per 100 patients, 2.7/100 of bloodstream infections, 26.3/100 of ventilator-associated pneumonia, and 23.6/100 of urinary tract infections. K. pneumoniae accounted for 33, 31, 23, and 25% among the agents of named infections, correspondingly [3]. The subject of this study was 41 resistant-to-antimicrobials K. pneumoniae isolates collected from 20 patients with severe postoperative infections (Table 1).

Table 1.
Clinical data of the patients: underlying disease, infections, antimicrobial treatment, and outcomes.
Depending on clinical manifestations, isolates were combined into two groups. Group A included 29 isolates collected from 13 patients (D, J, Q, P, M, S, L, K, N, R, A, I, and F) with the pronounced systemic inflammatory response (SIRS). Symptoms had been reported for the patients: fever >38 °C, leukocytosis, increased markers of systemic inflammation, multiple hemorrhages, thrombosis, or high risk of thromboembolism. Clinical forms had been detected: meningitis after the neurosurgical intervention, pansinusitis, pneumoniae, sepsis and septic shock, ventriculitis, and brain abscess. In this group, seven isolates (B-548/18, B-784/18, B-1154/18, B-1618/18, B-2625/18, B-14/19, and B-21/19) were associated with fatal incomes collected from the patients D, Q, and S died due to increased symptoms of inflammation, sepsis, septic shock, meningoencephalitis associated with K. pneumoniae infection. Other eight isolates (B-3002K/17, B-3060K/17, B-2016K/17, B-3299/17, B-1040/18-1, B-792/18, B-853/18-1, and B-1214/18-2) of Group A were associated with manifestations of vasculitis and microcirculation disorders (patients A, I, and F). Major isolates of Group A (17/29) were collected from the blood and cerebrospinal fluid, less (7/29)—from the endotracheal aspirate.
Group B consists of 12 strains isolated from the patients without increasing markers of systemic inflammation and septic reaction. Major isolates of Group B were from the endotracheal aspirate and urine (10/12) and the rest from the nervous system. One patient in this group died due to the underlying disease, videlicet multiple metastases of kidney cancer in the brain (Table 1 and Table 2). It should be noted that isolates of Group A were obtained from 2–3 body sites of one patient: three isolates—from blood, urine, and cerebrospinal fluid of the patient D; three isolates—from endotracheal aspirate and cerebrospinal fluid of patient S; and three isolates—from blood and cerebrospinal fluid of the patient L (Table 2).

Table 2.
Sources of isolation and antimicrobial resistance phenotypes of K. pneumoniae strains.
2.2. Susceptibility to Antimicrobials
According to Magiorakos et al. [11] criteria, 28 isolates were attributed to the XDR category (resistant to 6–7 functional groups of antimicrobials), 10 isolates to the MDR category (resistant to 3–4 functional groups), and 3 isolates to the R category (resistant to ampicillin only). XDR isolates were attributed to Groups A (n = 18) and B (n = 10); MDR isolates—to the Groups A (n = 8) and B (n = 2); R isolates—only to Group A (n = 3). All isolates were resistant to beta-lactams, 34 isolates—to nitrofurans, 33 isolates—to fluoroquinolones, 29 isolates—to chloramphenicol, 29 isolates—to sulfonamides, 27 isolates—to aminoglycosides, 26 isolates—to tetracyclines. Major isolates were susceptible to amikacin (n = 33) and imipenem (n = 27), all isolates were susceptible to colistin (Figure 1, Table S1).
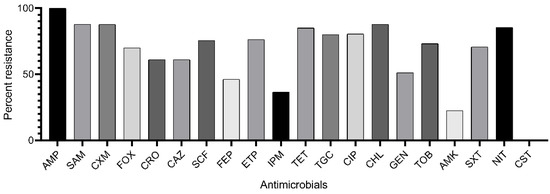
Figure 1.
Rate of K. pneumoniae isolates resistant to antimicrobials: AMP, ampicillin; SAM, ampicillin-sulbactam; CXM, cefuroxime; FOX, cefoxitin; CRO, ceftriaxone; CAZ, ceftazidime; SCF, cefoperazone-sulbactam; FEP, cefepime; ETP, ertapenem; IPM, imipenem; TET, tetracycline; TGC, tigecycline; CIP, ciprofloxacin; CHL, chloramphenicol; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; SXT, trimethoprim-sulfamethoxazole; NIT, nitrofurantoin; CST, colistin.
2.3. Beta-Lactamase Genes and Integrons
Total 113 beta-lactamase genes were identified in K. pneumoniae isolates, including blaSHV (n = 41), blaCTX-M (n = 28), blaTEM (n = 21), blaOXA-48 (n = 21), blaNDM (n = 1), and blaKPC (n = 1). The blaVIM and blaIMP genes were not detected. Number of beta-lactamase genes per strain varied from one to five. Single blaSHV gene was detected in 4 strains; two genes (blaSHV + blaCTX-M) or (blaSHV + blaOXA-48)—in 4 and 9 strains, respectively; three genes (blaSHV + blaCTX-M + blaOXA-48) or (blaSHV + blaCTX-M + blaTEM)—in 3 and 11 strains, respectively; four genes (blaSHV + blaCTX-M + blaTEM + blaNDM) or (blaSHV + blaCTX-M + blaTEM + blaOXA-48)—in 1 and 8 strains, respectively; five genes (blaSHV + blaCTX-M + blaTEM + blaOXA-48 + blaKPC) were identified in one strain. Moreover, 10 strains carried class 1 integrons, and one strain carried two integrons, class 1 and class 2 simultaneously (Table 3).

Table 3.
Molecular-genetic characteristics of K. pneumoniae strains.
