Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Setting
2.2. Isolates of E. coli
2.3. Molecular Typing Analyses and β-Lactamase Molecular Identification
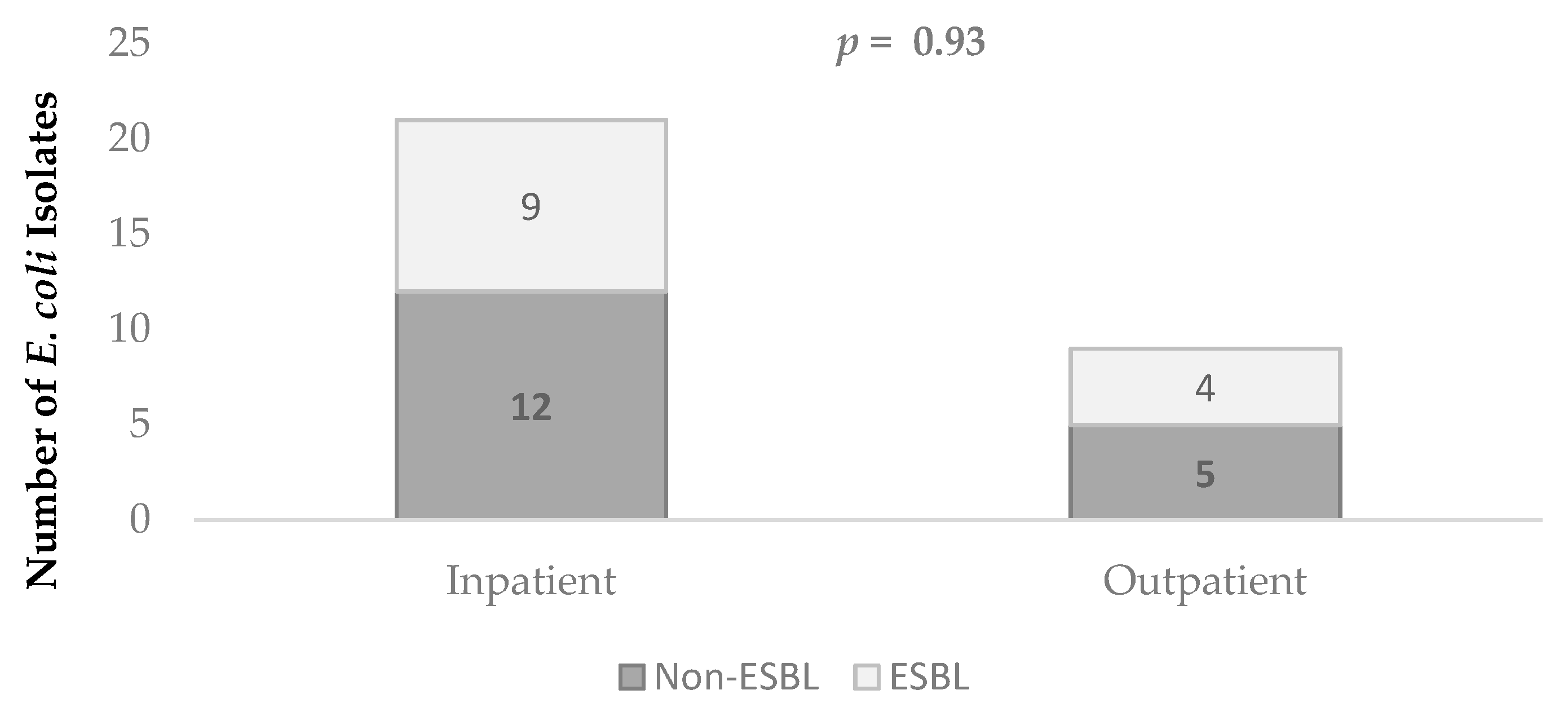
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Pitout, J.D.D. Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D. Infections with extended-spectrum β-lactamase-producing enterobacteriaceae: Changing epidemiology and drug treatment choices. Drugs 2010, 70, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Pitout, J.D.D. Extended-spectrum β-lactamase-producing Enterobacteriaceae: Update on molecular epidemiology and treatment options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Cassier, P.; Lallechère, S.; Aho, S.; Astruc, K.; Neuwirth, C.; Piroth, L.; Chavanet, P. Cephalosporin and fluoroquinolone combinations are highly associated with CTX-M β-lactamase-producing Escherichia coli: A case-control study in a French teaching hospital. Clin. Microbiol. Infect. 2011, 17, 1746–1751. [Google Scholar] [CrossRef]
- Calbo, E.; Romaní, V.; Xercavins, M.; Gómez, L.; Vidal, C.G.; Quintana, S.; Vila, J.; Garau, J. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2006, 57, 780–783. [Google Scholar] [CrossRef]
- Saely, S.; Kaye, K.S.; Fairfax, M.R.; Chopra, T.; Pogue, J.M. Investigating the impact of the definition of previous antibiotic exposure related to isolation of extended spectrum β-lactamase-producing Klebsiella pneumoniae. Am. J. Infect. Control. 2011, 39, 390–395. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Available online: http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 3 December 2020).
