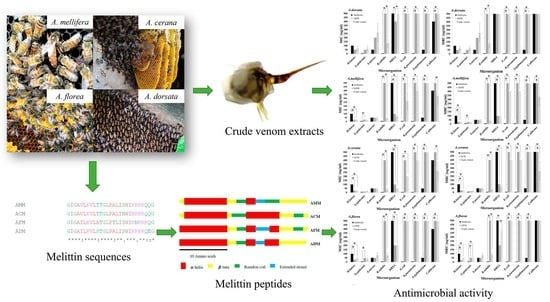
Comparative Study of Antimicrobial Properties of Bee Venom Extracts and Melittins of Honey Bees
Abstract
:1. Introduction
2. Results
2.1. Melittin Sequence
2.2. Amino Acid Composition
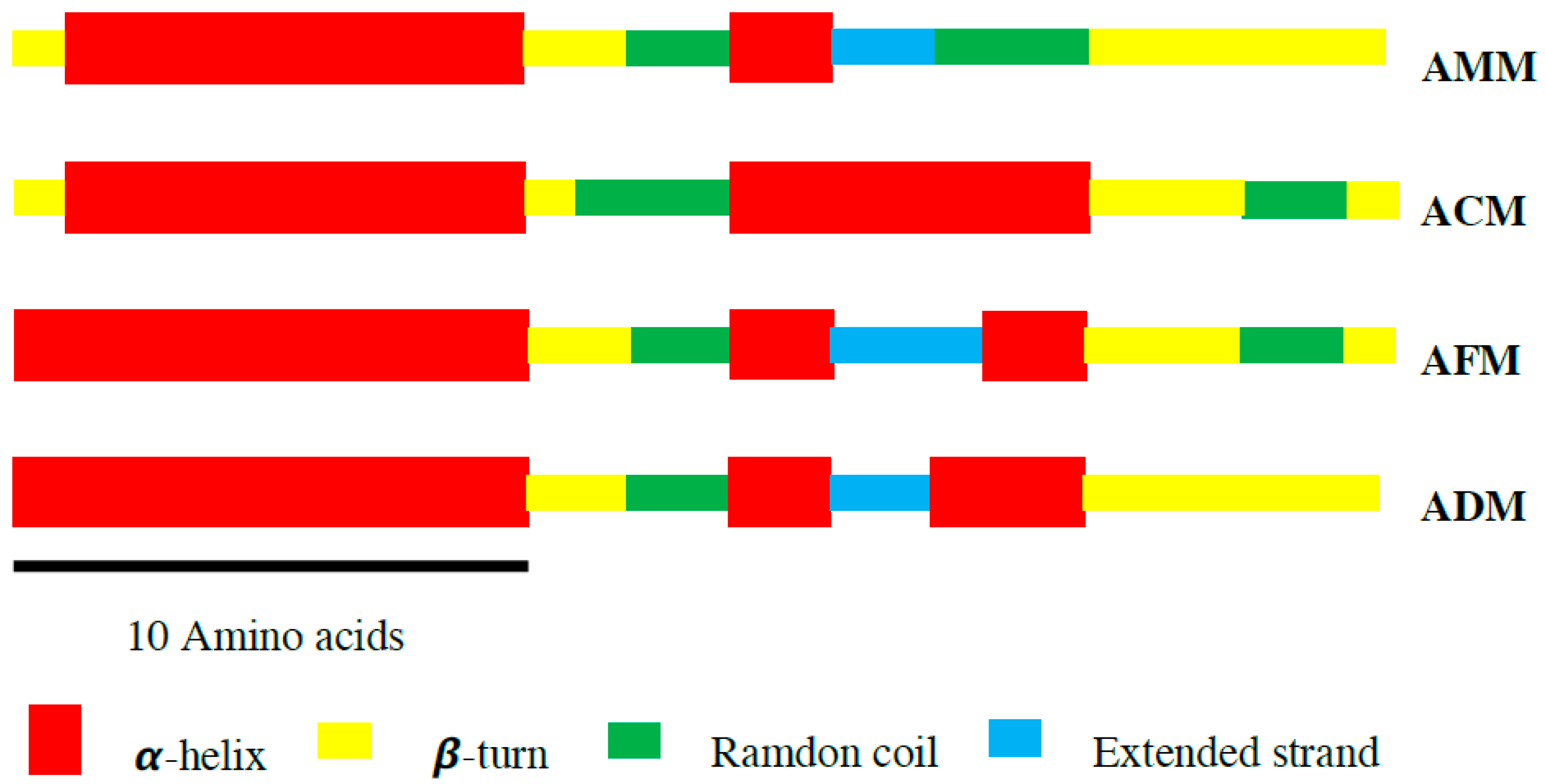
2.3. Secondary Structure
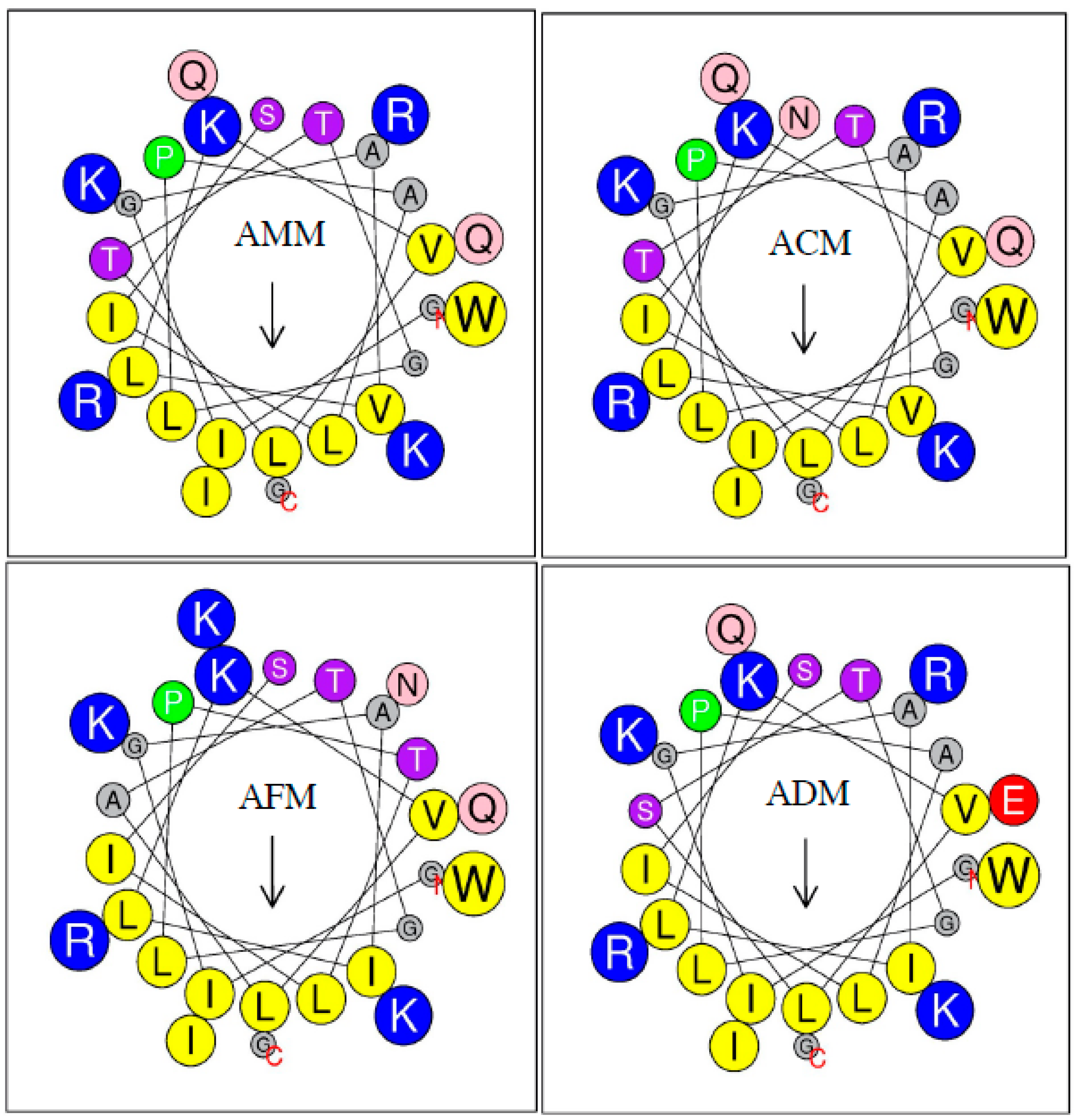
2.4. Primary Sequence Analysis
2.5. Physicochemical Parameters
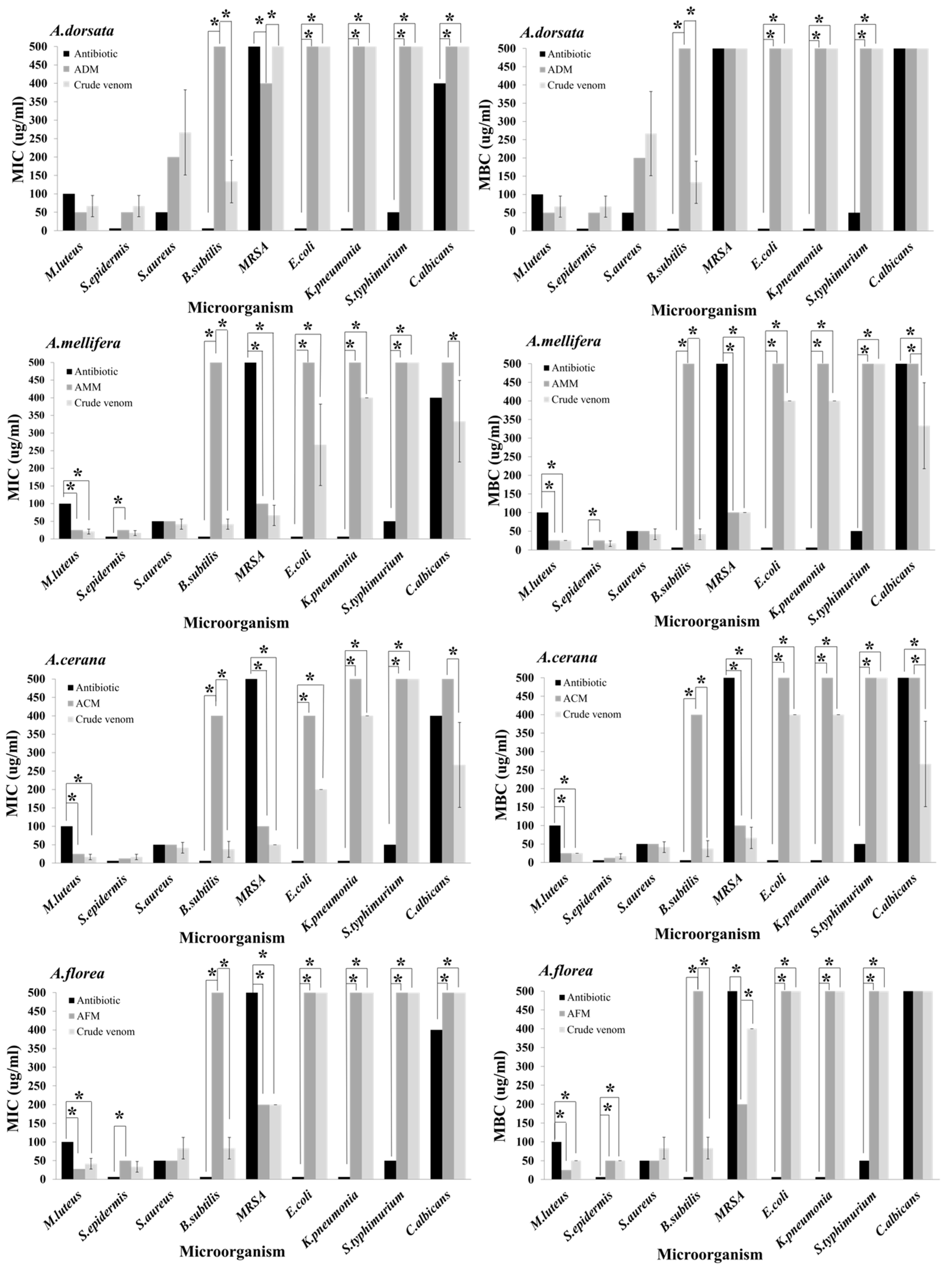
2.6. Antimicrobial Activities of Four Crude Honey Bee Venoms
2.7. Antimicrobial Activities of Four Melittin Peptides
2.8. Comparison of Antimicrobial Activity between Venoms and Their Melittin Peptides
3. Discussion
4. Materials and Methods
4.1. Collection and Preparation of the Bee Venom Extracts
4.2. Genomic DNA Isolation
4.3. Melittin Sequences
4.4. Peptide Synthesis
4.5. Tested Microorganisms
4.6. Detection of Minimum Inhibitory and Minimum Bactericidal Concentrations (MIC and MBC)
4.7. Bioinformatics Analysis for Melittin Structures
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Surendra, N.S.; Jayaram, G.N.; Reddy, M.R.S.; Ravikumar, H. Comparative morphometric studies of the sting apparatus of the worker bees of four different Apis species (Apis dorsata, Apis mellifera, Apis cerana and Apis florea). J. Apic. Res. 2013, 52, 74–80. [Google Scholar] [CrossRef]
- El-Seedi, H.; El-Wahed, A.; Yosri, N.; Musharraf, S.G.; Chen, L.; Moustafa, M.; Zou, X.; Al-Mousawi, S.; Guo, Z.; Khatib, A. Antimicrobial properties of Apis mellifera’s bee venom. Toxins 2020, 12, 451. [Google Scholar] [CrossRef] [PubMed]
- Son, D.J.; Lee, J.W.; Lee, Y.H.; Song, H.S.; Lee, C.K.; Hong, J.T. Therapeutics. Therapeutic application of anti-arthritis, pain-releasing, and anti-cancer effects of bee venom and its constituent compounds. Pharmacol. Ther. 2007, 115, 246–270. [Google Scholar] [CrossRef]