2.4. Virulence Genes
The prevalent virulence genes detected in 41 K. pneumoniae clinical isolates were wabG (n = 41), fimH (n = 41), allS (n = 41), and uge (n = 34). Other virulence genes were rare and attributed only to the Group A: rmpA (n = 8), kfu (n = 6), iroN (n = 5), and iroD (n = 5). Eight combinations of 3 to 8 virulence genes were identified. Combination of three genes (wabG + fimH + allS) was detected in three isolates; combinations of four genes (uge + wabG + fimH + allS) and (rmpA + wabG + fimH + allS)—in 24 and 2 isolates, respectively; combinations of five genes (uge + wabG + kfu + fimH + allS) and (rmpA + uge + wabG + fimH + allS)—in 6 and 1 isolates, respectively; combinations of six genes (rmpA + iroN + iroD + wabG + fimH + allS)—in 2 isolates; combinations of seven genes (rmpA + iroN + iroD + uge + wabG + fimH + allS)—in 3 isolates (Table 3).
2.5. Sequence Types and Capsular Types
It was found that K. pneumoniae clinical isolates belonged to 12 sequence types: ST14 (n = 3), ST23 (n = 5), ST39 (n = 3), ST76 (n = 1), ST86 (n = 2), ST147 (n = 1), ST218 (n = 2), ST219 (n = 3), ST268 (n = 1), ST307 (n = 3), ST395 (n = 16) and ST2674 (n = 1). Nine sequence types (ST14, ST23, ST39, ST76, ST86, ST218, ST219, ST268, and ST2674) were identified only in Group A. One sequence type (ST147) was obtained only in Group B. Two sequence types (ST395 and ST307) were identified both in Group A (8 isolates associated with systemic manifestations of vasculitis and microcirculation disorders) and in Group B.
All studied K. pneumoniae isolates were attributed to nine capsular types: K2 (n = 20), K20 (n = 1), K23 (n = 4), KL39 (n = 1), K47 (n = 1), K57 (n = 7), K14/64 (n = 1), K102/149/155 (n = 3), and K125/114 (n = 3). It was interesting in that K. pneumoniae isolates of capsular type K2 belonged to three sequence types (ST14, ST86, and ST395), isolates of capsular type K23—to two sequence types (ST39 and ST76), and isolates of capsular type K57—to two sequence types (ST23 and ST218). The remaining capsular types were associated with only one sequence type: K20—ST268, KL39—ST395, K47—ST2674, K14/64—ST147, K102/149/155—ST307, and K125/114—ST219 (Table 3).
2.6. Phylogenetic Analysis
The phylogenetic tree was constructed on the base of combined gene sequences of MLST profiles; two clusters were revealed. Cluster I consisted of one ST147 referring to one isolate not associated with severe manifestations of systemic infections (Group B). Cluster II included two sub-clusters: IIa consisting of one ST86 associated with a pronounced systemic inflammatory response (Group A); IIb consisting of 10 genetic lineages including 8 sequence types (ST14, ST23, ST39, ST76, ST218, ST219, ST268, and ST2674) referred to the strains of Group A, and 2 sequence types (ST307 and ST396) including isolates both Group A and Group B (Figure 2).
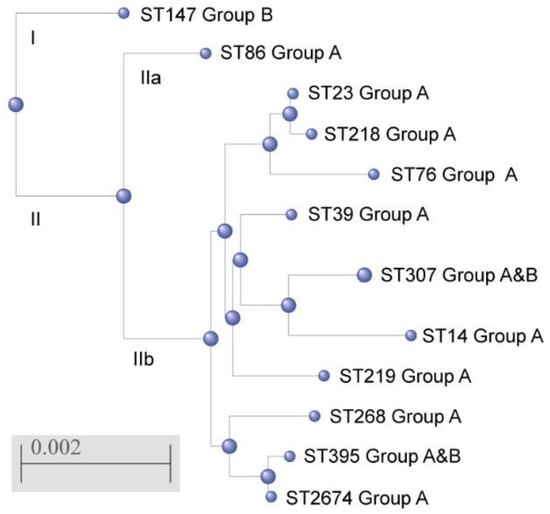
Figure 2.
Phylogenetic tree of K. pneumoniae sequence types identified in the study generated by using a web resource NCBI “Blastn” and “Blast Tree View”, based on combined gene sequences of MLST profiles.
2.7. Whole-Genome Sequencing
Whole-genome sequencing was done for nine isolates including eight isolates of Group A belonging to sequence types/capsular types ST2674/K47, ST23/K57, ST39/K23, ST219/K125, ST218/K57, ST76/K23, ST86/K2, and ST307/K102 and one isolate of Group B belonging to ST395/K39. From 86 to 164 contigs for each genome were obtained, the genome size ranged from 5.42 to 5.86 Mb, with the GC content ranging from 55.1 to 57.9%, and the number of genes from 5121 to 5845. All genomes carried 1–6 beta-lactamase genes including blaSHV, blaTEM, blaCTX-M, blaOXA, and blaKPC types. Six alleles were identified of the blaSHV gene (26, 28, 33, 40, 59, and 182), two alleles of blaOXA genes (1 and 48), and other beta-lactamase genes were presented as only one allele (blaTEM-1B, blaCTX-M-15, and blaKPC-2). The genetic environments of carbapenemase genes blaKPC-2 and blaOXA-48 and cefalosporinase gene blaCTX-M-15 were similar to previous descriptions of these genes. The gene coding of a putative transposition helper protein ISKpn7 was identified upstream blaKPC-2 gene, and ISKpn6 transposase gene downstream, that was similar to the genetic structure in the plasmid pBK32179 (JX430448). The environment for blaOXA-48 genes contained upstream, the transcriptional regulator LysR gene, and the transposase tir gene downstream, as in the plasmid pOXA-48 (JN626286). The surrounding genetic conditions for blaCTX-M-15 genes in clinical isolates were likewise similar to the plasmid pKp848CTX (NC_024992): ISEcp1 family transposase upstream and gene coding WbuC family cupin fold metalloprotein downstream. Aminoglycoside modifying enzyme genes were identified in all but one isolate: 1–3 genes per isolate (aac, aad, ant, aph, arm, and rmt). All genomes carried the fosA gene conferring resistance to fosfomycin. Phenicol resistance genes (4 alleles of cat gene) were detected in seven genomes. Six isolates carried 2–4 quinolone resistance genes (aac, qnr, oqx, and qep). Two alleles of sul gene (sulfonamide resistance) were identified in six genomes, tet gene (tetracycline resistance) in 5 genomes, four alleles of dfrA gene (trimethoprim resistance) in 4 genomes, and mph gene (macrolide resistance) in only one genome. The genes encoding efflux pumps were revealed in all isolates, seven of them carried 10 genes, two isolates—9 genes. Moreover, all genomes carried 1–4 genes encoding heavy metal resistance (to copper, lead, silver, arsenic, and tellurium) (Table 4).

