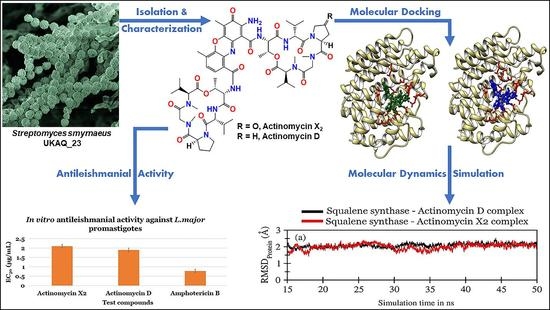
In Vitro and In Silico Approaches for the Antileishmanial Activity Evaluations of Actinomycins Isolated from Novel Streptomyces smyrnaeus Strain UKAQ_23
Abstract
1. Introduction
2. Results
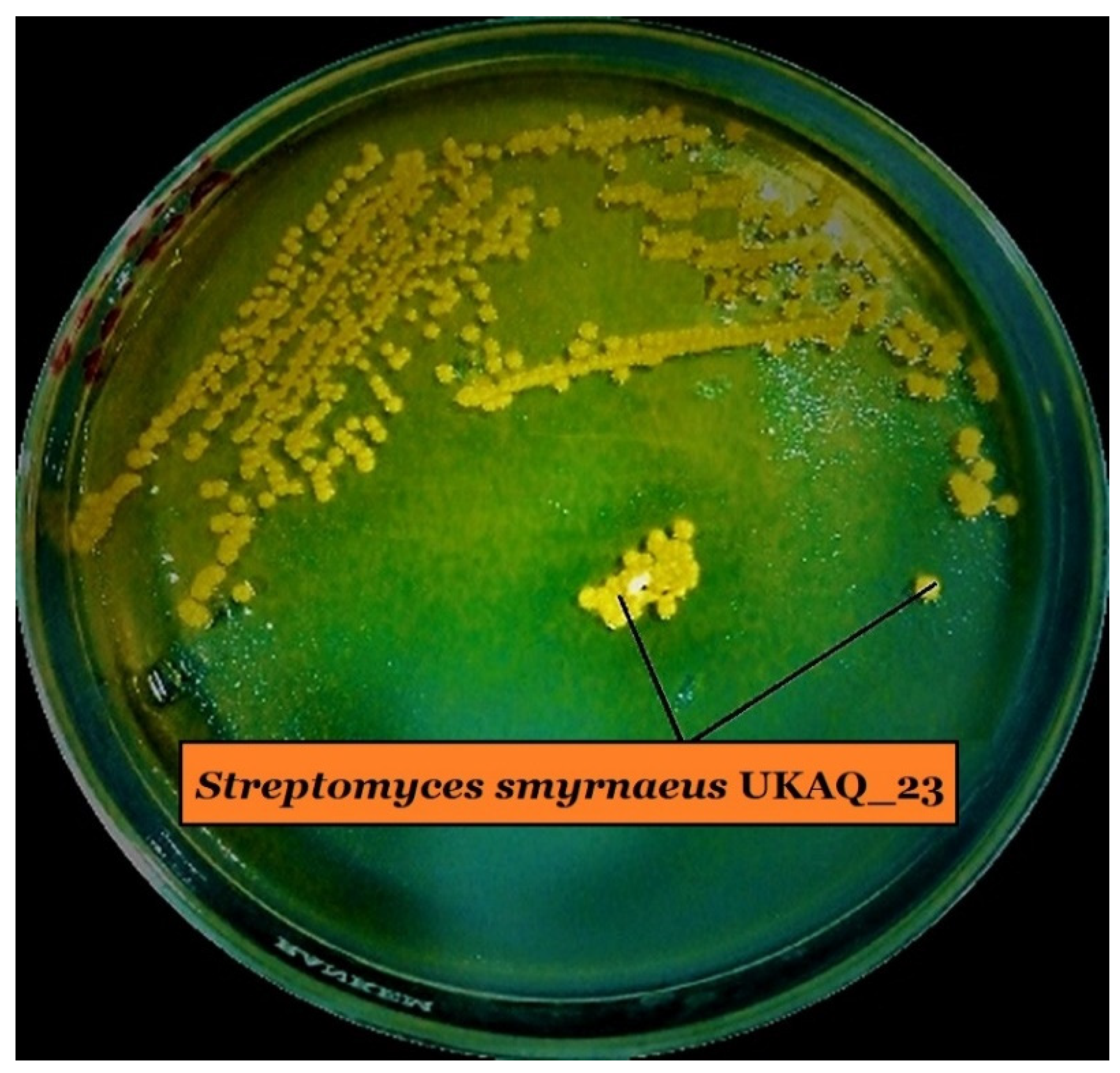
2.1. Isolation, Identification, and Characterization of Strain UKAQ_23
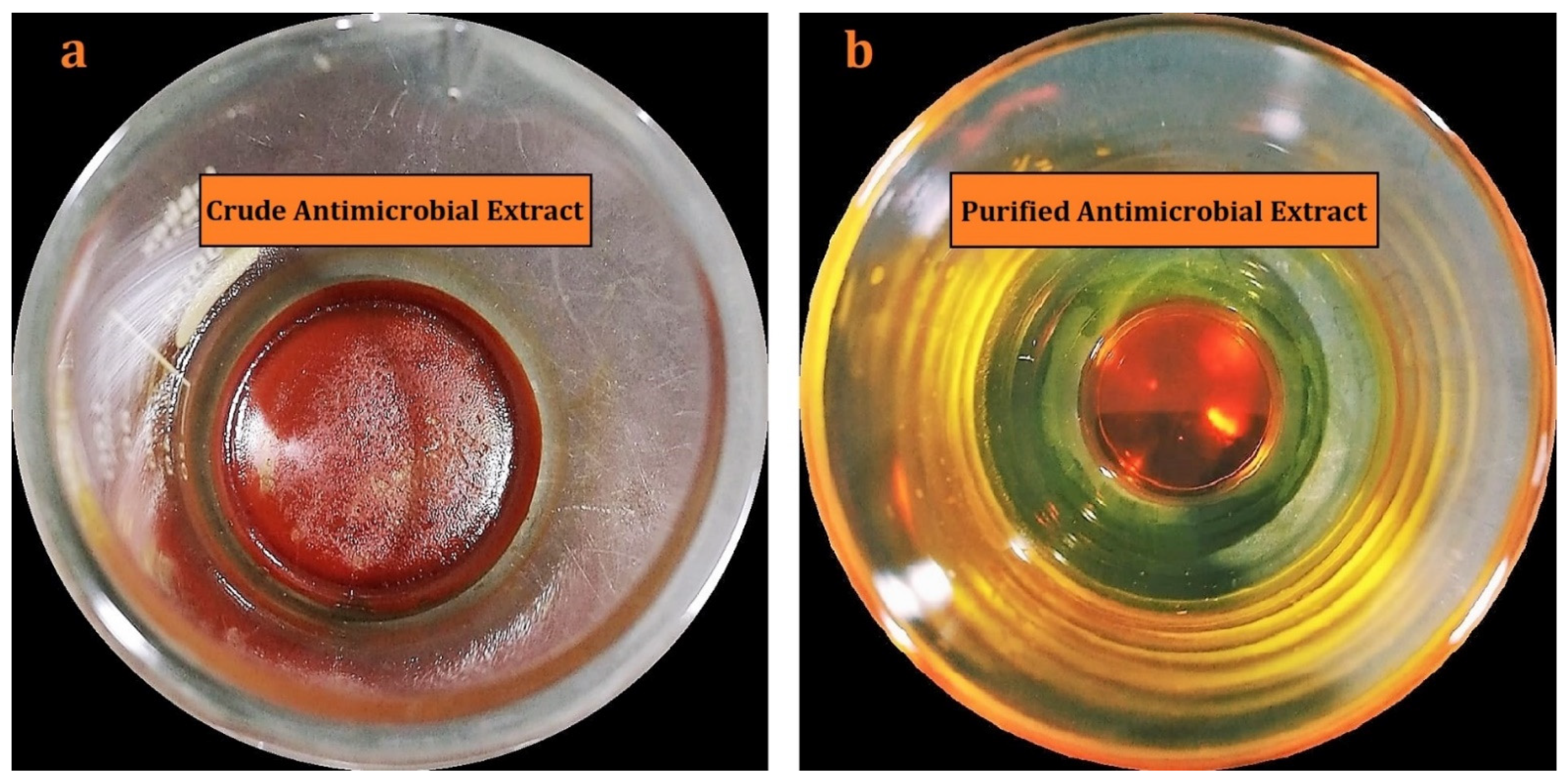
2.2. Isolation, Purification, and Characterization of Actinomycins X2 and D
2.3. Antileishmanial Activity Evaluations
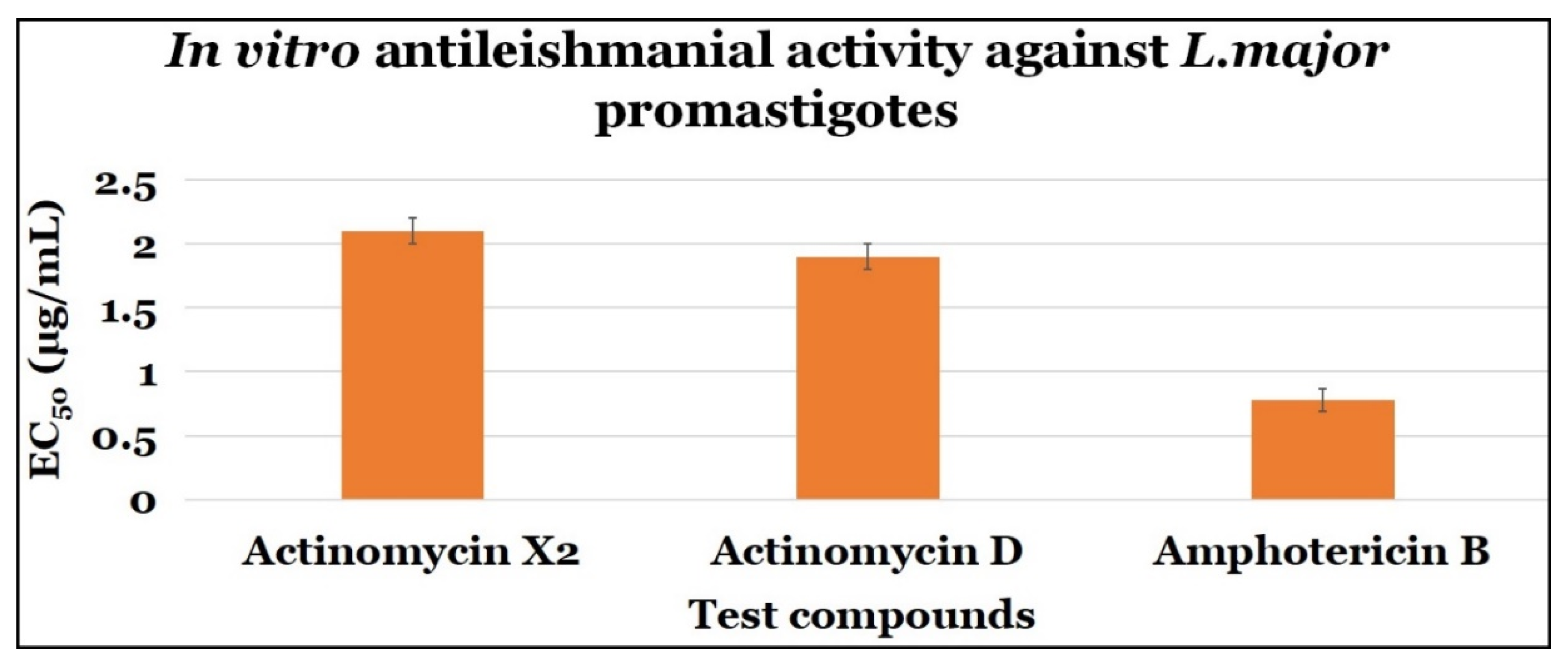
2.3.1. Anti-Promastigotes Evaluations
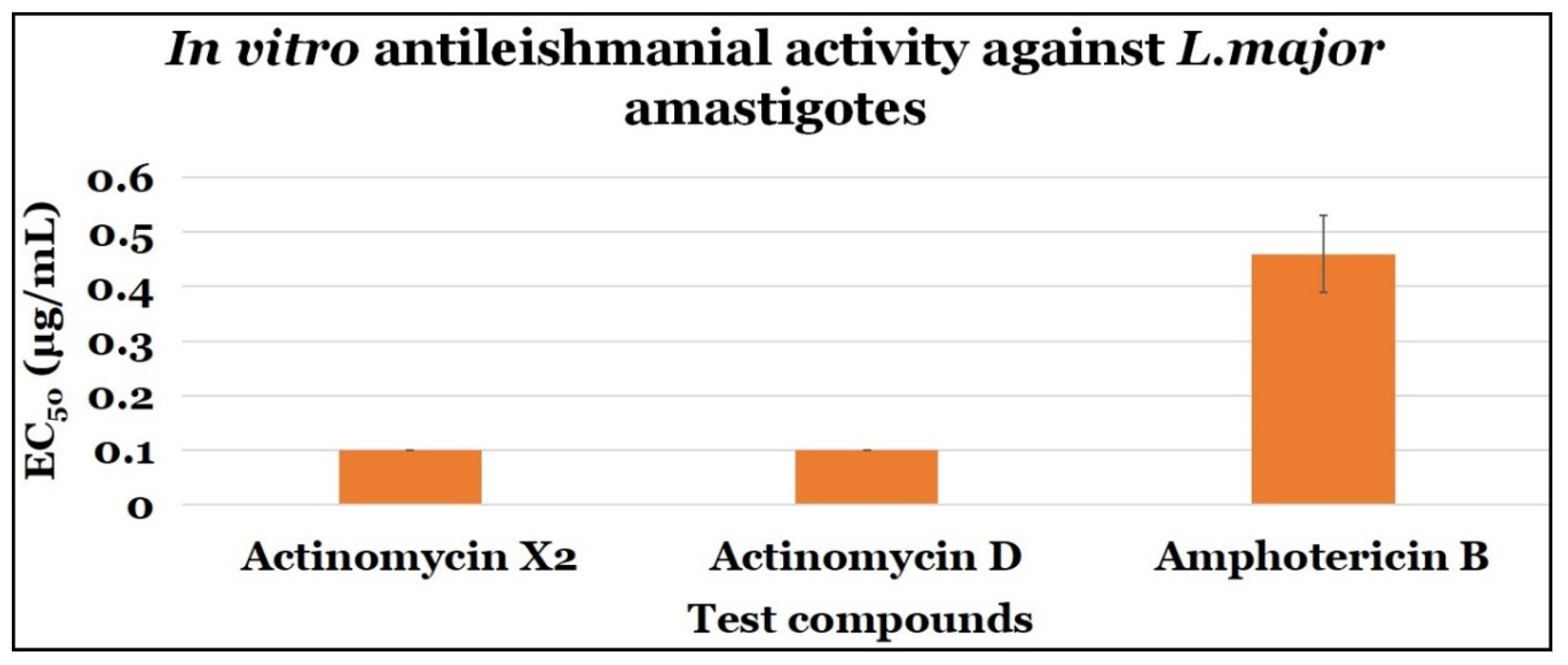