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015, 161 Pt 5, 980–988. [Google Scholar] [CrossRef]
- Johnson, J.R.; Clermont, O.; Johnston, B.; Clabots, C.; Tchesnokova, V.; Sokurenko, E.; Junka, A.F.; Maczynska, B.; Denamur, E. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J. Clin. Microbiol. 2014, 52, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; De Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio 2016, 7, e02162-15. [Google Scholar] [CrossRef] [PubMed]
- Ben Zakour, N.L.; Alsheikh-Hussain, A.S.; Ashcroft, M.M.; Khanh Nhu, N.T.; Roberts, L.W.; Stanton-Cook, M.; Schembri, M.A.; Beatson, S.A. Sequential Acquisition of Virulence and Fluoroquinolone Resistance Has Shaped the Evolution of Escherichia coli ST131. MBio 2016, 7, e00347-16. [Google Scholar] [CrossRef]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 2013, 4, e00377-13. [Google Scholar] [CrossRef]
- Petty, N.K.; Ben Zakour, N.L.; Stanton-Cook, M.; Skippington, E.; Totsika, M.; Forde, B.M.; Phan, M.D.; Gomes Moriel, D.; Peters, K.M.; Davies, M.; et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA 2014, 111, 5694–5699. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef]
- Blanco, V.M.; Maya, J.J.; Correa, A.; Perenguez, M.; Muñoz, J.S.; Motoa, G.; Pallares, C.J.; Rosso, F.; Matta, L.; Celis, Y.; et al. Prevalence and risk factors for extended-spectrum β-lactamase-producing Escherichia coli causing community-onset urinary tract infections in Colombia. Enferm. Infecc. Microbiol. Clin. 2016, 34, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Lahlaoui, H.; Ben Haj Khalifa, A.; Ben Moussa, M. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL). Med. Mal. Infect. 2014, 44, 400–404. [Google Scholar] [CrossRef]
- den Reijer, P.M.; van Burgh, S.; Burggraaf, A.; Ossewaarde, J.M.; van der Zee, A. The widespread presence of a multidrug-resistant Escherichia coli ST131 clade among community-associated and hospitalized patients. PLoS ONE 2016, 11, e0150420. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.L.; Cortés, J.A.; Arias, G.; Ovalle, M.V.; Saavedra, S.Y.; Buitrago, G.; Escobar, J.A.; Castro, B.E.; GREBO. Emergence of resistance to third generation cephalosporins by Enterobacteriaceae causing community-onset urinary tract infections in hospitals in Colombia. Enferm. Infecc. Microbiol. Clin. 2013, 31, 298–303. [Google Scholar] [CrossRef]
- Peirano, G.; Asensi, M.D.; Pitondo-Silva, A.; Pitout, J.D.D. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin. Microbiol. Infect. 2011, 17, 1039–1043. [Google Scholar] [CrossRef]
- Ruiz, S.J.; Montealegre, M.C.; Ruiz-Garbajosa, P.; Correa, A.; Briceño, D.F.; Martinez, E.; Rosso, F.; Muñoz, M.; Quinn, J.P.; Cantón, R.; et al. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J. Clin. Microbiol. 2011, 49, 1993–1996. [Google Scholar] [CrossRef]
- Chiluisa-Guacho, C.; Escobar-Perez, J.; Dutra-Asensi, M. First detection of the CTXM-15 producing Escherichia coli O25-ST131 pandemic clone in Ecuador. Pathogens 2018, 7, 42. [Google Scholar] [CrossRef]