Table 4.
Molecular-genetic characteristics of K. pneumoniae isolates based on Whole-Genome Sequencing.
Analysis of virulence genetic determinants revealed mrk gene coding type 3 adhesine—in all 9 genomes, irp gene of yersiniabactin biosynthesis and ybt gene of yersiniabactin transcriptional regulator—in 7 genomes, fyu gene of siderophore yersiniabactin receptor—in 6 genomes, iut gene of ferric aerobactin receptor—in 6 genomes, iuc gene of aerobactin siderophore synthesis—in 5 genomes, kvg gene of capsular polysaccharide synthesis regulator—in 2 genomes. Generally, each genome contained 1–7 virulence genes. Plasmids of eight incompatibility groups (IncC, IncFIA, IncFIB, IncFII, IncHI1B, IncM1, IncM2, and IncR) were identified in the K. pneumoniae isolate genomes, specifically 1–3 plasmids per genome. Molecular systems protecting bacteria from the foreign DNA, Type I Restriction-Modification systems, were detected in four genomes, Type II systems in all nine genomes, but CRISPR-Cas systems were not detected in the genomes (Table 4).
3. Discussion
K. pneumoniae isolates that caused severe postoperative infections in patients of neuro-ICU between 2017 and 2019 were divided into two groups, A and B, depending on observed clinical manifestations: associated and not associated with the pronounced systemic inflammatory response. Previously, the association of K. pneumoniae with severe systemic infections was described as the modern trend for ICU; for example, K. pneumoniae were the most frequent infecting species (47%) determined meningitis/encephalitis and 30-day mortality rates, 15% in post-neurosurgical patients [12]. In our study, as in the reports from other countries, the overwhelming majority of K. pneumoniae isolates obtained from neurosurgery patients were MDR and XDR [13]. Moreover, MDR and XDR K. pneumoniae isolates in our study were associated with severe systemic infections, including vasculitis and microcirculation disorders. It was reported previously that K. pneumoniae was associated with leukocytoclastic vasculitis [4,5,6], rapidly progressive retinal vasculitis [14], and acute vasculitis at respiratory infection [15]. In this study, we first identified K. pneumoniae of ST395 and ST307 as bacterial pathogen associated with vasculitis and microcirculation disorders.
A total of twelve sequence types and nine capsular types of K. pneumoniae were identified in this study, which possibly reflects a continuous influx of new genetic lineages into neuro-ICU from other hospitals and regions. The prevalent ST in the ICU was ST395 (16/41 isolates), which is similarly rated to that previously published in the studies from Poland, France, Italy, and Russia [16,17,18,19,20]. Nine STs in our study (ST14, ST23, ST39, ST76, ST86, ST218, ST219, ST268, and ST2674) were identified only for the isolates that caused severe bloodstream infections. Previously K. pneumoniae of ST23, ST86, ST76, and ST218 were described as a hypervirulent pathogen causing bacteremia, sepsis, and liver abscess in India, France, China, Taiwan, and Russia [21,22,23,24,25]. Three STs (ST14, ST39, and ST268) in our study were associated with fatal outcomes. These STs have been reported previously as the agents of severe bloodstream infections in ICU and surgery wards in other countries [26,27]. Two sequence types, ST219, and ST2674 were first identified in this study as the agent of severe sepsis in the patients of ICU. Recently, the ST219 was reported for the environmental MDR K. pneumoniae strains collected from hospital wastewater in Southern Romania [28]; the ST2674 was identified for the environmental isolate from Pakistan (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?page=info&db=pubmlst_klebsiella_isolates&id=5256 access date 8 August 2021).
The high prevalence of polyresistant isolates in our study was associated with antimicrobial resistance genes. The isolates of Group A carried ESBL genes blaCTX-M (22/29), carbapenemase genes blaOXA-48 (12/29), and blaKPC-2 (1/29), which correspond to the reports from Greece and China [27,29,30]. Moreover, four isolates in Group A carrying the only blaSHV were identified (ST218 and ST86). These isolates, as well as four additional isolates of ST268, ST23, and ST76, carried virulence gene rmpA coding regulator of hypermucoid phenotype. It was reported previously that overexpression of rmpA could enhance the virulence of K. pneumoniae isolates in the mouse model [31]. Virulence genes iroN and iroD associated with utilization of trivalent iron were detected in K. pneumoniae of ST23, ST218, ST76, ST86, and ST268 (isolates associated with severe bloodstream infections) which agrees with reports from other countries [22,23,29]. It is known that the majority of pathogenic bacteria including K. pneumoniae possess the iron-acquisition system with a higher affinity for iron than the host, which serves as one of the strategies for increasing bacterial virulence [32]. The kfu gene coding iron uptake system was identified in K. pneumoniae of ST14 and ST219; the latter is a novel genetic lineage carrying the kfu gene [33,34]. Kfu was shown to be a potential virulence factor in the intragastrical murine model, which indicates that kfu might contribute to intestinal colonization [35]. Therefore, the identified virulence genes indicate the high potential of studied isolates for hypervirulence.
