Long-Lasting Stable Expression of Human LL-37 Antimicrobial Peptide in Transgenic Barley Plants
Abstract
:1. Introduction
2. Results
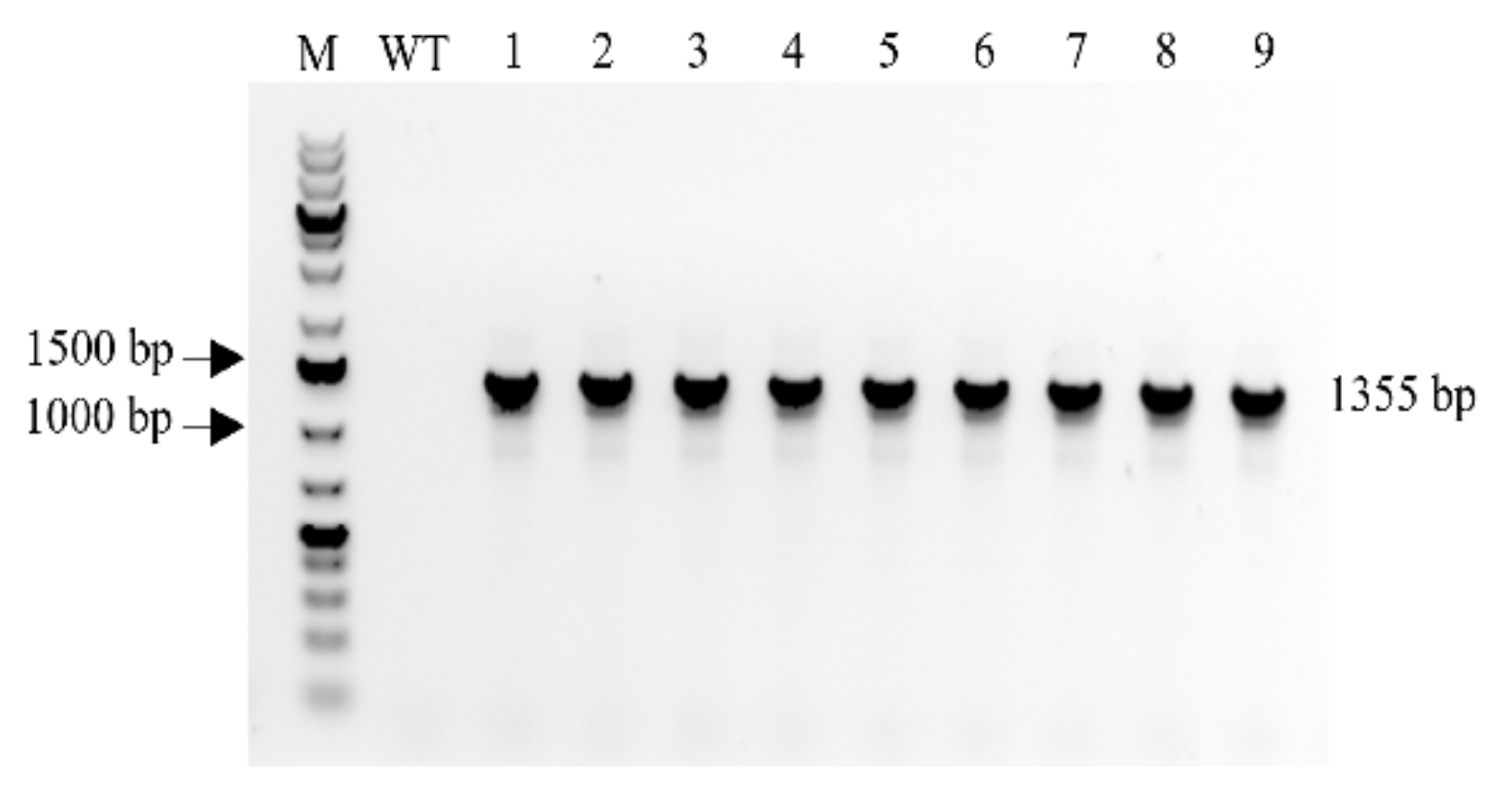
2.1. Confirmation of MBP::rhLL-37 Gene Insertion in Transgenic Progeny Barley Plants
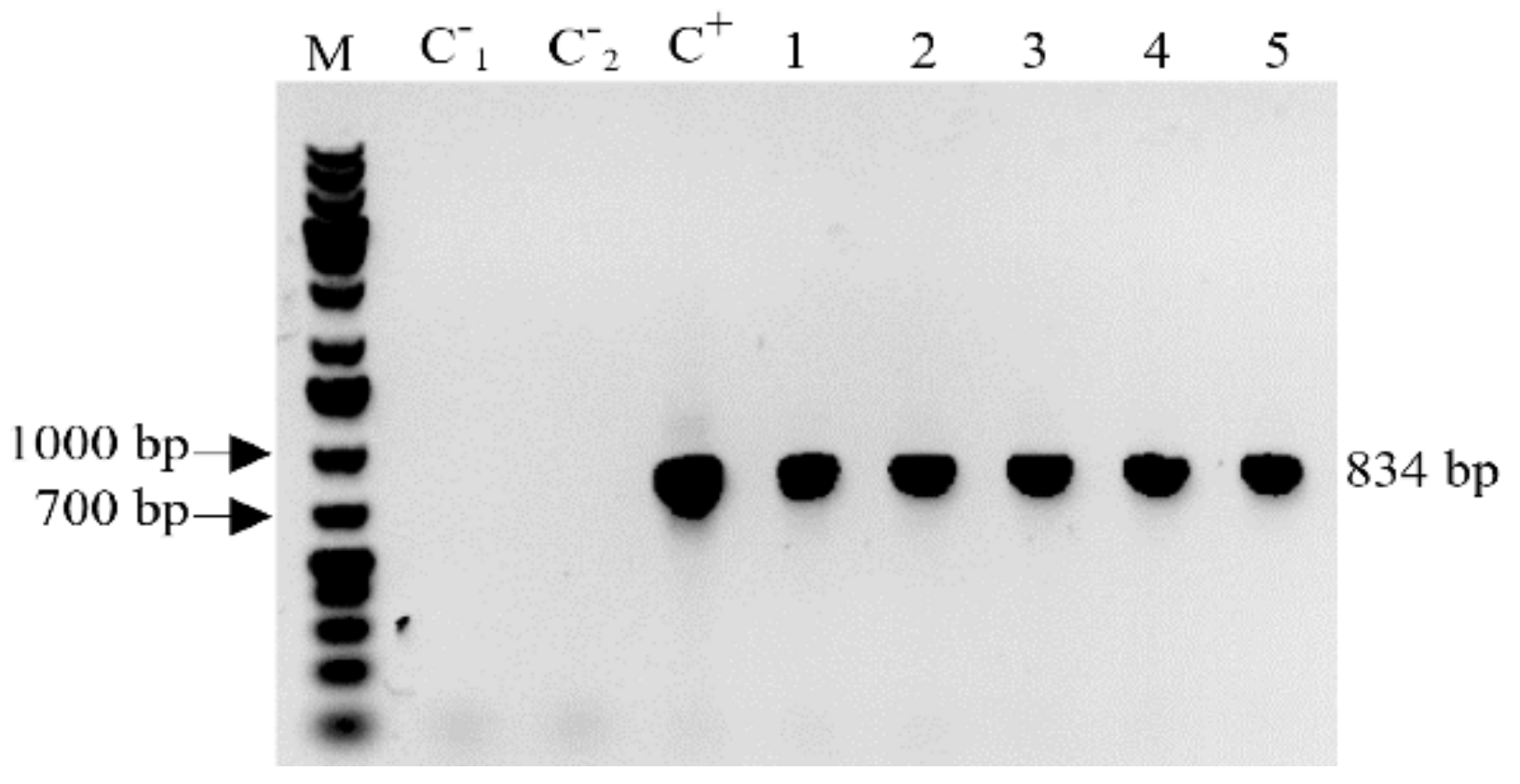
2.2. Detection of the MBP::rhLL-37 Gene Transcripts in Progeny Barley Grains
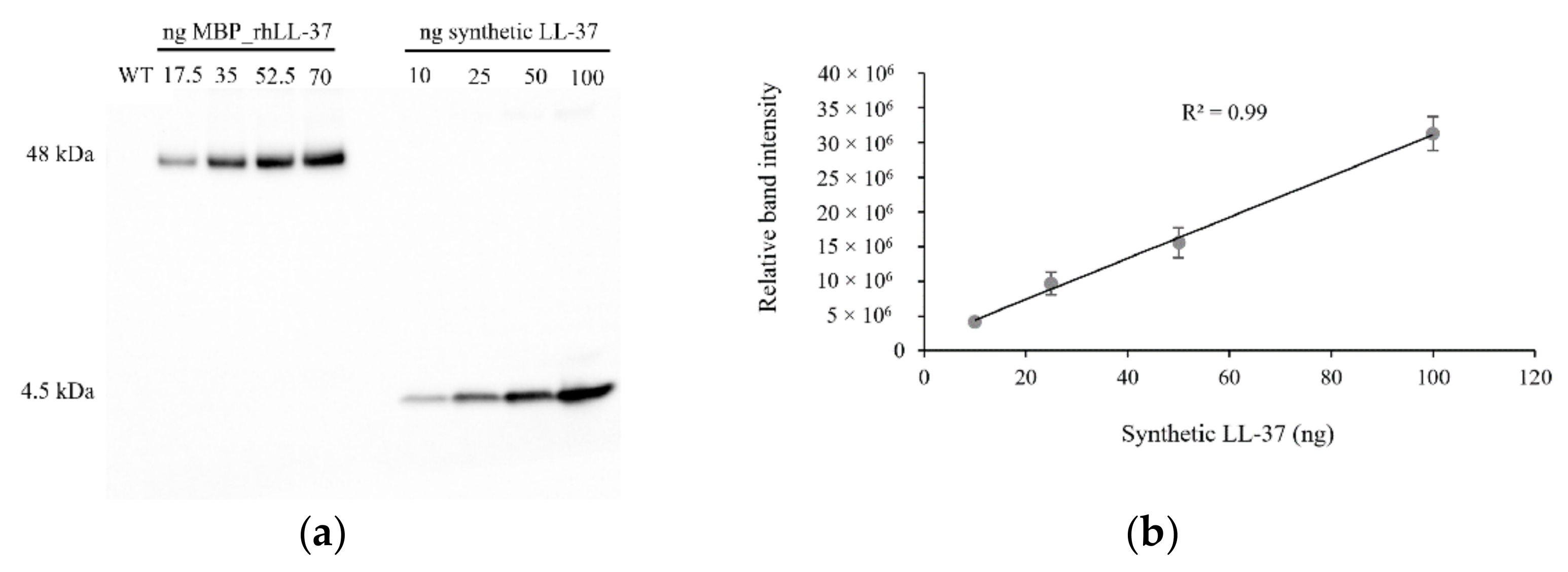
2.3. Detection and Quantification of the MBP::rhLL-37 Fusion Protein in Field-Grown Barley Plants
2.4. Analysis of the Antibacterial Activity of rhLL-37 from Barley
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Polymerase Chain Reaction (PCR) Analysis of the Transgene Insertion
4.3. Reverse Transcription-PCR of the Transgene Specific mRNA
4.4. Extraction and Quantification of the Recombinant Protein
4.5. Antibacterial Activity Assay
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wimley, W.C.; Hristova, K. Antimicrobial Peptides: Successes, Challenges and Unanswered Questions. J. Membr. Biol. 2011, 239, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Cunha, N.B.; Cobacho, N.B.; Viana, J.F.; Lima, L.A.; Sampaio, K.B.; Dohms, S.S.; Ferreira, A.C.; de la Fuente-Núñez, C.; Costa, F.F.; Franco, O.L. The next generation of antimicrobial peptides (AMPs) as molecular therapeutic tools for the treatment of diseases with social and economic impacts. Drug Discov. Today 2017, 22, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Torres, M.D.T.; de la Fuente-Nunez, C. Reprogramming biological peptides to combat infectious diseases. Chem. Commun. 2019, 55, 15020–15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for the delivery of antimicrobial peptides (Amps). Nanomaterials 2020, 10, 560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Kahlenberg, J.M.; Kaplan, M.J. Little peptide, big effects: The role of LL-37 in inflammation and autoimmune disease. J. Immunol. 2013, 191, 4895–4901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabisiak, A.; Murawska, N.; Fichna, J. LL-37: Cathelicidin-related antimicrobial peptide with pleiotropic activity. Pharmacol. Rep. 2016, 68, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Burton, M.F.; Steel, P.G. The chemistry and biology of LL-37. Nat. Prod. Rep. 2009, 26, 1572–1584. [Google Scholar] [CrossRef]