- Sobral, F.; Sampaio, A.; Falcão, S.; Queiroz, M.J.R.; Calhelha, R.C.; Vilas-Boas, M.; Ferreira, I.C.F.R. Chemical characterization, antioxidant, anti-inflammatory and cytotoxic properties of bee venom collected in Northeast Portugal. Food Chem. Toxicol. 2016, 94, 172–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.-M.; Lee, K.-G.; Yeo, J.-H.; Kweon, H.-Y.; Kim, B.-S.; Kim, J.-M.; Baek, H.-J.; Kim, S.-T. Antibacterial activity of the honey bee venom against bacterial mastitis pathogens infecting dairy cows. Int. J. Ind. Entomol. 2007, 14, 137–142. [Google Scholar]
- Lee, S.-B. Antifungal activity of bee venom and sweet bee venom against clinically isolated Candida albicans. J. Pharmacopunct. 2016, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Mata, É.C.G.d.; Mourão, C.B.F.; Rangel, M.; Schwartz, E.F.J.J.o.V.A.; Diseases, T.i.T. Antiviral activity of animal venom peptides and related compounds. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somwongin, S.; Chantawannakul, P.; Chaiyana, W. Antioxidant activity and irritation property of venoms from Apis species. Toxicon 2018, 145, 32–39. [Google Scholar] [CrossRef]
- Popplewell, J.; Swann, M.; Freeman, N.; McDonnell, C.; Ford, R.C. Quantifying the effects of melittin on liposomes. Biochim. Biophys. Acta BBA-Biomembr. 2007, 1768, 13–20. [Google Scholar] [CrossRef]
- Brown, L.R.; Lauterwein, J.; Wüthrich, K. High-resolution 1H-NMR studies of self-aggregation of melittin in aqueous solution. Biochim. Biophys. Acta BBA-Protein Struct. 1980, 622, 231–244. [Google Scholar] [CrossRef]
- Jamasbi, E.; Batinovic, S.; Sharples, R.A.; Sani, M.-A.; Robins-Browne, R.M.; Wade, J.D.; Separovic, F.; Hossain, M.A. Melittin peptides exhibit different activity on different cells and model membranes. Amino Acids 2014, 46, 2759–2766. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Jung, J.W.; Lee, M.O.; Lee, S.Y.; Kim, B.; Jin, H.J.; Kim, J.; Ahn, Y.