The isolates that were not associated with severe manifestations of systemic infections in our study were attributed to ST395, ST307, and ST147. Among them, the isolates of ST395 carrying the blaOXA-48 carbapenemase gene were prevalent. Such K. pneumoniae strains were reported earlier from Hungary and Russia [36,37]. In our study, one isolate of ST147 carried the blaNDM-1 carbapenemase gene, and the same strains have been described worldwide [38,39]. It should be noted that ST307 and ST147 have been estimated as K. pneumoniae High-Risk Clones (HRC) because of worldwide distribution, ability to cause serious infections, and association with polyresistance [40].
The whole-genome study of selected K. pneumoniae isolates belonged to nine sequence types, ST23, ST39, ST76, ST86, ST218, ST219, ST307, ST395, and ST2674, showing the great diversity of these isolates in the combination of virulence genes, antimicrobial resistance genes, heavy metal resistance, and plasmids. Analysis of their resistomes showed that the genes of beta-lactamases blaCTX-M-15, blaTEM-1B, blaNDM-1, blaOXA-48, blaKPC-2, and blaOXA-1 are represented by a single allele. These alleles were recently reported for K. pneumoniae isolated in Russia [41]. On the contrary, six alleles were identified of blaSHV-type genes, which were not reported in Russia before our study: blaSHV-26, blaSHV-28, blaSHV-33, blaSHV-40, blaSHV-59, and blaSHV-182. In general, all isolates in our work carried blaSHV genes, 68% isolates—blaCTX-M genes, 51%—blaTEM genes, 51%—blaOXA-48 genes, but only one isolate carried blaNDM-1 gene, and only one isolate blaKPC-2 gene. A similar representation of beta-lactamase genes was reported from the European countries and Russia [41,42]. Major isolates in our study were susceptible to amikacin and imipenem, which is consistent with recently published data from Saudi Arabia and Indonesia [43,44]. Interestingly, the rmtB gene encoding 16S rRNA methylase providing resistance to aminoglycosides in the K. pneumoniae ST23 isolate was detected in this study for the first time. This gene was reported earlier for ST258 and ST16 of KPC-producing K. pneumoniae [45]. Moreover, armA gene coding 16S rRNA methyltransferase was detected in K. pneumoniae of ST395 in this study; recently, this gene was detected in K. pneumoniae ST23, ST2502, and ST11 in Italy, Spain, and China, respectively [46,47].
The virulence determinants detected in the genomes were the following: mannose-resistant Klebsiella-like (mrk) hemagglutinin gene critical for K pneumoniae biofilm development in all 9 genomes; aerobactin siderophore locus (iuc, iut) in 6 genomes; regulator of capsular polysaccharide synthesis (kvg) in 2 isolates. A similar distribution of virulence genes was reported recently from Brazil [48]. The yersiniabactin locus (irp, ybt, and fyu) was detected in 7/9 genomes in our study, compared with 40% of K. pneumoniae genomes, particularly amongst those associated with invasive infections [49]. This data confirmed the high virulence potential of the studied clinical isolates. The presence in the genomes of the plasmids of different incompatibility groups indicates a possible role of ‘hybrid’ plasmids in the formation of K. pneumoniae strains, simultaneously carrying a large number of antibiotic resistance and virulence genes. [9,50].
Future research will focus on studying the structure of plasmids carrying genes for antimicrobial resistance and virulence, as well as the expression of these genes under conditions of different genetic environments and selective pressure of antibiotics. We believe that further study of the microbiology, molecular biology, physiology, and interactions with the host of K. pneumoniae will provide important knowledge to control K. pneumoniae infection in ICUs.
4. Materials and Methods
4.1. Bioethical Requirements and Patients
Each patient signed informed voluntary consent to treatment and laboratory examination, under the requirements of the Russian Federation Bioethical Committee. The study did not contain personal data of patients; the clinical bacterial isolates information did not include name, date of birth, address, and disease history. The study was a retrospective observational study. The study was done in the neuro-ICU department in a specialized Neurosurgical Hospital in Moscow, Russia, with 300 beds that care for approximately 8000 patients per year, 95% of whom undergo surgery. The surveillance software was designed in-house and integrated into the hospital’s electronic health record system [51]. Four types of health-associated infections were surveilled: bloodstream, respiratory and urinary tract infections, and healthcare-associated ventriculitis and meningitis [3].
4.2. Bacterial Isolates and Growing
K. pneumoniae isolates (n = 41) were collected from clinical samples (endotracheal aspirate, cerebrospinal fluid, blood, wound exudate, and urine) of the patients (n = 20) in the Neuro-ICU in the period from October 2017 to January 2019. Bacterial identification was performed using a Vitek-2 Compact (BioMerieux, Paris, France) and a MALDI-TOF Biotyper (Bruker Daltonics, Bremen, Germany) instruments. Bacterial isolates were grown at 37 °C on Nutrient Medium No. 1 (SRCAMB, Obolensk, Moscow region, Russia), Luria-Bertani broth (Difco Laboratories, Detroit, MI, USA), and Muller-Hinton broth (Himedia, Mumbai, Maharashtra, India). Bacterial isolates were stored in 15% glycerol at minus 80 °C.
4.3. Antimicrobial Susceptibility
Susceptibility to 20 antimicrobials (AMs) of 8 functional groups: beta-lactams (ampicillin, ampicillin-sulbactam, cefuroxime, cefoxitin, ceftriaxone, ceftazidime, cefoperazone-sulbactam, cefepime, ertapenem, imipenem), tetracyclines (tetracycline, tigecycline), fluoroquinolones (ciprofloxacin), phenicols (chloramphenicol), aminoglycosides (gentamicin, tobramycin, amikacin), sulfonamides (trimethoprim-sulfamethoxazole), nitrofurans (nitrofurantoin), and polymyxins (colistin) were determined using Vitek-2 Compact (BioMerieux, Paris, France). The results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing, Version 11.0, 2021 (http://www.eucast.org access date 8 August 2021). Reference strains Escherichia coli ATCC 25922 and ATCC 35218 were used as quality controls. The isolates were categorized as resistant (R), multidrug-resistant (MDR), or extensively drug-resistant (XDR) according to the criteria proposed by Magiorakos et al. [11].