- Chen, X.; Zou, X.; Qi, G.; Tang, Y.; Guo, Y.; Si, J.; Liang, L. Roles and mechanisms of human cathelicidin LL-37 in cancer. Cell Physiol. Biochem. 2018, 47, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Pahar, B.; Madonna, S.; Das, A.; Albanesi, C.; Girolomoni, G. Immunomodulatory role of the antimicrobial ll-37 peptide in autoimmune diseases and viral infections. Vaccines 2020, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Struyfs, C.; Cammue, B.; Thevissen, K. Membrane-Interacting Antifungal Peptides. Front. Cell Dev. Biol. 2021, 9, 706. [Google Scholar] [CrossRef]
- Promore Pharma P. Ropocamptide–Healing of Chronic Wounds. Available online: https://www.promorepharma.com/en/ll-37-healing-of-chronic-wounds/ (accessed on 22 June 2021).
- Doran, P.M. Foreign protein production in plant tissue cultures. Curr. Opin. Biotechnol. 2000, 11, 199–204. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-made vaccines and reagents for the One Health initiative. Hum. Vaccin. Immunother. 2017, 13, 2912–2917. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Buyel, J.F. Molecular farming–the slope of enlightenment. Biotechnol. Adv. 2020, 40, 107519. [Google Scholar] [CrossRef]
- Moon, J.-Y.; Henzler-Wildman, K.A.; Ramamoorthy, A. Expression and purification of a recombinant LL-37 from Escherichia coli. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, X.; Li, H.; Lockridge, O.; Wang, G. A novel method for purifying recombinant human host defense cathelicidin LL-37 by utilizing its inherent property of aggregation. Protein Expr. Purif. 2007, 54, 157–165. [Google Scholar] [CrossRef]
- Krahulec, J.; Hyršová, M.; Pepeliaev, S.; Jílková, J.; Černý, Z.; Machálková, J. High level expression and purification of antimicrobial human cathelicidin LL-37 in Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 88, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Production of human antimicrobial peptide LL-37 in Escherichia coli using a thioredoxin–SUMO dual fusion system. Protein Expr. Purif. 2013, 87, 72–78. [Google Scholar] [CrossRef]
- Wei, X.; Wu, R.; Zhang, L.; Ahmad, B.; Si, D.; Zhang, R. Expression, purification, and characterization of a novel hybrid peptide with potent antibacterial activity. Molecules 2018, 23, 1491. [Google Scholar] [CrossRef] [Green Version]
- Twyman, R.M.; Stoger, E.; Schillberg, S.; Christou, P.; Fischer, R. Molecular farming in plants: Host systems and expression technology. Trends Biotechnol. 2003, 21, 570–578. [Google Scholar] [CrossRef]