- Guzmán-Blanco, M.; Labarca, J.A.; Villegas, M.V.; Gotuzzo, E.; Latin American Working Group on Bacterial Resistance. Extended spectrum β-lactamase producers among nosocomial Enterobacteriaceae in Latin America. Braz. J. Infect. Dis. 2014, 18, 421–433. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, C.M.; van Duijkeren, E.; Schoormans, A.H.W.; van Essen Zandbergen, A.; Veldman, K.; Kant, A.; Huijsdens, X.W.; van der Zwaluw, K.; Wagenaar, J.A.; Mevius, D.J. Occurrence and characteristics of extended-spectrum-β-lactamase and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 2012, 67, 1368–1374. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Chen, W.-C.; Hung, C.-H.; Chen, Y.-S.; Cheng, J.-S.; Lee, S.S.-J.; Tseng, F.-C.; Cheng, M.-F.; Wang, J.-L. Bloodstream infections caused by extended-spectrum β-lactamase-producing Escherichia coli in patients with liver cirrhosis. Pathogens 2021, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Institut Pasteur. Escherichia coli Sequence Typing. Available online: https://bigsdb.pasteur.fr/ecoli/ecoli.html (accessed on 9 December 2020).
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Barnes, A.I.; Paraje, M.G. Break-down of antibiotic prescription in health centres by infection concerned. Aten Primaria 2006, 37, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Johnson, J.R. A new clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 2014, 58, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- De La Cadena, E.; Mojica, M.F.; Castillo, N.; Correa, A.; Appel, T.M.; García-Betancur, J.C.; Pallares, C.J.; Villegas, M.V. Genomic analysis of CTX-M-group-1-producing extraintestinal pathogenic E. coli (ExPEc) from patients with urinary tract infections (UTI) from Colombia. Antibiotics 2020, 9, 899. [Google Scholar] [CrossRef]
- Pavez, M.; Troncoso, C.; Osses, I.; Salazar, S.; Illescac, V.; Reydet, P.; Rodríguezc, C.; Chahind, C.; Concha, C.; Barrientos, L. High prevalence of CTX-M-1 group in ESBL-producing enterobacteriaceae infection in intensive care units in southern Chile. Braz. J. Infect. Dis. 2019, 23, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.L.; Volcão, L.M.; Klafke, G.B.; Vieira, R.S.; Gonçalves, C.V.; Ramis, I.V.; Almeida da Silva, P.E.; von Groll, A. Antimicrobial resistance and molecular characterization of extended-spectrum β-lactamases of Escherichia coli and Klebsiella spp. Isolates from urinary tract infections in southern Brazil. Microb. Drug Resist. 2019, 25, 173–181. [Google Scholar] [CrossRef]
- Peirano, G.; van der Bij, A.K.; Freeman, J.L.; Poirel, L.; Nordmann, P.; Costello, M.; Tchesnokova, V.L.; Pitout, J.D. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: Global distribution of the H30-Rx sublineage. Antimicrob. Agents Chemother. 2014, 58, 3762–3767. [Google Scholar] [CrossRef] [PubMed]