4.4. Detection of Antimicrobial Resistance Genes
Beta-lactamase genes blaSHV, blaCTX-M, blaTEM, blaOXA-48, blaKPC, blaVIM, blaIMP, and blaNDM, and class 1 and 2 integrons were detected by PCR using previously described specific primers [52,53,54,55,56,57,58,59,60].
4.5. Detection of Virulence Genes and Capsular Type Identification
Eight genes associated with K. pneumoniae virulence, rmpA (hypermucoid phenotype regulator), aer (aerobactin), kfu (ferric absorption system), uge (uridine diphosphate-galacturonate-4-epimerase), wabG (glucosyltransferase), fimH (fimbria type I), allS and allR (allantoin regulon), were detected by PCR using specific primers [35,61,62,63]. The capsular serotypes of the K. pneumoniae isolates were determined by wzi gene amplification using specific primers [64], sequencings, and allele identification using the Institute Pasteur, Paris, France, BIGS database (https://bigsdb.pasteur.fr/ access date 8 August 2021).
4.6. Multilocus Sequence Typing
Sequence types (STs) of K. pneumoniae isolates were identified based on allelic profiles of seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB), according to the Institute Pasteur, Paris, France, BIGS database protocol (https://bigsdb.pasteur.fr/klebsiella/primers_used.html access date 8 August 2021). Sequencing of DNA fragments was carried out at the SINTOL Center for collective use (Moscow, Russia). DNA sequences were analyzed using Vector NTI9 (Invitrogen, Carlsbad, CA, USA) and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi access date 8 August 2021).
4.7. Phylogenetic Analysis
The phylogenetic tree of K. pneumoniae STs was constructed using a web resource NCBI “Blastn” and “Blast Tree View” (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome access date 8 August 2021), based on combined gene sequences of MLST profiles.
4.8. Whole-Genome Sequencing
Whole-genome sequencing was done on the Illumina MiSeq platform using Nextera DNA Library Preparation Kit (Illumina, Carlsbad, CA, USA) and MiSeq Reagent Kits v3 (Illumina, Carlsbad, CA, United States). The obtained single reads were collected into contigs using the SPAdes 3.9.0 software (Petersburg State University, St-Petersburg, Russia). De novo assembled genomes were annotated in the GenBank database (https://github.com/ncbi/pgap access date 8 August 2021). AM resistance genes, STs, and plasmids were identified using the web resources of the Center for Genomic Epidemiology: ResFinder, KmerResistance (90% identity threshold and 10% threshold for depth corr.), MLST, and PlasmidFinder (95% threshold for minimum identity and 60% minimum coverage) (http://www.genomicepidemiology.org/ access date 8 August 2021). Virulence genes, capsular type, and efflux pumps were identified by the Institut Pasteur, Paris, France, BIGS database web-resource of (https://bigsdb.pasteur.fr/ access date 8 August 2021).
Supplementary Materials
The following are available online at www.mdpi.com/2079-6382/10/8/979/s1, Table S1: Minimal inhibitory concentrations (MICs) of antimicrobials for K. pneumoniae clinical isolates.
Author Contributions
Conceptualization, N.K.F., O.N.E. and I.A.D.; methodology, E.I.A., I.A.A., I.A.S.; software, A.A.K., M.V.F.; validation, T.S.N., M.V.F. and E.S.K.; formal analysis, O.N.E., N.K.F., S.F.B.; investigation, I.A.A., E.I.A., T.S.N., G.N.F., A.A.K., E.S.K.; resources, A.A.K., M.V.F.; data curation, A.A.K., M.V.F.; writing—original draft preparation, N.K.F., E.I.A.; writing—review and editing, O.N.E.; visualization, I.A.S., E.I.A.; supervision, N.K.F., O.N.E.; project administration, S.F.B., I.A.D.; funding acquisition, I.A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation, grant number 075-15-2019-1671 (agreement dated 31 October 2019).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, the study was designed as a retrospective observational study.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Following Whole genome sequences were deposited in the GenBank database: JAGRZJ000000000, JAGRZI000000000, JAFFJK000000000, JAGRZH000000000, JAFFJI000000000, JAGRZG000000000, JAGRZF000000000, JAGRZE000000000, and JAFFJJ000000000.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1–9. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Z.; Sun, L.; Wang, Z.; Sun, L.; Xu, J.; Zeng, L.; Sun, T. Clinical Observation and Prognostic Analysis of Patients With Klebsiella pneumoniae Bloodstream Infection. Front. Cell. Infect. Microbiol. 2020, 10, 577244. [Google Scholar] [CrossRef]
- Ershova, K.; Savin, I.; Kurdyumova, N.; Wong, D.; Danilov, G.; Shifrin, M.; Alexandrova, I.; Sokolova, E.; Fursova, N.; Zelman, V.; et al. Implementing an infection control and prevention program decreases the incidence of healthcare-associated infections and antibiotic resistance in a Russian neuro-ICU. Antimicrob. Resist. Infect. Control. 2018, 7, 94. [Google Scholar] [CrossRef]
- Lum, P.N.; Woo, P.C.Y.; Wong, S.S.; Yuen, K.-Y. Leukocytoclastic vasculitis complicating Klebsiella pneumoniae bacteremia. Diagn. Microbiol. Infect. Dis. 2000, 37, 275–277. [Google Scholar] [CrossRef]
- Lloret, P.; Redondo, P.; Molano, E. Klebsiella pneumoniae and leukocytoclastic vasculitis. Lancet 2002, 360, 1062. [Google Scholar] [CrossRef]