- Yusibov, V.; Streatfield, S.J.; Kushnir, N. Clinical development of plant-produced recombinant pharmaceuticals: Vaccines, antibodies and beyond. Hum. Vaccines 2011, 7, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ritala, A.; Leelavathi, S.; Oksman-Caldentey, K.-M.; Reddy, V.; Laukkanen, M.-L. Recombinant barley-produced antibody for detection and immunoprecipitation of the major bovine milk allergen, β-lactoglobulin. Transgenic Res. 2014, 23, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Mirzaee, M.; Jalali-Javaran, M.; Moieni, A.; Zeinali, S.; Behdani, M. Expression of VGRNb-PE immunotoxin in transplastomic lettuce (Lactuca sativa L.). Plant Mol. Biol. 2018, 97, 103–112. [Google Scholar] [CrossRef]
- Magnusdottir, A.; Vidarsson, H.; Björnsson, J.M.; Örvar, B.L. Barley grains for the production of endotoxin-free growth factors. Trends Biotechnol. 2013, 31, 572–580. [Google Scholar] [CrossRef]
- Cary, J.W.; Rajasekaran, K.; Jaynes, J.M.; Cleveland, T.E. Transgenic expression of a gene encoding a synthetic antimicrobial peptide results in inhibition of fungal growth in vitro and in planta. Plant Sci. 2000, 154, 171–181. [Google Scholar] [CrossRef]
- Patiño-Rodríguez, O.; Ortega-Berlanga, B.; Llamas-González, Y.Y.; Flores-Valdez, M.A.; Herrera-Díaz, A.; Montes-de-Oca-Luna, R.; Korban, S.S.; Alpuche-Solís, Á.G. Transient expression and characterization of the antimicrobial peptide protegrin-1 in Nicotiana tabacum for control of bacterial and fungal mammalian pathogens. Plant Cell Tissue Organ Cult. (PCTOC) 2013, 115, 99–106. [Google Scholar] [CrossRef]
- Zeitler, B.; Bernhard, A.; Meyer, H.; Sattler, M.; Koop, H.-U.; Lindermayr, C. Production of a de-novo designed antimicrobial peptide in Nicotiana benthamiana. Plant Mol. Biol. 2013, 81, 259–272. [Google Scholar] [CrossRef]
- Chahardoli, M.; Fazeli, A.; Ghabooli, M. Recombinant production of bovine Lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol. Biochem. 2018, 123, 414–421. [Google Scholar] [CrossRef]
- Mirzaee, H.; Peralta, N.L.N.; Carvalhais, L.C.; Dennis, P.G.; Schenk, P.M. Plant-produced bacteriocins inhibit plant pathogens and confer disease resistance in tomato. New Biotechnol. 2021, 63, 54–61. [Google Scholar] [CrossRef]
- Almasia, N.I.; Bazzini, A.A.; Hopp, H.E.; Vazquez-Rovere, C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008, 9, 329–338. [Google Scholar] [CrossRef]