-J.; Lee, K.W.; Song, Y.S.; et al. Functional characterization of naturally occurring melittin peptide isoforms in two honey bee species, Apis mellifera and Apis cerana. Peptides 2014, 53, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Garnery, L.; Vautrin, D.; Cornuet, J.M.; Solignac, M. Phylogenetic relationships in the genus Apis inferred from mitochondrial DNA sequence data. Apidologie 1991, 22, 87–92. [Google Scholar] [CrossRef] [Green Version]
- Surendra, N.S.; Jayaram, G.N.; Reddy, M.S. Antimicrobial activity of crude venom extracts in honeybees (Apis cerana, Apis dorsata, Apis florea) tested against selected pathogens. Afr. J. Microbiol. Res. 2011, 5, 2765–2772. [Google Scholar] [CrossRef] [Green Version]
- Owen, M.D.; Pfaff, L.A. Melittin synthesis in the venom system of the honey bee (Apis mellifera L.). Toxicon 1995, 33, 1181–1188. [Google Scholar] [CrossRef]
- Banks, B.E.C.; Shipolini, R.A. 7—Chemistry and Pharmacology of Honey-bee Venom A2-Piek, Tom. In Venoms of the Hymenoptera; Academic Press: Cambridge, MA, USA, 1986; pp. 329–416. [Google Scholar]
- Dotimas, E.M.; Hider, R.C. Honeybee Venom. Bee World 1987, 68, 51–70. [Google Scholar] [CrossRef]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A Membrane-active Peptide with Diverse Functions. Biosci. Rep. 2007, 27, 189. [Google Scholar] [CrossRef]
- Feigin, A.M.; Teeter, J.H.; Brand, J.G. The Influence of Sterols on the Sensitivity of Lipid Bilayers to Melittin. Biochem. Biophys. Res. Commun. 1995, 211, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Zinser, E.; Sperka-Gottlieb, C.D.; Fasch, E.V.; Kohlwein, S.D.; Paltauf, F.; Daum, G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991, 173, 2026–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, S.N.; Chaudhuri, K. (Eds.) Gram-Negative Bacteria: The cell Membranes. In Outer Membrane Vesicles of Bacteria; Springer: Berlin/Heidelberg, Germany, 2012; pp. 15–34. [Google Scholar]