- Huang, H.Y.; Wu, Y.-H.; Kuo, C.F. Klebsiella pneumoniaesepsis with unusual cutaneous presentation of generalized pustulosis. Clin. Exp. Dermatol. 2013, 38, 626–629. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchêne, S.; Jenney, A.; Holt, K. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [PubMed]
- El-Domany, R.A.; Awadalla, O.A.; Shabana, S.A.; El-Dardir, M.A.; Emara, M. Analysis of the Correlation Between Antibiotic Resistance Patterns and Virulence Determinants in Pathogenic Klebsiella pneumoniae Isolates from Egypt. Microb. Drug Resist. 2021, 27, 727–739. [Google Scholar] [CrossRef]
- Lan, P.; Jiang, Y.; Zhou, J.; Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 25, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-L.; Ko, W.-C.; Cheng, K.-C.; Lee, C.-C.; Lai, C.-C.; Chuang, Y.-C. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 2008, 62, 1–6. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Shi, Y.-J.; Zheng, G.-H.; Qian, L.-Y.; Qsman, R.A.; Li, G.-G.; Zhang, G.-J. Longitudinal Analysis of Risk Factors for Clinical Outcomes of Enterobacteriaceae Meningitis/Encephalitis in Post-Neurosurgical Patients: A Comparative Cohort Study During 2014–2019. Infect. Drug Resist. 2020, 13, 2161–2170. [Google Scholar] [CrossRef]
- Yang, W.; Wu, X.; Li, Z.; Yuan, Q.; Wu, G.; Yu, J.; Wu, X.; Du, Z.; Hu, J.; Zhou, L. Trends of Intra-Cranial Bacterial Infection in Patients Requiring Emergency Neurosurgery. Surg. Infect. 2020, 21, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.W.; Kim, E.; Kim, H.; Bae, S.H. Klebsiella Endophthalmitis as Retinal Vasculitis with Prostatic Abscess. Optom. Vis. Sci. 2015, 92, e158–e160. [Google Scholar] [CrossRef]
- Castan, P.; Maigne, G.; Boutemy, J.; Silva, N.M.; De Boysson, H.; Aouba, A.; Audemard-Verger, A. Vascularite à IgA satellite d’une pneumopathie abcédée à Klebsiella pneumoniae IgA vasculitis secondary to Klebsiella pneumoniae infection. Rev. Mal. Respir. 2020, 37, 417–421. [Google Scholar] [CrossRef]
- Izdebski, R.; Baraniak, A.; Żabicka, D.; Machulska, M.; Urbanowicz, P.; Fiett, J.; Literacka, E.; Bojarska, K.; Kozińska, A.; Zieniuk, B.; et al. Enterobacteriaceae producing OXA-48-like carbapenemases in Poland, 2013–January 2017. J. Antimicrob. Chemother. 2017, 73, 620–625. [Google Scholar] [CrossRef]
- Muggeo, A.; Guillard, T.; Klein, F.; Reffuveille, F.; François, C.; Babosan, A.; Bajolet, O.; Bertrand, X.; de Champs, C. Spread of Klebsiella pneumoniae ST395 non-susceptible to carbapenems and resistant to fluoroquinolones in North-Eastern France. J. Glob. Antimicrob. Resist. 2018, 13, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Maida, C.M.; Bonura, C.; Geraci, D.M.; Graziano, G.; Carattoli, A.; Rizzo, A.; Torregrossa, M.V.; Vecchio, D.; Giuffrè, M. Outbreak of ST395 KPC-Producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in Palermo, Italy. Infect. Control. Hosp. Epidemiol. 2018, 39, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Kuptsov, N.; Kornienko, M.; Gorodnichev, R.; Danilov, D.; Malakhova, M.; Parfenova, T.; Makarenko, G.; Shitikov, E.; Ilina, E. Efficacy of commercial bacteriophage products against ESKAPE pathogens. Bull. Russ. St. Med. Univer. 2020, 3, 18–24. [Google Scholar] [CrossRef]
- Fursova, N.K.; Astashkin, E.I.; Gabrielyan, N.I.; Novikova, T.S.; Fedyukina, G.N.; Kubanova, M.K.; Esenova, N.M.; Sharapchenko, S.O.; Volozhantsev, N.V. Emergence of Five Genetic Lines ST395NDM-1, ST13OXA-48, ST3346OXA-48, ST39CTX-M-14, and Novel ST3551OXA-48 of Multidrug-Resistant Clinical Klebsiella pneumoniae in Russia. Microb. Drug Resist. 2020, 26, 924–933. [Google Scholar] [CrossRef]
- Shankar, C.; Jacob, J.J.; Vasudevan, K.; Biswas, R.; Manesh, A.; Sethuvel, D.P.M.; Varughese, S.; Biswas, I.; Veeraraghavan, B. Emergence of Multidrug Resistant Hypervirulent ST23 Klebsiella pneumoniae: Multidrug Resistant Plasmid Acquisition Drives Evolution. Front. Cell. Infect. Microbiol. 2020, 10, 575289. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.A.; Pascale, L.-M.; Million, M.; Briantais, A.; Durand, J.-M.; Hadjadj, L.; Rolain, J.-M. Whole genome sequencing to decipher the virulence phenotype of hypervirulent Klebsiella pneumoniae responsible for liver abscess, Marseille, France. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1073–1077. [Google Scholar] [CrossRef]
- Su, S.; Li, C.; Zhao, Y.; Yu, L.; Wang, Y.; Wang, Y.; Bao, M.; Fu, Y.; Zhang, J.; Zhang, X. Outbreak of KPC-2–Producing Klebsiella pneumoniae ST76 Isolates in an Intensive Care Unit and Neurosurgery Unit. Microb. Drug Resist. 2020, 26, 1009–1018. [Google Scholar] [CrossRef]
- Liao, C.-H.; Huang, Y.T.; Chang, C.Y.; Hsu, H.S.; Hsueh, P.-R. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 33, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Lev, A.I.; Astashkin, E.I.; Kislichkina, A.A.; Solovieva, E.V.; Kombarova, T.I.; Korobova, O.V.; Ershova, O.N.; Alexandrova, I.A.; Malikov, V.E.; Bogun, A.G.; et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog. Glob. Health 2018, 112, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Perilli, M.; Piccirilli, V.; Segatore, B.; Amicosante, G.; Maccacaro, L.; Bazaj, A.; Naso, L.; Cascio, G.L.; Cornaglia, G. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae ST14 and ST512 causing bloodstream infections in ICU and surgery wards of a tertiary university hospital of Verona (northern Italy): Co-production of KPC-3, OXA-48, and CTX-M-15 β-lactamases. Diagn. Microbiol. Infect. Dis. 2020, 96, 114968. [Google Scholar] [CrossRef]