- Rivero, M.; Furman, N.; Mencacci, N.; Picca, P.; Toum, L.; Lentz, E.; Bravo-Almonacid, F.; Mentaberry, A. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J. Biotechnol. 2012, 157, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Hancock, R.E.; Mattoo, A.K.; Misra, S. Expression of an engineered heterologous antimicrobial peptide in potato alters plant development and mitigates normal abiotic and biotic responses. PLoS ONE 2013, 8, e77505. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.-M.; Xie, C.-J.; Wang, D.; Wei, Y.-M.; Cai, J.; Cheng, S.-S.; Yang, X.-Y.; Sui, A.-P. Psc-AFP from Psoralea corylifolia L. overexpressed in Pichia pastoris increases antimicrobial activity and enhances disease resistance of transgenic tobacco. Appl. Microbiol. Biotechnol. 2017, 101, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Lee, S.-Y.; Moon, Y.-S.; Kang, K.-K. Enhanced resistance to bacterial and fungal pathogens by overexpression of a human cathelicidin antimicrobial peptide (hCAP18/LL-37) in Chinese cabbage. Plant Biotechnol. Rep. 2012, 6, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.-J. Enhanced resistance to bacterial pathogen in transgenic tomato plants expressing cathelicidin antimicrobial peptide. Biotechnol. Bioprocess Eng. 2013, 18, 615–624. [Google Scholar] [CrossRef]
- Lee, I.H.; Jung, Y.-J.; Cho, Y.G.; Nou, I.S.; Huq, M.A.; Nogoy, F.M.; Kang, K.-K. SP-LL-37, human antimicrobial peptide, enhances disease resistance in transgenic rice. PLoS ONE 2017, 12, e0172936. [Google Scholar] [CrossRef] [Green Version]
- Holásková, E.; Galuszka, P.; Mičúchová, A.; Šebela, M.; Öz, M.T.; Frébort, I. Molecular farming in barley: Development of a novel production platform to produce human antimicrobial peptide LL-37. Biotechnol. J. 2018, 13, 1700628. [Google Scholar] [CrossRef]
- Ramessar, K.; Sabalza, M.; Capell, T.; Christou, P. Maize plants: An ideal production platform for effective and safe molecular pharming. Plant Sci. 2008, 174, 409–419. [Google Scholar] [CrossRef]
- Moustafa, K.; Makhzoum, A.; Trémouillaux-Guiller, J. Molecular farming on rescue of pharma industry for next generations. Crit. Rev. Biotechnol. 2016, 36, 840–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegel, H.; Stöger, E.; Twyman, R.M.; Buyel, J.F. Current status and perspectives of the molecular farming landscape. In Molecular Pharming: Applications, Challenges and Emerging Areas; Kermode, A.R., Jiang, L., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 1–23. [Google Scholar] [CrossRef]
- Buyel, J.F. Plant molecular farming–Integration and exploitation of side streams to achieve sustainable biomanufacturing. Front. Plant Sci. 2019, 9, 1893. [Google Scholar] [CrossRef]