- Navarre, W.W.; Schneewind, O. Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef] [Green Version]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, α-helical antimicrobial peptides. Pept. Sci. 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Pept. Sci. 2008, 90, 369–383. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Nakamura, A.; Murase, O.; Sugishita, K.-i.; Fujii, N.; Miyajima, K. Modulation of Magainin 2−Lipid Bilayer Interactions by Peptide Charge. Biochemistry 1997, 36, 2104–2111. [Google Scholar] [CrossRef]
- Thành, M.X. Effect of Secondary Structure on Biological Activities of Antimicrobial Peptides. VNU J. Sci. Nat. Sci. Technol. 2015, 31, 44–53. [Google Scholar]
- Ringstad, L.; Andersson Nordahl, E.; Schmidtchen, A.; Malmsten, M. Composition Effect on Peptide Interaction with Lipids and Bacteria: Variants of C3a Peptide CNY21. Biophys. J. 2007, 92, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieprecht, T.; Dathe, M.; Beyermann, M.; Krause, E.; Maloy, W.L.; MacDonald, D.L.; Bienert, M. Peptide Hydrophobicity Controls the Activity and Selectivity of Magainin 2 Amide in Interaction with Membranes. Biochemistry 1997, 36, 6124–6132. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Lev, N.; Shai, Y. Effect of the Hydrophobicity to Net Positive Charge Ratio on Antibacterial and Anti-Endotoxin Activities of Structurally Similar Antimicrobial Peptides. Biochemistry 2010, 49, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of Peptide Hydrophobicity in the Mechanism of Action of α-Helical Antimicrobial Peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevalainen, T.J.; Graham, G.G.; Scott, K.F. Antibacterial actions of secreted phospholipases A2. Review. Biochim. Et Biophys. Acta BBA-Mol. Cell Biol. Lipids 2008, 1781, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, A.P.; Wu, Y.-Z.; Paya, M.; Delclaux, C.; Touqui, L.; Goossens, P.L. High bactericidal efficiency of type IIA phospholipase A2 against Bacillus anthracis and inhibition of its secretion by the lethal toxin. J. Immunol. 2004, 173, 521. [Google Scholar] [CrossRef] [PubMed]