- Galani, I.; Karaiskos, I.; Angelidis, E.; Papoutsaki, V.; Galani, L.; Souli, M.; Antoniadou, A.; Giamarellou, H. Emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in KPC-2-producing Klebsiella pneumoniae of sequence type 39 during treatment. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 219–224. [Google Scholar] [CrossRef]
- Surleac, M.; Barbu, I.C.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef]
- Shen, P.; Berglund, B.; Chen, Y.; Zhou, Y.; Xiao, T.; Xiao, Y.; Zhou, K. Hypervirulence Markers Among Non-ST11 Strains of Carbapenem- and Multidrug-Resistant Klebsiella pneumoniae Isolated From Patients With Bloodstream Infections. Front. Microbiol. 2020, 11, 1199. [Google Scholar] [CrossRef]
- Xie, S.; Fu, S.; Li, M.; Guo, Z.; Zhu, X.; Ren, J.; Hu, F. Microbiological Characteristics of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates Collected from County Hospitals. Infect. Drug Resist. 2020, 13, 1163–1169. [Google Scholar] [CrossRef]
- Lin, Z.-W.; Zheng, J.-X.; Bai, B.; Xu, G.-J.; Lin, F.-J.; Chen, Z.; Sun, X.; Qu, D.; Yu, Z.-J.; Deng, Q.-W. Characteristics of Hypervirulent Klebsiella pneumoniae: Does Low Expression of rmpA Contribute to the Absence of Hypervirulence? Front. Microbiol. 2020, 11, 436. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 642484. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhou, J.-W.; Qiu, C.-N.; Wang, M.-M.; Wang, X.-J.; Ruan, Z.; Fan, J.-Z.; Qiong, C.; Jia-Wei, Z.; Chun-Ning, Q.; et al. Antimicrobial susceptibility and microbiological and epidemiological characteristics of hypermucoviscous Klebsiella pneumoniae strains in a tertiary hospital in Hangzhou, China. J. Glob. Antimicrob. Resist. 2018, 15, 61–64. [Google Scholar] [CrossRef]
- Nava, R.G.; Oliveira-Silva, M.; Nakamura-Silva, R.; Pitondo-Silva, A.; Vespero, E.C. New sequence type in multidrug-resistant Klebsiella pneumoniae harboring the blaNDM-1-encoding gene in Brazil. Int. J. Infect. Dis. 2019, 79, 101–103. [Google Scholar] [CrossRef]
- Ma, L.-C.; Fang, C.-T.; Lee, C.-Z.; Shun, C.-T.; Wang, J.-T. Genomic Heterogeneity in Klebsiella pneumoniae Strains Is Associated with Primary Pyogenic Liver Abscess and Metastatic Infection. J. Infect. Dis. 2005, 192, 117–128. [Google Scholar] [CrossRef]
- Kovács, K.; Nyul, A.; Mestyán, G.; Melegh, S.; Fenyvesi, H.; Jakab, G.; Szabó, H.; Jánvári, L.; Damjanova, I.; Tóth, Á. Emergence and interhospital spread of OXA-48-producing Klebsiella pneumoniae ST395 clone in Western Hungary. Infect. Dis. 2016, 49, 231–233. [Google Scholar] [CrossRef]
- Alekseeva, E.A.; Brusnigina, N.F.; Solntsev, A.L.; Gordinskaya, A.N. The molecular typing of clinical isolates Klebsiella pneumoniae producing beta-lactamases of extended specter of action. Klin. Lab. Diagn. 2017, 62, 699–704. [Google Scholar]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Arena, F.; Di Pilato, V.; Vannetti, F.; Fabbri, L.; Antonelli, A.; Coppi, M.; Pupillo, R.; Macchi, C.; Rossolini, G.M. Population structure of KPC carbapenemase-producing Klebsiella pneumoniae in a long-term acute-care rehabilitation facility: Identification of a new lineage of clonal group 101, associated with local hyperendemicity. Microb. Genom. 2020, 6, e000308. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging Antimicrobial-Resistant High-Risk Klebsiella pneumoniae Clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef] [PubMed]
- Skachkova, T.; Shipulina, O.; Shipulin, G.; Shelenkov, A.; Yanushevich, Y.; Mikhaylova, Y.; Zamyatin, M.; Gusarov, V.; Petrova, N.; Lashenkova, N.; et al. Characterization of genetic diversity of the Klebsiella pneumoniae strains in a Moscow tertiary care center using next-generation sequencing. Clin. Microbiol. Antimicrob. Chemother. 2019, 21. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef]
- Azim, N.S.A.; Nofal, M.Y.; Alharbi, M.A.; Al-Zaban, M.I.; Somily, A. Molecular-diversity, Prevalence and Antibiotic Susceptibility of Pathogenic Klebsiella pneumoniae under Saudi Condition. Pak. J. Biol. Sci. 2019, 22, 174–179. [Google Scholar] [CrossRef][Green Version]
- Ishii, A.; Shigemura, K.; Kitagawa, K.; Harada, M.; Kan, Y.; Hayashi, F.; Osawa, K.; Kuntaman, K.; Shirakawa, T.; Fujisawa, M. Cross-Resistance and the Mechanisms of Cephalosporin-Resistant Bacteria in Urinary Tract Infections Isolated in Indonesia. Curr. Microbiol. 2021, 78, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Roch, M.; Sierra, R.; Sands, K.; Martins, W.M.; Schrenzel, J.; Walsh, T.R.; Gales, A.C.; Andrey, D.O. Vertical and horizontal dissemination of an IncC plasmid harbouring rmtB 16S rRNA methylase gene, conferring resistance to plazomicin, among invasive ST258 and ST16 KPC-producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 24, 183–189. [Google Scholar] [CrossRef]