2.3.2. Anti-Amastigotes Evaluations
2.3.3. Statistical Analysis
2.4. In Silico Antileishmanial Activity Evaluations
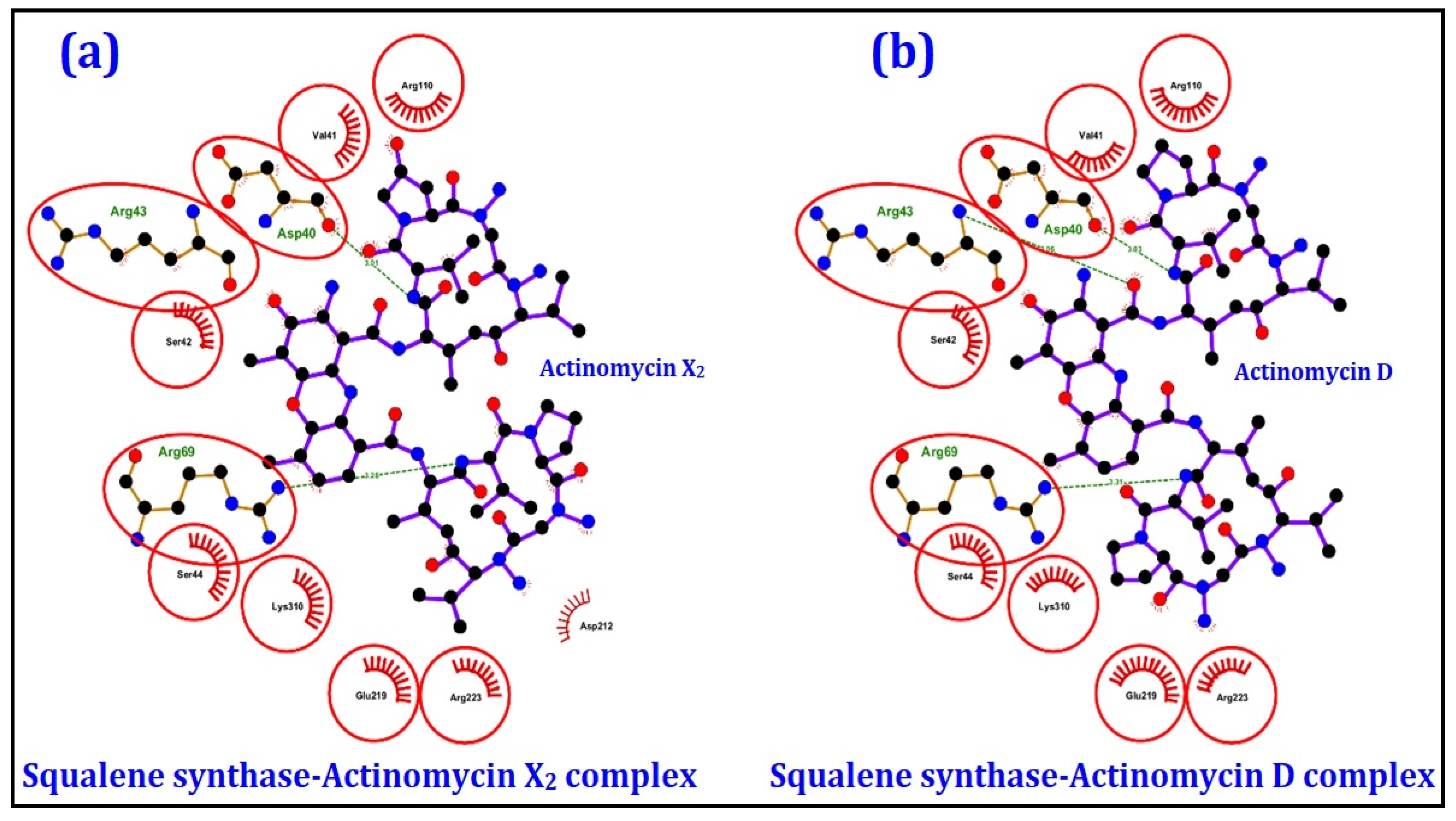
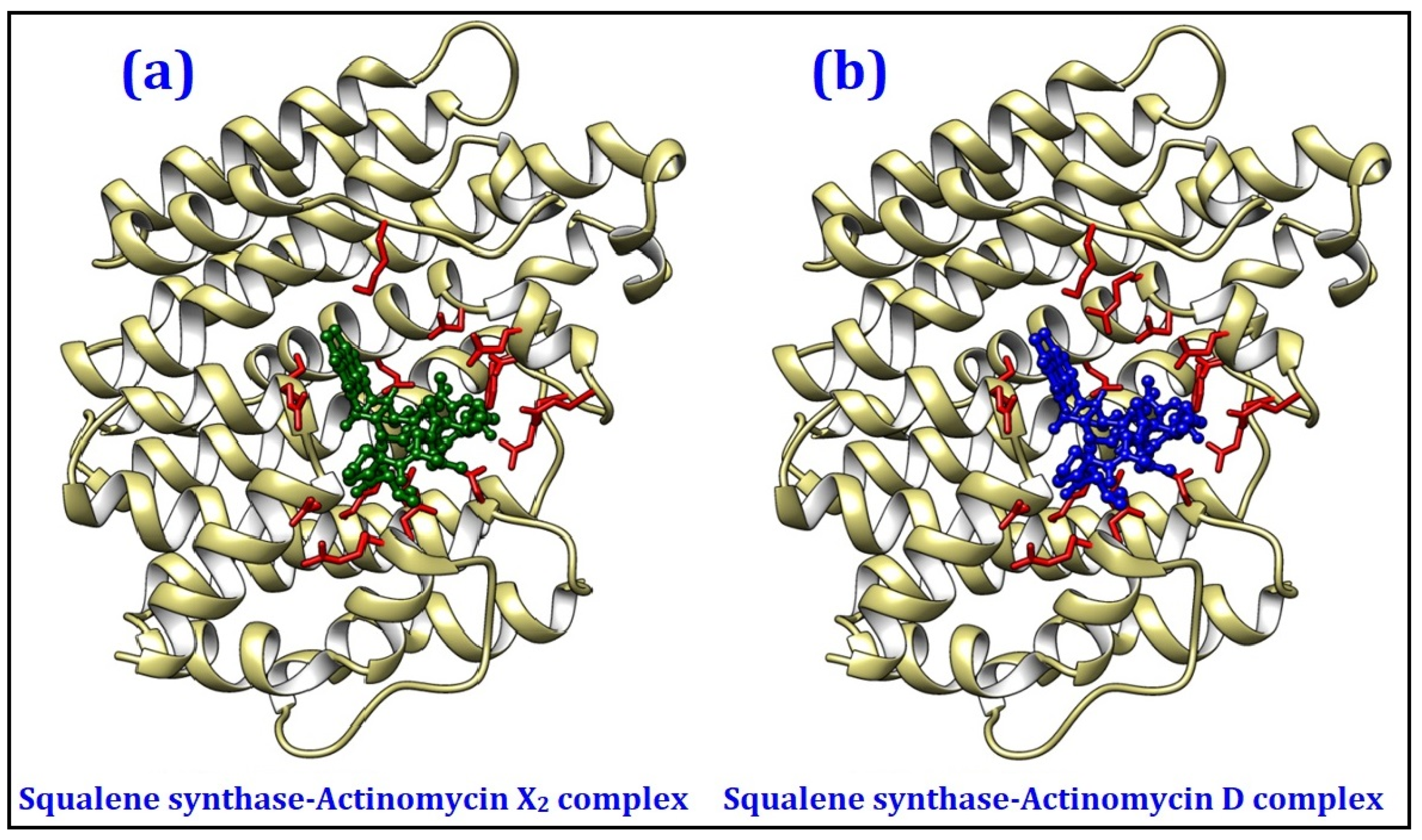
2.4.1. Molecular Docking Studies
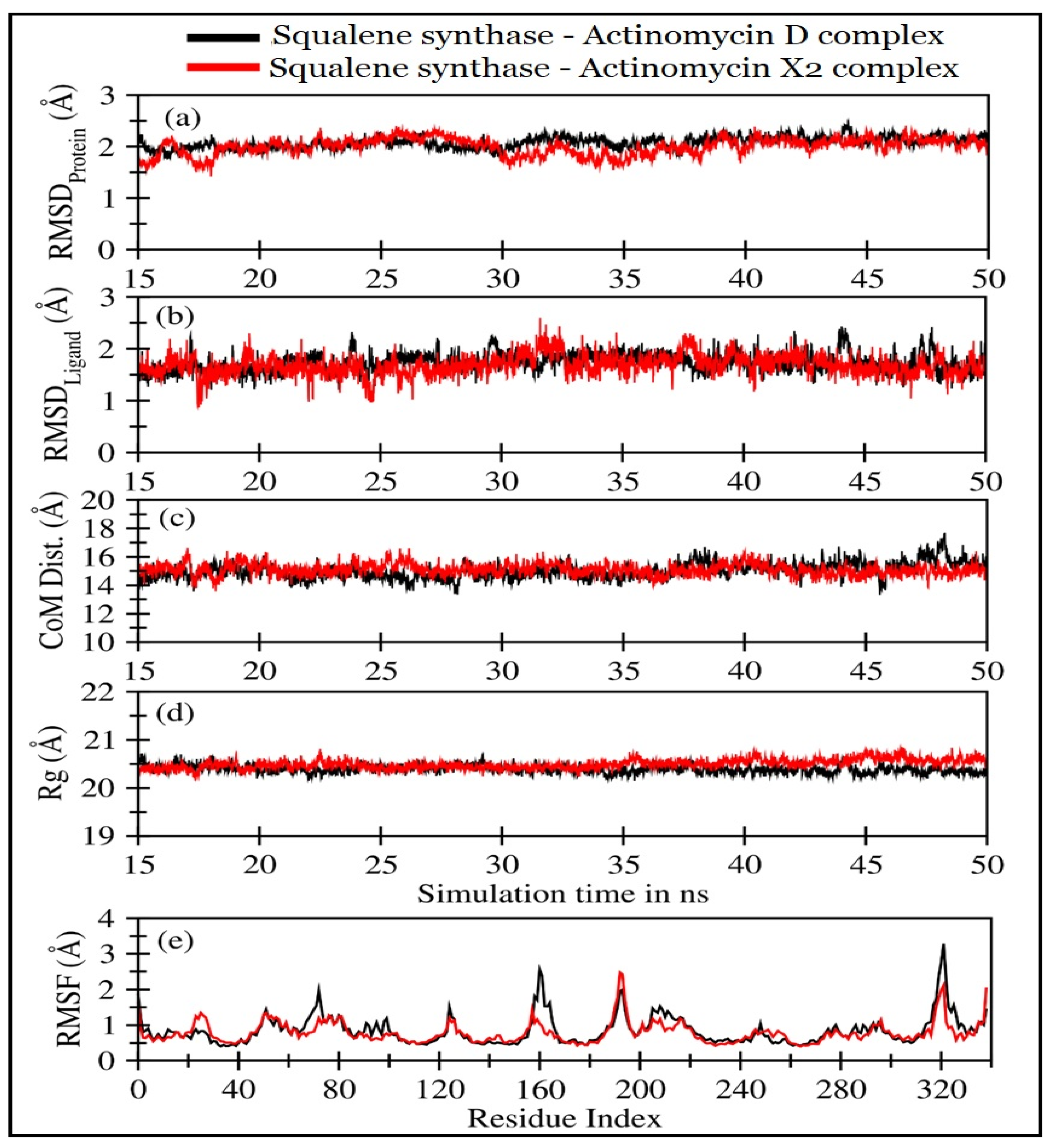
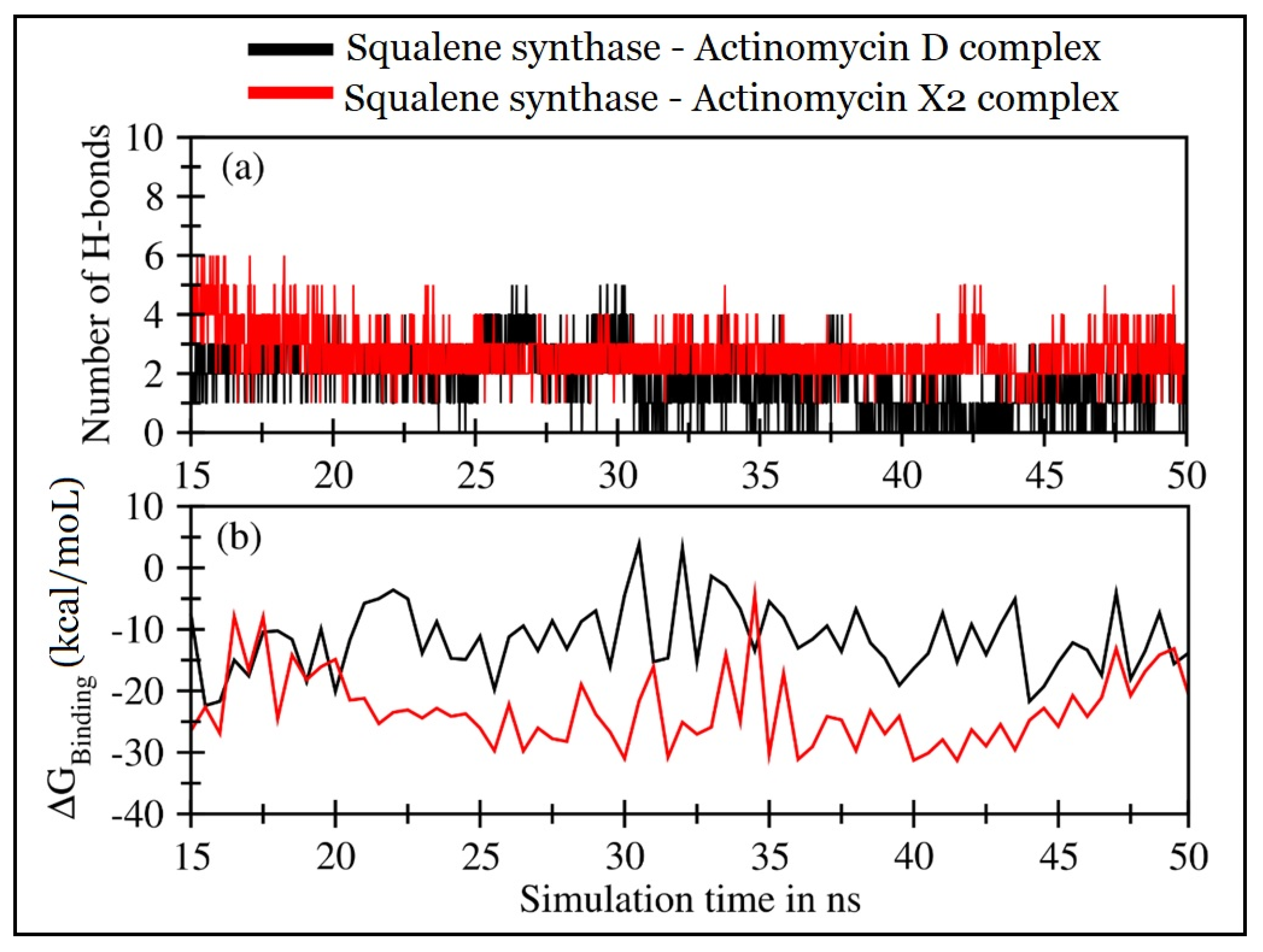
2.4.2. Molecular Dynamics (MD) Simulations
3. Discussions
4. Materials and Methods
4.1. Isolation, Identification, and Characterization of Strain UKAQ_23
4.2. Production, Purification, and Characterization of Actinomycins
4.3. In Vitro Antileishmanial Evaluations