- Grönroos, J.O.; Laine, V.J.O.; Janssen, M.J.W.; Egmond, M.R.; Nevalainen, T.J. Bactericidal Properties of Group IIA and Group V Phospholipases A2. J. Immunol. 2001, 166, 4029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Third Informational Supplement M27-S3; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Wikler, M.A. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; CLSI: Wayne, PA, USA, 2006; Volume 26, p. M7-A7. [Google Scholar]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]

| No. | Species | Accession No. | Amino Acid Sequences | Reference |
|---|---|---|---|---|
| 1 | A. mellifera | AFI40556 | GIGAVLKVLTTGLPALISWIKRKRQQG | [12] |
| 2 | A. cerana | P0DPR9.1 | GIGAVLKVLTTGLPALINWIKRKRQQG | |
| 3 | A. florea | AMP82000 | GIGAILKVLATGLPTLISWIKNKRKQG | This study |
| 4 | A. dorsata | AMP81999 | GIGAILKVLSTGLPALISWIKRKRQEG |
| AMM | ACM | AFM | ADM | |
|---|---|---|---|---|
| No. of AA | 27 | 27 | 27 | 27 |
| Mw (Da) | 2904.54 | 2931.56 | 2876.52 | 2905.52 |
| pI | 12.02 | 12.02 | 11.33 | 11.10 |
| GRAVY | 0.248 | 0.148 | 0.281 | 0.256 |
| Instability index (II) | 43.44 | 39.60 | 34.64 | 50.58 |
| Aliphatic index | 130 | 130 | 133.7 | 133.7 |
| Extinction coefficients | 5500 | 5500 | 5500 | 5500 |
| R− | 0 | 0 | 0 | 1 |
| R+ | 5 | 5 | 5 | 5 |
| Hydrophobicity <H> | 0.492 | 0.471 | 0.500 | 0.487 |
| Hydrophobic moment <μH> | 0.380 | 0.400 | 0.412 | 0.401 |
| Net Charge (z) | 5 | 5 | 5 | 4 |
| Ala | 7.4 | 7.4 | 7.4 | 7.4 |
| Arg | 7.4 | 7.4 | 3.7 | 7.4 |
| Asn | 0 | 3.7 | 3.7 | 0 |
| Asp | 0 | 0 | 0 | 0 |
| Cys | 0 | 0 | 0 | 0 |
| Gln | 7.4 | 7.4 | 3.7 | 3.7 |
| Glu | 0 | 0 | 0 | 3.7 |
| Gly | 14.8 | 14.8 | 14.8 | 14.8 |
| His | 0 | 0 | 0 | 0 |
| Ile | 11.1 | 11.1 | 14.8 | 14.8 |
| Leu | 14.8 | 14.8 | 14.8 | 14.8 |
| Lys | 11.1 | 11.1 | 14.8 | 11.1 |
| Met | 0 | 0 | 0 | 0 |
| Phe | 0 | 0 | 0 | 0 |