- Martin, G.E.; Timko, M.P.; Wilks, H.M. Purification and kinetic analysis of pea (Pisum sativum L.) NADPH: Protochlorophyllide oxidoreductase expressed as a fusion with maltose-binding protein in Escherichia coli. Biochem. J. 1997, 325, 139–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Dzineku, F.; Blackshear, P.J. Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-α mRNA and serve as a substrate for mitogen-activated protein kinases. Arch. Biochem. Biophys. 2003, 412, 106–120. [Google Scholar] [CrossRef]
- Momin, A.A.; Hameed, U.F.S.; Arold, S.T. passenger sequences can promote interlaced dimers in a common variant of the maltose-binding protein. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Johansson, J.; Gudmundsson, G.H.; Rottenberg, M.E.; Berndt, K.D.; Agerberth, B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 1998, 273, 3718–3724. [Google Scholar] [CrossRef] [Green Version]
- Oren, Z.; Lerman, J.C.; Gudmundsson, G.H.; Agerberth, B.; Shai, Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: Relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 1999, 341, 501–513. [Google Scholar] [CrossRef]
- Sancho-Vaello, E.; François, P.; Bonetti, E.-J.; Lilie, H.; Finger, S.; Gil-Ortiz, F.; Gil-Carton, D.; Zeth, K. Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species. Sci. Rep. 2017, 7, 1–11. [Google Scholar]
- Christou, P.; Stoger, E.; Twyman, R.M. Monocot expression systems for molecular farming. Mol. Farming 2004, 55–67. [Google Scholar]
- Arcalis, E.; Ibl, V.; Peters, J.; Melnik, S.; Stoger, E. The dynamic behavior of storage organelles in developing cereal seeds and its impact on the production of recombinant proteins. Front. Plant Sci. 2014, 5, 439. [Google Scholar] [CrossRef] [Green Version]
- Hensel, G. Genetic transformation of Triticeae cereals–Summary of almost three-decade’s development. Biotechnol. Adv. 2020, 40, 107484. [Google Scholar] [CrossRef]
- Ritala, A.; Wahlström, E.; Holkeri, H.; Hafren, A.; Mäkeläinen, K.; Baez, J.; Mäkinen, K.; Nuutila, A.-M. Production of a recombinant industrial protein using barley cell cultures. Protein Expr. Purif. 2008, 59, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Hensel, G.; Himmelbach, A.; Chen, W.; Douchkov, D.K.; Kumlehn, J. Transgene expression systems in the Triticeae cereals. J. Plant Physiol. 2011, 168, 30–44. [Google Scholar] [CrossRef]