4.3.1. Isolation of L. major and Culture Conditions
4.3.2. Anti-Promastigotes Evaluations
4.3.3. Anti-Amastigotes Evaluations
4.3.4. In Vitro Cytotoxicity Evaluations
4.3.5. Statistical Analysis
4.4. In Silico Antileishmanial Evaluations
4.4.1. Molecular Docking Evaluations
4.4.2. Molecular Dynamics Simulations
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chawla, B.; Madhubala, R. Drug targets in Leishmania. J. Parasit. Dis. 2010, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Al Nasr, I.; Jentzsch, J.; Winter, I.; Schobert, R.; Ersfeld, K.; Koko, W.S.; Mujawah, A.A.H.; Khan, T.A.; Biersack, B. Antiparasitic activities of new lawsone Mannich bases. Arch. Pharm. 2019, 352, 1–9. [Google Scholar] [CrossRef]
- Verma, R.K.; Prajapati, V.K.; Verma, G.K.; Chakraborty, D.; Sundar, S.; Rai, M.; Dubey, V.K.; Singh, M.S. Molecular docking and in vitro antileishmanial evaluation of chromene-2-thione analogues. ACS Med. Chem. Lett. 2012, 3, 243–247. [Google Scholar] [CrossRef]
- Khan, T.A.; Al Nasr, I.S.; Mujawah, A.H.; Koko, W.S. Assessment of Euphorbia retusa and Pulicaria undulata activity against Leishmania major and Toxoplasma gondii. Trop. Biomed. 2021, 38, 135–141. [Google Scholar] [CrossRef]