| Pro | 3.7 | 3.7 | 3.7 | 3.7 |
| Ser | 3.7 | 0 | 3.7 | 7.3 |
| Thr | 7.4 | 7.4 | 7.4 | 3.7 |
| Trp | 3.7 | 3.7 | 3.7 | 3.7 |
| Tyr | 0 | 0 | 0 | 0 |
| Val | 7.4 | 7.4 | 3.7 | 3.7 |
| Pyl | 0 | 0 | 0 | 0 |
| Sec | 0 | 0 | 0 | 0 |
| α-Helix | β-Turn | Random Coil | Extended Strand | |
|---|---|---|---|---|
| AMM | 40.74 | 33.33 | 18.52 | 7.41 |
| ACM | 59.26 | 22.22 | 18.52 | 0 |
| AFM | 51.85 | 22.22 | 14.82 | 11.11 |
| ADM | 55.55 | 29.63 | 7.41 | 7.41 |
| Microorganism | MIC (μg/mL) | MBC/MFC (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| A. dorsatac | A. melliferaa | A. ceranaa | A. floreab | A. dorsatac | A. melliferaa | A. ceranaa | A. floreab | |
| Gram-negative bacteria | ||||||||
| E. coli | >400 | 266.7 ± 115.5 | 200.0 ± 0.0 | >400 | >400 | 400.0 ± 0.0 | 400.0 ± 0.0 | >400 |
| K. pneumonia | >400 | 400.0 ± 0.0 | 400.0 ± 0.0 | >400 | >400 | 400.0 ± 0.0 | 400.0 ± 0.0 | >400 |
| S. typhimurium | >400 | >400 | >400 | >400 | >400 | >400 | >400 | >400 |
| Gram-positive bacteria | ||||||||
| B. subtilis | 133.3 ± 57.7 | 41.7 ± 14.4 | 37.5 ± 21.7 | 83.3 ± 28.9 | 133.3 ± 57.7 | 41.7 ± 14.4 | 37.5 ± 21.7 | 83.3 ± 28.9 |
| M. luteus | 66.7.0 ± 28.9 | 20.8 ± 7.2 | 16.7 ± 7.2 | 41.7 ± 14.4 | 66.7 ± 28.9 | 25.0 ± 0.0 | 25.0 ± 0.0 | 50.0 ± 14.4 |
| S. aureus | 266.7 ± 115.5 | 41.7 ± 14.4 | 41.7 ± 14.4 | 83.3 ± 28.9 | 266.7 ± 115.5 | 41.7 ± 14.4 | 41.7 ± 14.4 | 83.3 ± 28.9 |
| MRSA | >400 | 66.7 ± 28.9 | 50.0 ± 0.0 | 200.0 ± 0.0 | >400 | 100.0 ± 0.0 | 66.7 ± 28.9 | 400.0 ± 0.0 |
| S. epidermidis | 66.7.0 ± 28.9 | 16.7 ± 7.2 | 16.7 ± 7.2 | 33.3 ± 14.4 | 66.7 ± 28.9 | 16.7 ± 7.2 | 16.7 ± 7.2 | 50.0 ± 0.0 |
| Fungus | ||||||||
| C. albicans | >400 | 333.3 ± 115.5 | 266.7 ± 115.5 | >400 | >400 | 333.3 ± 115.5 | 266.7 ± 115.5 | >400 |
| Microorganism | MIC (μg/mL) | MBC/MFC (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| ADM c | AMM b | ACM a | AFM b,c | ADM c | AMM a,b | ACM a | AFM b | |
