Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food
Abstract
:1. Introduction
2. Results
2.1. Collected Salmonella Strains
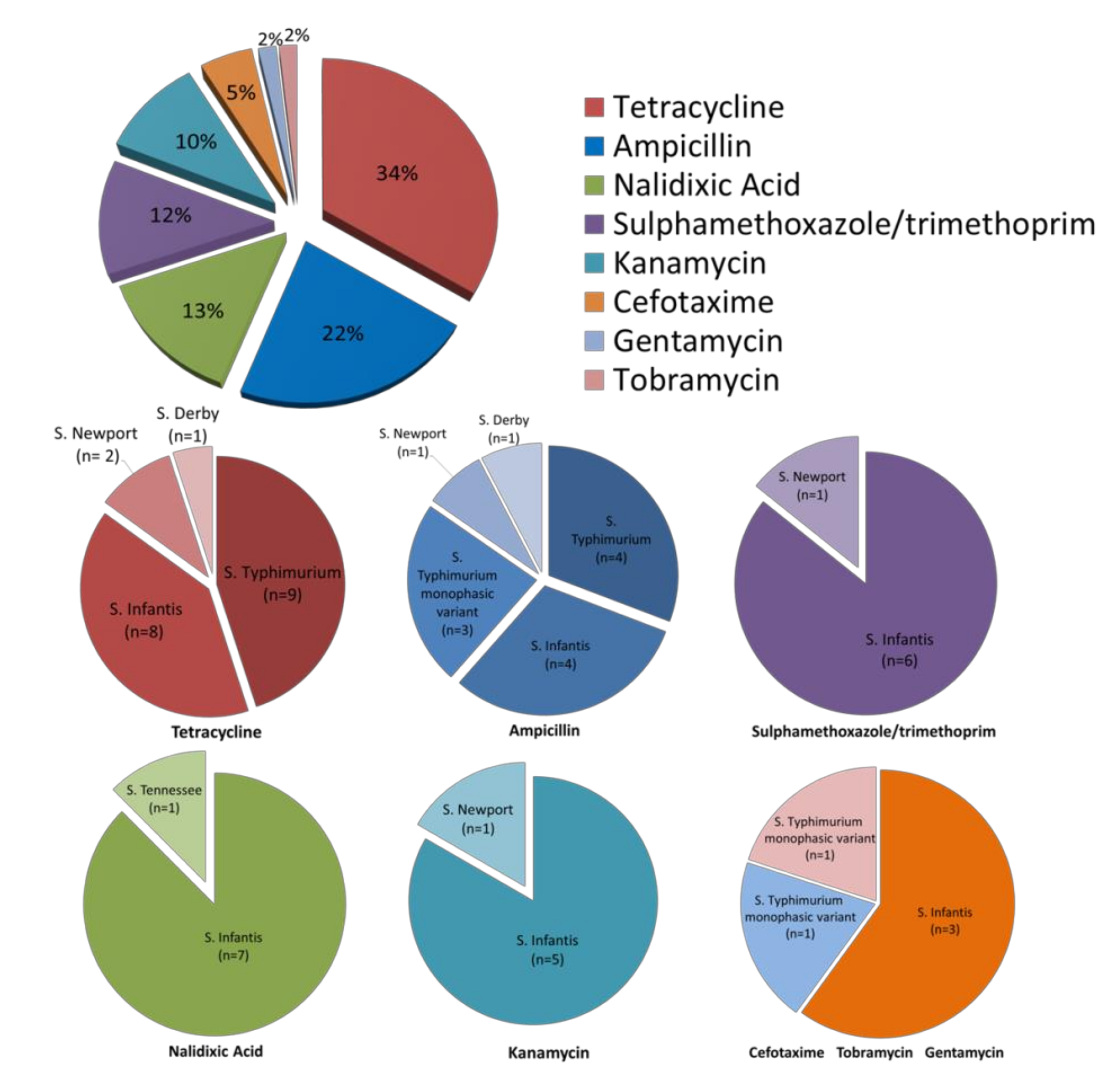
2.2. Antimicrobial Susceptibility
2.3. Detection of Tetracycline Resistance Genes and Class-1 Integron
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of Salmonella spp.