- Mrízová, K.; Holasková, E.; Öz, M.T.; Jiskrová, E.; Frébort, I.; Galuszka, P. Transgenic barley: A prospective tool for biotechnology and agriculture. Biotechnol. Adv. 2014, 32, 137–157. [Google Scholar] [CrossRef]
- Panting, M.; Holme, I.B.; Björnsson, J.M.; Brinch-Pedersen, H. Modulation of Barley (Hordeum vulgare L.) Grain Protein Sink-Source Relations Towards Human Epidermal Growth Factor Instead of B-hordein Storage Protein. Mol. Biotechnol. 2021, 63, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Stahl, R.; Horvath, H.; Van Fleet, J.; Voetz, M.; von Wettstein, D.; Wolf, N. T-DNA integration into the barley genome from single and double cassette vectors. Proc. Natl. Acad. Sci. USA 2002, 99, 2146–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanasienko, I.; Yemets, A.; Pirko, Y.; Korhkovyy, V.; Abumhadi, N.; Blume, Y.B. Generation of transgenic barley lines producing human lactoferrin using mutant alpha-tubulin gene as the selective marker. Cytol. Genet. 2011, 45, 1–6. [Google Scholar] [CrossRef]
- Kamenarova, K.; Gecheff, K.; Stoyanova, M.; Muhovski, Y.; Anzai, H.; Atanassov, A. Production of recombinant human lactoferin in transgenic barley. Biotechnol. Biotechnol. Equip. 2007, 21, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Eskelin, K.; Ritala, A.; Suntio, T.; Blumer, S.; Holkeri, H.; Wahlström, E.H.; Baez, J.; Mäkinen, K.; Maria, N.A. Production of a recombinant full-length collagen type I α-1 and of a 45-kDa collagen type I α-1 fragment in barley seeds. Plant Biotechnol. J. 2009, 7, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Erlendsson, L.S.; Muench, M.O.; Hellman, U.; Hrafnkelsdóttir, S.M.; Jonsson, A.; Balmer, Y.; Mäntylä, E.; Örvar, B.L. Barley as a green factory for the production of functional Flt3 ligand. Biotechnol. J. 2010, 5, 163–171. [Google Scholar] [CrossRef]
- Schünmann, P.H.; Coia, G.; Waterhouse, P.M. Biopharming the SimpliRED™ HIV diagnostic reagent in barley, potato and tobacco. Mol. Breed. 2002, 9, 113–121. [Google Scholar] [CrossRef]
- Hensel, G.; Floss, D.M.; Arcalis, E.; Sack, M.; Melnik, S.; Altmann, F.; Rutten, T.; Kumlehn, J.; Stoger, E.; Conrad, U. Transgenic production of an anti HIV antibody in the barley endosperm. PLoS ONE 2015, 10, e0140476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czubacka, A.; Sacco, E.; Olszak-Przybyś, H.; Doroszewska, T. Inheritance and effectiveness of two transgenes determining PVY resistance in progeny from crossing independently transformed tobacco lines. J. Appl. Genet. 