- Lodi, G.; Sannino, M.; Caterino, P.; Cannarozzo, G.; Bennardo, L.; Nisticò, S.P. Fractional CO2 laser-assisted topical rifamycin drug delivery in the treatment of pediatric cutaneous leishmaniasis. Pediatr. Dermatol. 2021, 38, 717–720. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.A.; Adlem, E.; Aert, R.; et al. The genome of the kinetoplastid parasite. Leishmania major. Sci. 2005, 309, 436–442. [Google Scholar] [CrossRef]
- Tanaka, Y.; Omura, S. Agroactive compounds of microbial origin. Annu. Rev. Microbiol. 1993, 47, 57–87. [Google Scholar] [CrossRef]
- Singh, L.S.; Baruah, I.; Bora, T.C. Actinomycetes of Loktak habitat: Isolation and screening for antimicrobial activities. Biotechnology 2006, 5, 217–221. [Google Scholar] [CrossRef]
- Sanasam, S.; Ningthoujam, D.S. Screening of local actinomycete isolates in Manipur for anticandidal activity. Asian J. Biotechnol. 2010, 2, 139–145. [Google Scholar] [CrossRef][Green Version]
- Jamal, Q. Antileishmanial, Cytotoxic and Genotoxic Effects of Actinomycin D, Z3, Z5 and Hydrazine Derivatives of Isosteviol; University of Peshawar: Peshawar, Pakistan, 2020; Available online: http://prr.hec.gov.pk/jspui/handle/123456789/12101 (accessed on 17 June 2021).