4.2. Antimicrobial Susceptibility by the Disk Diffusion Method
4.3. DNA Extraction
4.4. Detection of Antibiotic Resistance Genes and Class-1 Integron
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tindall, B.J.; Grimont, P.A.D.; Garrity, G.M.; Euzéby, J.P. Nomenclature and Taxonomy of the Genus Salmonella. Int. J. Syst. Evol. Microbiol. 2005, 55, 521–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Post, A.S.; Diallo, S.N.; Guiraud, I.; Lompo, P.; Tahita, M.C.; Maltha, J.; Van Puyvelde, S.; Mattheus, W.; Ley, B.; Thriemer, K.; et al. Supporting Evidence for a Human Reservoir of Invasive Non-Typhoidal Salmonella from Household Samples in Burkina Faso. PLoS Negl. Trop. Dis. 2019, 13, e0007782. [Google Scholar] [CrossRef] [Green Version]
- International Office of Epizootics; Biological Standards Commission. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: (Mammals, Birds and Bees); OIE: Paris, France, 2018; ISBN 978-92-95108-18-9.
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A Review on Pathogenesis, Epidemiology and Antibiotic Resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef] [Green Version]
- Guardabassi, L. Pet Animals as Reservoirs of Antimicrobial-Resistant Bacteria: Review. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef]
- Damborg, P.; Broens, E.M.; Chomel, B.B.; Guenther, S.; Pasmans, F.; Wagenaar, J.A.; Weese, J.S.; Wieler, L.H.; Windahl, U.; Vanrompay, D.; et al. Bacterial Zoonoses Transmitted by Household Pets: State-of-the-Art and Future Perspectives for Targeted Research and Policy Actions. J. Comp. Pathol. 2016, 155, S27–S40. [Google Scholar] [CrossRef] [Green Version]
- Vilela, F.P.; Gomes, C.N.; Passaglia, J.; Rodrigues, D.P.; Costa, R.G.; Tiba Casas, M.R.; Fernandes, S.A.; Falcão, J.P.; Campioni, F. Genotypic Resistance to Quinolone and Tetracycline in Salmonella Dublin Strains Isolated from Humans and Animals in Brazil. Microb. Drug Resist. 2018, 25, 143–151. [Google Scholar] [CrossRef] [PubMed]
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic Use in Plant Agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–465. [Google Scholar] [CrossRef]
- Frech, G.; Schwarz, S. Molecular Analysis of Tetracycline Resistance in Salmonella Enterica Subsp. Enterica Serovars Typhimurium, Enteritidis, Dublin, Choleraesuis, Hadar and Saintpaul: Construction and Application of Specific Gene Probes. J. Appl. Microbiol. 2000, 89, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Elnekave, E.; Hong, S.L.; Lim, S.; Boxrud, D.; Rovira, A.; Mather, A.E.; Perez, A.; Alvarez, J. Transmission of Multidrug-Resistant Salmonella Enterica Subspecies Enterica 4,[5],12:I:- Sequence Type 34 between Europe and the United States. Emerg. Infect. Dis. 2020, 26, 3034–3038. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority; European Centre for Disease Prevention and Control The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017/2018. EFSA J. 2020, 18. [CrossRef] [Green Version]
- Vitale, M.; Scatassa, M.L.; Cardamone, C.; Oliveri, G.; Piraino, C.; Alduina, R.; Napoli, C. Staphylococcal Food Poisoning Case and Molecular Analysis of Toxin Genes in Staphylococcus Aureus Strains Isolated from Food in Sicily, Italy. Foodborne Pathog. Dis. 2015, 12, 21–23. [Google Scholar] [CrossRef]