2017, 58, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Shahar, N.; Landman, S.; Weiner, I.; Elman, T.; Dafni, E.; Feldman, Y.; Tuller, T.; Yacoby, I. The integration of multiple nuclear-encoded transgenes in the green alga Chlamydomonas reinhardtii results in higher transcription levels. Front. Plant Sci. 2020, 10, 1784. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-W.; Yu, X.-H.; Lemaux, P.G.; Cho, M.-J. Stability and inheritance of endosperm-specific expression of two transgenes in progeny from crossing independently transformed barley plants. Plant Cell Rep. 2009, 28, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Schulze, J. Barley. In Alien Gene Transfer in Crop Plants; Springer: New York, NY, USA, 2014; Volume 2, pp. 85–120. [Google Scholar]
- Kihara, M.; Okada, Y.; Kuroda, H.; Saeki, K.; Yoshigi, N.; Ito, K. Improvement of β-amylase thermostability in transgenic barley seeds and transgene stability in progeny. Mol. Breed. 2000, 6, 511–517. [Google Scholar] [CrossRef]
- Horvath, H.; Huang, J.; Wong, O.; Kohl, E.; Okita, T.; Kannangara, C.G.; von Wettstein, D. The production of recombinant proteins in transgenic barley grains. Proc. Natl. Acad. Sci. USA 2000, 97, 1914–1919. [Google Scholar] [CrossRef] [Green Version]
- Horvath, H.; Jensen, L.; Wong, O.; Kohl, E.; Ullrich, S.; Cochran, J.; Kannangara, C.; Von Wettstein, D. Stability of transgene expression, field performance and recombination breeding of transformed barley lines. Theor. Appl. Genet. 2001, 102, 1–11. [Google Scholar] [CrossRef]
- Cho, M.J.; Choi, H.W.; Jiang, W.; Ha, C.D.; Lemaux, P.G. Endosperm-specific expression of green fluorescent protein driven by the hordein promoter is stably inherited in transgenic barley (Hordeum vulgare) plants. Physiol. Plant. 2002, 115, 144–154. [Google Scholar] [CrossRef]
- Bregitzer, P.; Tonks, D. Inheritance and expression of transgenes in barley. Crop Sci. 2003, 43, 4–12. [Google Scholar] [CrossRef]
- Choi, H.; Yu, X.; Lemaux, P.; Cho, M. Stability and inheritance of endosperm-specific expression of two transgenes in progeny of crosses of independently transformed barley (Hordeum vulgare L.) plants. Vitr. Cell. Dev. Biol. 2004, 40, 68A. [Google Scholar]
- Desai, P.N.; Shrivastava, N.; Padh, H. Production of heterologous proteins in plants: Strategies for optimal expression. Biotechnol. Adv. 2010, 28, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Saberianfar, R.; Menassa, R. Strategies to increase expression and accumulation of recombinant proteins. Mol. Pharming Appl. Chall. Emerg. Areas 2018, 119–135. [Google Scholar]