| Gram-negative bacteria | ||||||||
| E. coli | >400 | >400 | 400 ± 0.0 | >400 | >400 | >400 | >400 | >400 |
| K. pneumonia | >400 | >400 | >400 | >400 | >400 | >400 | >400 | >400 |
| S. typhimurium | >400 | >400 | >400 | >400 | >400 | >400 | >400 | >400 |
| Gram-positive bacteria | ||||||||
| B. subtilis | >400 | >400 | 400 ± 0.0 | >400 | >400 | >400 | 400 ± 0.0 | >400 |
| M. luteus | 50 ± 0.0 | 25 ± 0.0 | 25 ± 0.0 | 25 ± 0.0 | 50 ± 0.0 | 25 ± 0.0 | 25 ± 0.0 | 25 ± 0.0 |
| S. aureus | 200 ± 0.0 | 50 ± 0.0 | 50 ± 0.0 | 50 ± 0.0 | 200 ± 0.0 | 50 ± 0.0 | 50 ± 0.0 | 50 ± 0.0 |
| MRSA | 400 ± 0.0 | 100 ± 0.0 | 100 ± 0.0 | 200 ± 0.0 | >400 | 100 ± 0.0 | 100 ± 0.0 | 200 ± 0.0 |
| S. epidermidis | 50 ± 0.0 | 25 ± 0.0 | 12.5 ± 0.0 | 50 ± 0.0 | 50 ± 0.0 | 25 ± 0.0 | 12.5 ± 0.0 | 50 ± 0.0 |
| Fungus | ||||||||
| C. albicans | >400 | >400 | >400 | >400 | >400 | >400 | >400 | >400 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maitip, J.; Mookhploy, W.; Khorndork, S.; Chantawannakul, P. Comparative Study of Antimicrobial Properties of Bee Venom Extracts and Melittins of Honey Bees. Antibiotics 2021, 10, 1503. https://doi.org/10.3390/antibiotics10121503
Maitip J, Mookhploy W, Khorndork S, Chantawannakul P. Comparative Study of Antimicrobial Properties of Bee Venom Extracts and Melittins of Honey Bees. Antibiotics. 2021; 10(12):1503. https://doi.org/10.3390/antibiotics10121503
Chicago/Turabian StyleMaitip, Jakkrawut, Wannapha Mookhploy, Supharerk Khorndork, and Panuwan Chantawannakul. 2021. "Comparative Study of Antimicrobial Properties of Bee Venom Extracts and Melittins of Honey Bees" Antibiotics 10, no. 12: 1503. https://doi.org/10.3390/antibiotics10121503
APA StyleMaitip, J., Mookhploy, W., Khorndork, S., & Chantawannakul, P. (2021). Comparative Study of Antimicrobial Properties of Bee Venom Extracts and Melittins of Honey Bees. Antibiotics, 10(12), 1503. https://doi.org/10.3390/antibiotics10121503