- Larramendy, S.; Deglaire, V.; Dusollier, P.; Fournier, J.P.; Caillon, J.; Beaudeau, F.; Moret, L. Risk factors of extended-spectrum β-lactamases-producing Escherichia coli community acquired urinary tract infections: A systematic review. Infect. Drug Resist. 2020, 13, 3945–3955. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Choi, S.M.; Lee, D.G.; Cho, S.Y.; Lee, H.J.; Choi, J.K.; Choi, J.H.; Yoo, J.H. Impact of extended-spectrum β-lactamase production on treatment outcomes of acute pyelonephritis caused by Escherichia coli in patients without health care-associated risk factors. Antimicrob. Agents Chemother. 2015, 59, 1962–1968. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Hujer, A.M.; Marshall, S.H.; Domitrovic, T.N.; Rudin, S.D.; Zheng, X.; Qureshi, N.K.; Hayden, M.K.; Scaggs, F.A.; Karadkhele, A.; et al. Analysis of β-lactamase resistance determinants in Enterobacteriaceae from Chicago children: A multicenter survey. Antimicrob. Agents Chemother. 2016, 60, 3462–3469. [Google Scholar] [CrossRef]
| Antimicrobial Agent | MIC Breakpoint (µg/mL) | Escherichia coli Isolates by Resistance, n (%) | ||
|---|---|---|---|---|
| Total (n = 30) | ESBL (n = 13) | Non-ESBL (n = 17) | ||
| Ampicillin | ≥32 | 27 (90) | 13 (100) | 14 (82) |
| Piperacillin-tazobactam | ≥128/4 | 1 (3) | 0 (0) | 1 (6) |
| Cephalothin | ≥64 | 20 (67) | 13 (100) | 7 (41) |
| Cefuroxime | ≥64 | 16 (53) | 13 (100) | 3 (18) |
| Cefotaxime | ≥64 | 13 (43) | 13 (100) | 0 (0) |
| Ceftazidime | ≥64 | 13 (43) | 13 (100) | 0 (0) |
| Cefepime | ≥64 | 13 (43) | 13 (100) | 0 (0) |
| Amikacin | ≥16 | 2 (7) | 1 (8) | 1 (6) |
| Gentamicin | ≥16 | 8 (27) | 5 (38) | 3 (18) |
| Nalidixic acid | ≥32 | 17 (57) | 5 (38) | 12 (71) |
| Ciprofloxacin | ≥4 | 23 (77) | 11 (85) | 12 (71) |
| Trimethoprim-sulfamethoxazole | ≥320 | 19 (63) | 10 (77) | 9 (53) |
| Variables, n (%) | ESBL (n = 13) | Non-ESBL (n = 17) | p Value |
|---|---|---|---|
| Age Groups, Years | 0.09 | ||
| 1–19 | 2 (15) | 3 (18) | |
| 20–59 | 1 (8) | 3 (18) | |
| 60–79 | 3 (23) | 9 (53) | |
| ≥80 | 7 (54) | 2 (12) | |
| Sex | 0.42 | ||
| Female | 9 (69) | 10 (59) | |
| Male | 4 (31) | 7 (41) | |
| Risk Factors | |||
| Hospitalized ≥2 d in the prior 90 d | 8 (62) | 7 (41) | 0.28 |
| Antibiotic treatment in the prior 90 d | 7 (54) | 5 (29) | 0.50 |
| Wound care at home | 2 (15) | 0 (0) | 0.10 |
| Outpatient chemotherapy | 1 (8) | 1 (6) | 0.85 |
| Personal history of immunosuppressive therapy | 1 (8) | 0 (0) | 0.25 |
| Hemodialysis in the prior 90 d | 1 (8) | 0 (0) | 0.25 |
| No known risk factors | 0 (0) | 6 (35) | 0.021 |
| Isolate | Sequence Typing | ESBL | β-Lactamases | Originating Site | Source | Phenotypic Profile |
|---|---|---|---|---|---|---|
| 378 | 3 | + | CTX-M-group 9, TEM | H (Surg) | Wound | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, STX |
| 2724 | 3 | - | Outpatient | Urine | AMP, CEF | |
| 439 | 4 | - | H (Peds) | Urine | AMP, STX | |
| 1232 | 43 | + | CTX-M-group-1 (CTX-M-15) | H (Ort) | Wound | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, GEN, CIP |
| 2690 | 43 | + | CTX-M-group 9, TEM | Outpatient | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, NAL, CIP, STX |