- Gao, L.; Tian, Y.; Chen, M.-C.; Wei, L.; Gao, T.-G.; Yin, H.-J.; Zhang, J.-L.; Kumar, T.; Liu, L.-B.; Wang, S.-M. Cloning and functional characterization of epidermis-specific promoter MtML1 from Medicago truncatula. J. Biotechnol. 2019, 300, 32–39. [Google Scholar] [CrossRef]
- Li, Y.; Yu, L.; Wang, Q.; Zhao, X.; Li, X.; Qi, B. Analysis of the Promoter of Emb5 from Zea mays Identifies a Region of 523 bp Responsible for Its Embryo-Specific Activity. Plant Mol. Biol. Rep. 2020, 39, 288–300. [Google Scholar] [CrossRef]
- Takaiwa, F.; Wakasa, Y.; Ozawa, K.; Sekikawa, K. Improvement of production yield and extraction efficacy of recombinant protein by high endosperm-specific expression along with simultaneous suppression of major seed storage proteins. Plant Sci. 2021, 302, 110692. [Google Scholar] [CrossRef]
- Pallotta, M.; Graham, R.; Langridge, P.; Sparrow, D.; Barker, S. RFLP mapping of manganese efficiency in barley. Theor. Appl. Genet. 2000, 101, 1100–1108. [Google Scholar] [CrossRef]
- Holásková, E.; Galuszka, P.; Frébort, I.; Milek, J. A method of preparing barley plants that produce antimicrobial peptide. WO2019052588. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2019052588 (accessed on 21 March 2019).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirzaee, M.; Holásková, E.; Mičúchová, A.; Kopečný, D.J.; Osmani, Z.; Frébort, I. Long-Lasting Stable Expression of Human LL-37 Antimicrobial Peptide in Transgenic Barley Plants. Antibiotics 2021, 10, 898. https://doi.org/10.3390/antibiotics10080898
Mirzaee M, Holásková E, Mičúchová A, Kopečný DJ, Osmani Z, Frébort I. Long-Lasting Stable Expression of Human LL-37 Antimicrobial Peptide in Transgenic Barley Plants. Antibiotics. 2021; 10(8):898. https://doi.org/10.3390/antibiotics10080898
Chicago/Turabian StyleMirzaee, Malihe, Edita Holásková, Alžbeta Mičúchová, David J. Kopečný, Zhila Osmani, and Ivo Frébort. 2021. "Long-Lasting Stable Expression of Human LL-37 Antimicrobial Peptide in Transgenic Barley Plants" Antibiotics 10, no. 8: 898. https://doi.org/10.3390/antibiotics10080898
APA StyleMirzaee, M., Holásková, E., Mičúchová, A., Kopečný, D. J., Osmani, Z., & Frébort, I. (2021). Long-Lasting Stable Expression of Human LL-37 Antimicrobial Peptide in Transgenic Barley Plants. Antibiotics, 10(8), 898. https://doi.org/10.3390/antibiotics10080898