- Annang, F.; Pérez-Moreno, G.; García-Hernández, R.; Cordon-Obras, C.; Martín, J.; Tormo, J.R.; Rodríguez, L.; De Pedro, N.; Gómez-Pérez, V.; Valente, M.; et al. High-throughput screening platform for natural product-based drug discovery against 3 neglected tropical diseases: Human African trypanosomiasis, leishmaniasis, and chagas disease. J. Biomol. Screen. 2015, 20, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.; Guiguemde, W.A.; Hammill, J.T.; Carrillo, A.K.; Chen, Y.; Connlly, M.; Stalheim, K.; Elya, C.; Johnson, A.; Min, J.; et al. Discovery of novel, orally bioavailable, antileishmanial compounds using phenotypic screening. PLoS Negl. Trop. Dis. 2017, 11, e0006157. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Lebrun, C.; Duguet, T.; Talmont, F.; Beech, R.; Orlowski, S.; André, F.; Prichard, R.K.; Lespine, A. Structural model, functional modulation by ivermectin and tissue localization of Haemonchus contortus P-glycoprotein-13. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, K.A.; Bholay, A.D.; Rai, P.K.; Mohammed, H.A.; Khan, R.A.; Azam, F.; Jaremko, M.; Emwas, A.-H.; Stefanowicz, P.; Waliczek, M.; et al. Isolation, characterization, anti-MRSA evaluation, and in-silico multi-target anti-microbial validations of actinomycin X2 and actinomycin D produced by novel Streptomyces smyrnaeus UKAQ_23. Sci. Rep. 2021, 11, 14539. [Google Scholar] [CrossRef] [PubMed]
- Wadanambi, P.M.; Mannapperuma, U. Computational study to discover potent phytochemical inhibitors against drug target, squalene synthase from Leishmania donovani. Heliyon 2021, 7, e07178. [Google Scholar] [CrossRef]
- Urbina, J.A.; Concepcion, J.L.; Rangel, S.; Visbal, G.; Lira, R. Squalene synthase as a chemotherapeutic target in Trypanosoma cruzi and Leishmania mexicana. Mol. Biochem. Parasitol. 2002, 125, 35–45. [Google Scholar] [CrossRef]
- Rodrigues, J.C.F.; Urbina, J.A.; De Souza, W. Antiproliferative and ultrastructural effects of BPQ-OH, a specific inhibitor of squalene synthase, on Leishmania amazonensis. Exp. Parasitol. 2005, 111, 230–238. [Google Scholar] [CrossRef]
- Granthon, A.C.; Braga, M.V.; Rodrigues, J.C.F.; Cammerer, S.; Lorente, S.O.; Gilbert, I.H.; Urbina, J.A.; de Souza, W. Alterations on the growth and ultrastructure of Leishmania chagasi induced by squalene synthase inhibitors. Vet. Parasitol. 2007, 146, 25–34. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, F.; Wang, Q.; Abdel-Mageed, W.M.; Guo, H.; Fu, C.; Hou, W.; Dai, H.; Liu, X.; Yang, N.; et al. A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-Tuberculosis activity. Appl. Microbiol. Biotechnol. 2012, 95, 919–927. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wang, M.; Tan, Y.; Hu, X.; He, H.; Xiao, C.; You, X.; Wang, Y.; Gan, M. Neo-actinomycins A and B, natural actinomycins bearing the 5H-oxazolo[4,5-b]phenoxazine chromophore, from the marine-derived Streptomyces sp. IMB094. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- El-Naggar, M.Y.M.; El-Aassar, S.A.; Hashem, M.A.; Stoodley, R.J.; Raynor, C.M.; Sigee, D.C. Production of actinomycin X2 by immobilized Streptomyces nasri YG62 mycelia. Microbios 1998, 95, 165–179. [Google Scholar]
- Xiong, Z.Q.; Zhang, Z.P.; Li, J.H.; Wei, S.J.; Tu, G.Q. Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X2, fungichromin, and a new polyene macrolide antibiotic. Appl. Environ. Microbiol. 2012, 78, 589–592. [Google Scholar] [CrossRef]
- Bird, C.W.; Latif, M. Antibiotics from the newly isolated Streptomyces elizabethii. II. Isolation and characterisation of the antibiotics. J. Chem. Technol. Biotechnol. 2007, 31, 368–370. [Google Scholar] [CrossRef]
- Wei, Z.; Xu, C.; Wang, J.; Lu, F.; Bie, X.; Lu, Z. Identification and characterization of Streptomyces flavogriseus NJ-4 as a novel producer of actinomycin D and holomycin. PeerJ 2017, 5, e3601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kurosawa, K.; Bui, V.P.; VanEssendelft, J.L.; Willis, L.B.; Lessard, P.A.; Ghiviriga, I.; Sambandan, T.G.; Rha, C.K.; Sinskey, A.J. Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2-producer. Appl. Microbiol. Biotechnol. 2006, 72, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Praveen, V.; Tripathi, C.K.M. Studies on the production of actinomycin-D by Streptomyces griseoruber—A novel source. Lett. Appl. Microbiol. 2009, 49, 450–455. [Google Scholar] [CrossRef]
- Sharma, M.; Manhas, R.K. Purification and characterization of actinomycins from Streptomyces strain M7 active against methicillin resistant Staphylococcus aureus and vancomycin resistant Enterococcus. BMC Microbiol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Khieu, T.N.; Liu, M.J.; Nimaichand, S.; Quach, N.T.; Chu-Ky, S.; Phi, Q.T.; Vu, T.T.; Nguyen, T.D.; Xiong, Z.; Prabhu, D.M.; et al. Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front. Microbiol. 2015, 6, 574. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, C.; Gui, P.; Liu, H.; Khalaf, S.M.H.; Elsayed, E.A.; Wadaan, M.A.M.; Hozzein, W.N.; Zhu, W. Identification, bioactivity, and productivity of actinomycins from the marine-derived Streptomyces heliomycini. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Kulkarni, M.; Gorthi, S.; Banerjee, G.; Chattopadhyay, P. Production, characterization and optimization of actinomycin D from Streptomyces hydrogenans IB310, a(n antagonistic bacterium against phytopathogens. Biocatal. Agric. Biotechnol. 2017, 10, 69–74. [Google Scholar] [CrossRef]