- Vitale, M.; Gaglio, S.; Galluzzo, P.; Cascone, G.; Piraino, C.; Di Marco Lo Presti, V.; Alduina, R. Antibiotic Resistance Profiling, Analysis of Virulence Aspects and Molecular Genotyping of Staphylococcus Aureus Isolated in Sicily, Italy. Foodborne Pathog. Dis. 2018, 15, 177–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitale, M.; Galluzzo, P.; Buffa, P.G.; Carlino, E.; Spezia, O.; Alduina, R. Comparison of Antibiotic Resistance Profile and Biofilm Production of Staphylococcus Aureus Isolates Derived from Human Specimens and Animal-Derived Samples. Antibiotics 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sciortino, S.; Arculeo, P.; Alio, V.; Cardamone, C.; Nicastro, L.; Arculeo, M.; Alduina, R.; Costa, A. Occurrence and Antimicrobial Resistance of Arcobacter Spp. Recovered from Aquatic Environments. Antibiotics 2021, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Veterinary Drug Usage and Antimicrobial Resistance in Bacteria of Animal Origin. Basic Htmlent Glyphamp Asciiamp Clin. Pharmacol. Htmlent Glyphamp Asciiamp Toxicol. 2005, 96, 271–281. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Su, J.; Xu, Z. Antibiotic Susceptibility, Biofilm-Forming Ability, and Incidence of Class 1 Integron of Salmonella Spp., Escherichia Coli, and Staphylococcus Aureus Isolated from Various Foods in a School Canteen in China. Foodborne Pathog. Dis. 2020, 17, 269–275. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, W.; Xu, X.; Zhou, X.; Shi, C. Transmissible ST3-IncHI2 Plasmids Are Predominant Carriers of Diverse Complex IS26-Class 1 Integron Arrangements in Multidrug-Resistant Salmonella. Front. Microbiol. 2018, 9, 2492. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018, 16. [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19. [CrossRef]
- Connell, S.R.; Tracz, D.M.; Nierhaus, K.H.; Taylor, D.E. Ribosomal Protection Proteins and Their Mechanism of Tetracycline Resistance. Antimicrob. Agents Chemother. 2003, 47, 3675–3681. [Google Scholar] [CrossRef] [Green Version]
- Hall, R.M. Salmonella Genomic Islands and Antibiotic Resistance in Salmonella Enterica. Future Microbiol. 2010, 5, 1525–1538. [Google Scholar] [CrossRef]
- World Health Organization; Food and Agriculture Organization of the United Nations; World Organisation for Animal Health. Monitoring Global Progress On Addressing Antimicrobial Resistance; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella Persisters Promote the Spread of Antibiotic Resistance Plasmids in the Gut. Nature 2019, 573, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ștefan, M.G.; Loghin, F. Antibiotics in the Environment: Causes and Consequences. Med. Pharm. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gambino, D.; Persichetti, M.F.; Gentile, A.; Arculeo, M.; Visconti, G.; Currò, V.; Caracappa, G.; Crucitti, D.; Piazza, A.; Mancianti, F.; et al. First Data on Microflora of Loggerhead Sea Turtle (Caretta Caretta) Nests from the Coastlines of Sicily. Biol. Open 2020, 9, bio045252. [Google Scholar] [CrossRef] [Green Version]
- Blasi, M.F.; Migliore, L.; Mattei, D.; Rotini, A.; Thaller, M.C.; Alduina, R. Antibiotic Resistance of Gram-Negative Bacteria from Wild Captured Loggerhead Sea Turtles. Antibiotics 2020, 9, 162. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, J.; Lu, J.; Wu, J. Antibiotic Resistance Genes Might Serve as New Indicators for Wastewater Contamination of Coastal Waters: Spatial Distribution and Source Apportionment of Antibiotic Resistance Genes in a Coastal Bay. Ecol. Indic. 2020, 114, 106299. [Google Scholar] [CrossRef]