- Hernández, M.; López-Urrutia, L.; Abad, D.; Serna, M.D.F.; Ocampo-Sosa, A.; Eiros, J. First Report of an Extensively Drug-Resistant ST23 Klebsiella pneumoniae of Capsular Serotype K1 Co-Producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics 2021, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; De Wang, L.; Li, D.; Du, F.-L.; Long, D.; Liu, Y.; Ng, O.; Zhang, W. High Prevalence of 16s rRNA Methylase Genes Among Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Isolates in a Chinese Tertiary Hospital. Microb. Drug Resist. 2021, 27, 44–52. [Google Scholar] [CrossRef]
- Nakamura-Silva, R.; Oliveira-Silva, M.; Furlan, J.P.R.; Stehling, E.G.; Miranda, C.E.S.; Pitondo-Silva, A. Characterization of multidrug-resistant and virulent Klebsiella pneumoniae strains belonging to the high-risk clonal group 258 (CG258) isolated from inpatients in northeastern Brazil. Arch. Microbiol. 2021, 203, 4351–4359. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Wyres, K.L.; Gorrie, C.L.; Judd, L.M.; Jenney, A.W.J.; Brisse, S.; Holt, K.E. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 2018, 4, e000196. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, J.; Miró, E.; Brown-Jaque, M.; Hurtado, J.C.; Moreno, A.; Muniesa, M.; González-López, J.J.; Vila, J.; Espinal, P.; Navarro, F. Comparison of Commensal and Clinical Isolates for Diversity of Plasmids in Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2020, 64, e02064-19. [Google Scholar] [CrossRef]
- Shifrin, M.; Kurdumova, N.; Danilov, G.; Ershova, O.; Savin, I.; Alexandrova, I.; Sokolova, E.; Tabasaranskiy, T. Electronic patient records system as a monitoring tool. Stud. Heal. Technol. Inform. 2015, 210, 236–238. [Google Scholar]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2005, 57, 14–23. [Google Scholar] [CrossRef]
- Edelstein, M.; Pimkin, M.; Palagin, I.; Stratchounski, L. Prevalence and Molecular Epidemiology of CTX-MExtended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae in RussianHospitals. Antimicrob. Agents Chemother. 2003, 47, 3724–3732. [Google Scholar] [CrossRef]
- Priamchuk, S.D.; Fursova, N.K.; Abaev, I.; Kovalev, I.N.; Shishkova, A.N.; Pecherskikh, I.E.; Korobova, O.V.; Astashkin, I.E.; Pachkunov, D.M.; Kruglov, A.N.; et al. Genetic determinants of antibacterial resistance among nosocomial Escherichia coli, Klebsiella spp., and Enterobacter spp. isolates collected in Russia within 2003–2007. Antibiot. Khimioter. 2010, 55, 3–10. [Google Scholar] [PubMed]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic Features of the Widespread Plasmid Coding for the Carbapenemase OXA-48. Antimicrob. Agents Chemother. 2011, 56, 559–562. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Biddle, J.W.; Anderson, K.F.; Washer, L.; Chenoweth, C.; Perrin, J.; Newton, D.W.; Patel, J.B. Detection of the Klebsiella pneumoniae Carbapenemase Type 2 Carbapenem-Hydrolyzing Enzyme in Clinical Isolates of Citrobacter freundii and K. oxytoca Carrying a Common Plasmid. J. Clin. Microbiol. 2008, 46, 2066–2069. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hujer, K.M.; Hujer, A.M.; Hulten, E.A.; Bajaksouzian, S.; Adams, J.M.; Donskey, C.J.; Ecker, D.J.; Massire, C.; Eshoo, M.W.; Sampath, R.; et al. Analysis of Antibiotic Resistance Genes in Multidrug-Resistant Acinetobacter sp. Isolates from Military and Civilian Patients Treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006, 50, 4114–4123. [Google Scholar] [CrossRef]
- Martins, A.F.; Zavascki, A.P.; Gaspareto, P.B.; Barth, A.L. Dissemination of Pseudomonas aeruginosa Producing SPM-1-like and IMP-1-like Metallo-β-lactamases in Hospitals from Southern Brazil. Infection 2007, 35, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Y.; Jia, X.; Luo, Y.; Song, Q.; Zhao, W.; Wang, Y.; Liu, H.; Zheng, D.; Xia, Y.; et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 2012, 18, E506–E513. [Google Scholar] [CrossRef]
- Machado, E.; Cantόn, R.; Baquero, F.; Galán, J.-C.; Rollán, A.; Peixe, L.; Coque, T.M. Integron Content of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Strains over 12 Years in a Single Hospital in Madrid, Spain. Antimicrob. Agents Chemother. 2005, 49, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Nassrf, X.; Honoré, N.; Vasselon, T.; Cole, S.T.; Sansonetti, P.J. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Kiebsiella pneumoniae. Mol. Microbiol. 1989, 3, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Regué, M.; Hita, B.; Piqué, N.; Izquierdo, L.; Merino, S.; Fresno, S.; Benedí, V.J.; Tomás, J.M. A Gene, uge, Is Essential for Klebsiella pneumoniae Virulence. Infect. Immun. 2004, 72, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, L.; Coderch, N.; Piqué, N.; Bedini, E.; Corsaro, M.M.; Merino, S.; Fresno, S.; Tomás, J.M.; Regué, M. The Klebsiella pneumoniae wabG Gene: Role inBiosynthesis of the Core Lipopolysaccharide andVirulence. J. Bacteriol. 2003, 185, 7213–7221. [Google Scholar] [CrossRef]
- Brisse, S.; Passet, V.; Haugaard, A.B.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decré, D. wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella Strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).