- Kaplum, V.; Cogo, J.; Sangi, D.P.; Ueda-Nakamura, T.; Corrêa, A.G.; Nakamura, C.V. In vitro and in vivo activities of 2,3-diarylsubstituted quinoxaline derivatives against Leishmania amazonensis. Antimicrob. Agents Chemother. 2016, 60, 3433–3444. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, K.A.; Seroor, M.; AI-Masabi, A.; Saykhan, M.A.; Mutairi, Y.A.; Elhassan, G.O.; Khan, R.A. Bio-characterizations of some marine bacterial strains isolated from mangrove sediment samples of four major cities of Saudi Arabia. J. Environ. Biol. 2020, 41, 1003–1012. [Google Scholar] [CrossRef]
- Maiti, P.K.; Mandal, S. Majority of actinobacterial strains isolated from kashmir himalaya soil are rich source of antimicrobials and industrially important biomolecules. Adv. Microbiol. 2019, 09, 220–238. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Sheldrick, G.M.; Bahner, I.; Lackner, H. Crystal structures of actinomycin D and actinomycin Z3. Angew. Chem. Int. Ed. 1998, 37, 2381–2384. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]

| Compounds | Anti-Promastigotes Evaluation | ||
|---|---|---|---|
| EC50 (μg/mL) | CC50 (μg/mL) | SI | |
| Actinomycin X2 | 2.10 ± 0.10 | 0.10 ± 0.0 | 0.048 |
| Actinomycin D | 1.90 ± 0.10 | 0.10 ± 0.0 | 0.053 |
| Amphotericin B | 0.78 ± 0.09 | 7.40 ± 2.64 | 9.490 |
| Compounds | Anti-Amastigotes Evaluation | ||
|---|---|---|---|
| EC50 (μg/mL) | CC50 (μg/mL) | SI | |
| Actinomycin X2 | 0.10 ± 0.0 | 0.10 ± 0.0 | 1.0 |
| Actinomycin D | 0.10 ± 0.0 | 0.10 ± 0.0 | 1.0 |
| Amphotericin B | 0.46 ± 0.07 | 7.4 ± 2.64 | 16.09 |
| Enzymes | Pathway | Binding Energy (kcal/mol) | |
|---|---|---|---|
| Actinomycin X2 | Actinomycin D | ||
| Trypanothione reductase | Trypanothione pathway | −8.8 | −8.8 |
| Trypanothione synthetase-amidase | Trypanothione pathway | −8.5 | −8.1 |
| Tryparedoxin peroxidase | Trypanothione pathway | −8.0 | −8.1 |
| Squalene synthase | Sterol biogenetic pathway | −10.0 | −9.9 |
| Squalene monooxygenase | Sterol biogenetic pathway | −7.5 | −7.3 |
| Farnesyl pyrophosphate synthase | Sterol biogenetic pathway | −8.3 | −8.0 |
| Glyceraldehyde-3-phosphate dehydrogenase | Glycolytic pathway | −7.7 | −7.5 |
| Triosephosphate isomerase | Glycolytic pathway | −6.8 | −6.7 |
| Phosphoglycerate kinase | Glycolytic pathway | −8.0 | −8.1 |
| Pyruvate kinase | Glycolytic pathway | −8.0 | −7.7 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | Glycolytic pathway | −7.9 | −7.8 |
| Fructose-bisphosphate aldolase | Glycolytic pathway | −8.7 | −8.7 |
| Adenine phosphoribosyltransferase | Purine salvage pathway | −7.6 | −7.3 |
| Xanthine phosphoribosyltransferase | Purine salvage pathway | −8.8 | −5.9 |
| Deoxyhypusine hydroxylase | Hypusine biosynthetic pathway | −8.5 | −8.4 |
| Squalene Synthase Complexed with: | RMSDProtein (Å) | RMSDLigand (Å) | CoMProtein-Ligand (Å) | Rg (Å) |
|---|---|---|---|---|
| Actinomycin X2 | 1.92 ± 0.25 | 1.52 ± 0.30 | 14.80 ± 0.69 | 20.48 ± 0.1 |
| Actinomycin D | 1.96 ± 0.27 | 1.58 ± 0.31 | 15.05 ± 0.57 | 20.40 ± 0.09 |
| Squalene Synthase Complexed with: | ∆EVDW (Van der Waal’s Energy) | ∆Eelec (Coulombic Energy) | ∆GPB (Poisson-Boltzmann Polar Solvation Energy) | ∆ESASA (Non-Polar Solvation Energy) | ∆GMMPBSA (Protein-Ligand Binding Energy) |
|---|---|---|---|---|---|
| Actinomycin X2 | −45.14 ± 5.33 | −35.37 ± 10.16 | 62.93 ± 10.62 | −5.49 ± 0.38 | −23.07 ± 5.96 |
| Actinomycin D | −41.78 ± 5.217 | −12.31 ± 11.48 | 47.95 ± 11.05 | −5.42 ± 0.38 | −11.57 ± 5.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qureshi, K.A.; Al Nasr, I.; Koko, W.S.; Khan, T.A.; Fatmi, M.Q.; Imtiaz, M.; Khan, R.A.; Mohammed, H.A.; Jaremko, M.; Emwas, A.-H.; et al. In Vitro and In Silico Approaches for the Antileishmanial Activity Evaluations of Actinomycins Isolated from Novel Streptomyces smyrnaeus Strain UKAQ_23. Antibiotics 2021, 10, 887. https://doi.org/10.3390/antibiotics10080887
Qureshi KA, Al Nasr I, Koko WS, Khan TA, Fatmi MQ, Imtiaz M, Khan RA, Mohammed HA, Jaremko M, Emwas A-H, et al. In Vitro and In Silico Approaches for the Antileishmanial Activity Evaluations of Actinomycins Isolated from Novel Streptomyces smyrnaeus Strain UKAQ_23. Antibiotics. 2021; 10(8):887. https://doi.org/10.3390/antibiotics10080887
Chicago/Turabian StyleQureshi, Kamal A., Ibrahim Al Nasr, Waleed S. Koko, Tariq A. Khan, M. Qaiser Fatmi, Mahrukh Imtiaz, Riaz A. Khan, Hamdoon A. Mohammed, Mariusz Jaremko, Abdul-Hamid Emwas, and et al. 2021. "In Vitro and In Silico Approaches for the Antileishmanial Activity Evaluations of Actinomycins Isolated from Novel Streptomyces smyrnaeus Strain UKAQ_23" Antibiotics 10, no. 8: 887. https://doi.org/10.3390/antibiotics10080887
APA StyleQureshi, K. A., Al Nasr, I., Koko, W. S., Khan, T. A., Fatmi, M. Q., Imtiaz, M., Khan, R. A., Mohammed, H. A., Jaremko, M., Emwas, A.-H., Azam, F., Bholay, A. D., Elhassan, G. O., & Prajapati, D. K. (2021). In Vitro and In Silico Approaches for the Antileishmanial Activity Evaluations of Actinomycins Isolated from Novel Streptomyces smyrnaeus Strain UKAQ_23. Antibiotics, 10(8), 887. https://doi.org/10.3390/antibiotics10080887