- Yukawa, S.; Uchida, I.; Tamura, Y.; Ohshima, S.; Hasegawa, T. Characterisation of Antibiotic Resistance of Salmonella Isolated from Dog Treats in Japan. Epidemiol. Infect. 2019, 147, e102. [Google Scholar] [CrossRef] [Green Version]
- Nguyen Thi, H.; Pham, T.-T.-T.; Turchi, B.; Fratini, F.; Ebani, V.V.; Cerri, D.; Bertelloni, F. Characterization of Salmonella Spp. Isolates from Swine: Virulence and Antimicrobial Resistance. Animals 2020, 10, 2418. [Google Scholar] [CrossRef]
- Sucato, A.; Vecchioni, L.; Savoca, D.; Presentato, A.; Arculeo, M.; Alduina, R. A Comparative Analysis of Aquatic and Polyethylene-Associated Antibiotic-Resistant Microbiota in the Mediterranean Sea. Biology 2021, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Aldema, M.L.; McMurry, L.M.; Walmsley, A.R.; Levy, S.B. Purification of the Tn10-Specified Tetracycline Efflux Antiporter TetA in a Native State as a Polyhistidine Fusion Protein. Mol. Microbiol. 1996, 19, 187–195. [Google Scholar] [CrossRef]
- Roberts, M.C. Update on Acquired Tetracycline Resistance Genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef]
- Bryan, A.; Shapir, N.; Sadowsky, M.J. Frequency and Distribution of Tetracycline Resistance Genes in Genetically Diverse, Nonselected, and Nonclinical Escherichia Coli Strains Isolated from Diverse Human and Animal Sources. Appl. Environ. Microbiol. 2004, 70, 2503–2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawyabur, Md.; Islam, Md.S.; Sobur, Md.A.; Hossain, Md.J.; Mahmud, Md.M.; Paul, S.; Hossain, M.T.; Ashour, H.M.; Rahman, Md.T. Isolation and Characterization of Multidrug-Resistant Escherichia Coli and Salmonella Spp. from Healthy and Diseased Turkeys. Antibiotics 2020, 9, 770. [Google Scholar] [CrossRef]
- Koo, H.-J.; Woo, G.-J. Distribution and Transferability of Tetracycline Resistance Determinants in Escherichia Coli Isolated from Meat and Meat Products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.S.; Osborne, D.J.; Stanley, J. Enterobacterial Tetracycline Resistance in Relation to Plasmid Incompatibility. Mol. Cell. Probes 1992, 6, 313–317. [Google Scholar] [CrossRef]
- Roberts, M.C. Epidemiology of Tetracycline-Resistance Determinants. Trends Microbiol. 1994, 2, 353–357. [Google Scholar] [CrossRef]
- McDermott, P.F.; Zhao, S.; Tate, H. Antimicrobial Resistance in Nontyphoidal Salmonella. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Soares, F.B.; Camargo, C.H.; Cunha, M.P.V.; de Almeida, E.A.; de Jesus, A.M.B.; de Carvalho, E.; de Paiva, J.B.; Fernandes, S.A.; Tiba-Casas, M.R. Subtyping of Plasmid-Mediated Quinolone Resistance among Salmonella Serotypes by Whole Genome Sequencing. Diagn. Microbiol. Infect. Dis. 2019, 94, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Wang, X.; Liu, X.; Xia, J.; Fang, L.; Sun, J.; Liao, X.; Liu, Y. OqxAB-Positive IncHI2 Plasmid PHXY0908 Increase Salmonella Enterica Serotype Typhimurium Strains Tolerance to Ciprofloxacin. Front. Cell. Infect. Microbiol. 2019, 9, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Gambino, D.; Vicari, D.; Vitale, M.; Schirò, G.; Mira, F.; Giglia, M.L.; Riccardi, A.; Gentile, A.; Giardina, S.; Carrozzo, A.; et al. Study on Bacteria Isolates and Antimicrobial Resistance in Wildlife in Sicily, Southern Italy. Microorganisms 2021, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Alduina, R.; Gambino, D.; Presentato, A.; Gentile, A.; Sucato, A.; Savoca, D.; Filippello, S.; Visconti, G.; Caracappa, G.; Vicari, D.; et al. Is Caretta Caretta a Carrier of Antibiotic Resistance in the Mediterranean Sea? Antibiotics 2020, 9, 116. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zhou, R.; Yang, Y.; Chen, B.; Cheng, Z.; Zhang, M.; Li, J.; Zhang, G.; Zou, S. Characterizing the Antibiotic Resistance Genes in a River Catchment: Influence of Anthropogenic Activities. J. Environ. Sci. 2018, 69, 125–132. [Google Scholar] [CrossRef]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (No. 48) to the White–Kauffmann–Le Minor Scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals CLSI Supplement VET08, 4th ed.; Committee for Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 978-1-68440-010-2. [Google Scholar]
- Lynne, A.M.; Rhodes-Clark, B.S.; Bliven, K.; Zhao, S.; Foley, S.L. Antimicrobial Resistance Genes Associated with Salmonella Enterica Serovar Newport Isolates from Food Animals. Antimicrob. Agents Chemother. 2008, 52, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Molecular Characterization of Multidrug-Resistant Avian Pathogenic Escherichia Coli Isolated from Septicemic Broilers. Int. J. Med. Microbiol. 2013, 303, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Zambri, M.; Cloutier, M.; Adam, Z.; Lapen, D.R.; Wilkes, G.; Sunohara, M.; Topp, E.; Talbot, G.; Khan, I.U.H. Novel Virulence, Antibiotic Resistance and Toxin Gene-Specific PCR-Based Assays for Rapid Pathogenicity Assessment of Arcobacter Faecis and Arcobacter Lanthieri. BMC Microbiol. 2019, 19, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Origin | Serotypes Analyzed | Number of Strains |
|---|---|---|
| Livestock Animals (n = 16) | S. Derby | 6 |
| S. Typhimurium Monophasic Variant | 3 | |
| S. Muenster | 3 | |
| S. Typhimurium | 1 | |
| S. Elomrane | 1 | |
| S. Montevideo | 1 | |
| S. Kedougou | 1 | |
| Pets (n = 12) | S. Typhimurium | 6 |
| S. Goldcoast | 2 | |
| S. Typhimurium Monophasic Variant | 1 | |
| S. Enteritidis | 1 | |
| S. Elomrane | 1 | |
| S. Schleissheim | 1 | |
| Zoo Animals (n = 8) | S. Typhimurium | 2 |
| S. Richmond | 2 | |
| S. Potsdam | 1 | |
| S. Kiambu | 1 | |
| S. Infantis | 1 | |
| S. Bahrenfeld | 1 | |
| Wild Animals (n = 8) | S. Newport | 1 |
| S. Heron | 1 | |
| S. bongori | 1 | |
| S. Elomrane | 1 | |
| S. Veneziana | 1 | |
| S. Abony | 1 | |
| S. Tennessee | 1 | |
| S. Enteritidis | 1 | |
| Food from Animal Origin (n = 16) | S. Infantis | 7 |
| S. Bredeney | 2 | |
| S. Kentucky | 2 | |
| S. Typhimurium | 1 | |
| S. Derby | 1 | |
| S. Newport | 1 | |
| S. Enteritidis | 1 | |
| S. Cardoner | 1 |
| Id. Strain | Serotype | Phenotypic Resistance |
|---|---|---|
| Livestock Animals (n = 3/16) | ||
| S1 | S. Typhimurium Monophasic Variant | AMP |
| S2 | S. Typhimurium Monophasic Variant | AMP |
| S64 | S. Typhimurium Monophasic Variant | TE |
| Pets (n = 6/12) | ||
| S3 | S. Typhimurium | CN, TOB, AMP, TE |
| S19 | S. Typhimurium | TE |
| S20 | S. Typhimurium | TE |
| S24 | S. Typhimurium | AMP, TE |
| S25 | S. Typhimurium | AMP, TE |
| S32 | S. Typhimurium | TE |
| Zoo Animals (n = 3/8) | ||
| S26 | S. Typhimurium | AMP, TE |
| S27 | S. Typhimurium | AMP, TE |
| S43 | S. Infantis | K, AMP, NA, SXT, TE |
| Wild Animals (n = 2/8) | ||
| S4 | S. Newport | TE |
| S41 | S. Tennessee | NA |
| Food from Animal Origin (n = 9/16) | ||
| S58 | S. Newport | K, AMP, SXT, TE, |
| S62 | S. Infantis | K, AMP, CTX, NA, TE |
| S63 | S. Infantis | K, AMP, CTX, NA, SXT, TE |
| S48 | S. Infantis | AMP, CTX, NA, SXT, TE |
| S55 | S. Infantis | K, NA, SXT, TE |
| S56 | S. Infantis | TE |
| S68 | S. Infantis | NA, SXT, TE |