| 0-3630 | 43 | + | CTX-M-group 9, TEM | Outpatient | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, NAL, CIP, STX |
| 640 | 53 | + | CTX-M group 9 | H (Surg) | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, CIP |
| 370 | 53 | - | H (Ob-Gyn) | Urine | AMP, TZP, CEF, CXM, CAZ, AMK, NAL, CIP | |
| 2685 | 53 | - | Outpatient | Urine | AMP, CEF, NAL, CIP, STX | |
| 2710 | 458 | + | CTX-M-group 9 | Outpatient | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, NAL, CIP, STX |
| 19-2410 | 458 | - | H (IM) | Blood | AMP, CIP, STX | |
| 2699 | 458 | - | Outpatient | Urine | AMP, CEF, NAL, CIP | |
| 3627 | 479 | - | H (Surg) | Urine | AMP, GEN, NAL, CIP, STX | |
| 382 | 526 | + | CTX-M-group-1 (CTX-M-15) | H (IM) | Wound | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, STX |
| HRV-09 | 594 | + | ND | I (ICU) | Wound | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, GEN, CIP, STX |
| 361 | 621 | + | CTX-M-group-1 (CTX-M-15) | H (IM) | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, CIP, STX |
| 2676 | 833 | + | CTX-M-group-1, TEM | Outpatient | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, GEN, NAL, CIP, STX |
| 542 | 833 | - | H (IM) | Blood | AMP, NAL, CIP, STX | |
| 375 | N/A | + | CTX-M-group-1 (CTX-M-15), TEM | H (Surg) | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, NAL, GEN, CIP |
| 638 | N/A | + | CTX-M-group-1 (CTX-M-15) | H (Surg) | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, GEN, CIP, STX |
| CC2 | N/A | + | CTX-M-group 9 | H (IM) | Urine | AMP, CEF, CFZ, CXM, CTX, CAZ, FEP, CIP, STX |
| 435, 655 | N/A | - | H(IM) | Urine | AMP, NAL, CIP, STX | |
| 543, 544 | N/A | - | H (Ob-Gyn) | Blood | AMP, NAL, CIP, STX | |
| 545 | N/A | - | H (IM) | Blood | AMP, CIP, STX | |
| O-2115 | N/A | - | Outpatient | Urine | AMP, CIP, STX | |
| CC1 | N/A | - | H (IM) | Urine | NAL | |
| 519 | N/A | - | H (Ob-Gyn) | Wound | AMP, NAL | |
| 436 | N/A | - | Outpatient | Urine | AMP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Samudio, V.; Pecchio, M.; Pimentel-Peralta, G.; Quintero, Y.; Herrera, M.; Landires, I. Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama. Antibiotics 2021, 10, 899. https://doi.org/10.3390/antibiotics10080899
Núñez-Samudio V, Pecchio M, Pimentel-Peralta G, Quintero Y, Herrera M, Landires I. Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama. Antibiotics. 2021; 10(8):899. https://doi.org/10.3390/antibiotics10080899
Chicago/Turabian StyleNúñez-Samudio, Virginia, Maydelin Pecchio, Gumercindo Pimentel-Peralta, Yohana Quintero, Mellissa Herrera, and Iván Landires. 2021. "Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama" Antibiotics 10, no. 8: 899. https://doi.org/10.3390/antibiotics10080899
APA StyleNúñez-Samudio, V., Pecchio, M., Pimentel-Peralta, G., Quintero, Y., Herrera, M., & Landires, I. (2021). Molecular Epidemiology of Escherichia coli Clinical Isolates from Central Panama. Antibiotics, 10(8), 899. https://doi.org/10.3390/antibiotics10080899