| S69 | S. Infantis | K, NA, SXT, TE |
| S47 | S. Derby | AMP, TE |
| Id. | Source | Serovar | Tetracycline Resistance Genes | int1 | Tetracycline Phenotypic Resistance |
|---|---|---|---|---|---|
| S1 | Livestock | S. Typhimurium monofasic variant | tet (A) | int1 | S |
| S7 | S. Montevideo | tet (D) | - | S | |
| S11 | S. Derby | tet (G) | int1 | S | |
| S17 | S. Muenster | tet (D) | - | S | |
| S54 | S. Derby | tet (E), tet (D) | - | S | |
| S3 | Pets | S. Typhimurium monofasic variant | tet (B), tet (G) | - | R |
| S8 | S. Goldcoast | tet (A), tet (D) | int1 | S | |
| S9 | S. Goldcoast | tet (A) | int1 | S | |
| S19 | S. Typhimurium | tet (A), tet (E), tet (G), tet (C) | - | R | |
| S20 | S. Typhimurium | tet (E), tet (B), tet (C) | - | R | |
| S24 | S. Typhimurium | tet (A) | - | R | |
| S25 | S. Typhimurium | tet (A), tet (G) | - | R | |
| S30 | S. Enteritidis | tet (A), tet (E) | int1 | S | |
| S32 | S. Typhimurium | tet (A) | - | R | |
| S35 | S. Elomrane | tet (C) | - | S | |
| S15 | Zoo | S. Kiambu | tet (G) | - | S |
| S26 | S. Typhimurium | tet (A) | - | R | |
| S27 | S. Typhimurium | tet (A) | - | R | |
| S29 | S. Richmond | tet (A) | - | S | |
| S43 | S. Infantis | tet (A) | int1 | R | |
| S21 | S. bongori | tet (C) | - | S | |
| S47 | Wildlife | S. Derby | tet (G) | R | |
| S59 | Food | S. Cardoner | tet (B) | - | S |
| S67 | S. Enteritidis | tet (A) | int1 | S | |
| S69 | S. Infantis | tet (A) | int1 | R |
| Target Gene | Primer Sequence (5′–3′) | Function | Annealing Temperature (°C) | Amplicon Size (bp) | References |
|---|---|---|---|---|---|
| tet (A) | GCTACATCCTGCTTGCCTTC CATAGATCGCCGTGAAGAGG | Efflux | 60 | 210 | [51] |
| tet (B) | TTGGTTAGGGGCAAGTTTTG GTAATGGGCCAATAACACCG | Efflux | 60 | 659 | [51] |
| tet (C) | CTTGAGAGCCTTCAACCCAG ATGGTCGTCATCTACTGCC | Efflux | 60 | 418 | [51] |
| tet (D) | AAACCATTACGGCATTCTGC GACCGGATACACCATCCATC | Efflux | 60 | 787 | [52] |
| tet (E) | AAACCACATCCTCCATACGC AAATAGGCCACAACCGTCAG | Efflux | 60 | 278 | [52] |
| tet (G) | GCTCGGTGGTATCTCTGCTC AGCAACAGAATCGGGAACAC | Efflux | 60 | 844 | [52] |
| tet (O) | GGAGGGGTTCAACCACAAAG CTATGTAAATAAAATGGATAG | Ribosomal protection | 55 | 88 | [53] |
| tet (W) | ACATCATTGATACTCCAGGTCACG TTTCACTTTGTGGTTGAACCCCTC | Ribosomal protection | 60 | 142 | [53] |
| int1 | CCT CCC GCA CGA TGA TC TCC ACG CAT CGT CAG GC | Class 1 integron | 60 | 280 | [53] |
| 16S rDNA | CGGTGAATACGTTCYCGG GGHTACCTTGTTACGACTT | Positive Control | 55 | 142 | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gargano, V.; Sciortino, S.; Gambino, D.; Costa, A.; Agozzino, V.; Reale, S.; Alduina, R.; Vicari, D. Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics 2021, 10, 809. https://doi.org/10.3390/antibiotics10070809
Gargano V, Sciortino S, Gambino D, Costa A, Agozzino V, Reale S, Alduina R, Vicari D. Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics. 2021; 10(7):809. https://doi.org/10.3390/antibiotics10070809
Chicago/Turabian StyleGargano, Valeria, Sonia Sciortino, Delia Gambino, Antonella Costa, Vincenzo Agozzino, Stefano Reale, Rosa Alduina, and Domenico Vicari. 2021. "Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food" Antibiotics 10, no. 7: 809. https://doi.org/10.3390/antibiotics10070809
APA StyleGargano, V., Sciortino, S., Gambino, D., Costa, A., Agozzino, V., Reale, S., Alduina, R., & Vicari, D. (2021). Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics, 10(7), 809. https://doi.org/10.3390/antibiotics10070809









