Abstract
Globally, tuberculosis (TB) remains a prevalent threat to public health. In 2019, TB affected 10 million people and caused 1.4 million deaths. The major challenge for controlling this infectious disease is the emergence and spread of drug-resistant Mycobacterium tuberculosis, the causative agent of TB. The antibiotic streptomycin is not a current first-line anti-TB drug. However, WHO recommends its use in patients infected with a streptomycin-sensitive strain. Several mutations in the M. tuberculosis rpsL, rrs and gidB genes have proved association with streptomycin resistance. In this study, we performed a molecular analysis of these genes in clinical isolates to determine the prevalence of known or novel mutations. Here, we describe the genetic analysis outcome. Furthermore, a biocomputational analysis of the MtGidB L101F variant, the product of a novel mutation detected in gidB during molecular analysis, is also reported as a theoretical approach to study the apparent genotype-phenotype association.
1. Introduction
Worldwide, tuberculosis (TB) is among the top 10 causes of death and the leading disease caused by a single infectious agent, Mycobacterium tuberculosis, ranking above HIV/AIDS [1,2]. TB control has become a global challenge due to the continued emergence of multidrug-resistant (MDR) strains [3,4]. Precise identification of such strains demands bacterial confirmation and drug resistance assessment using culture methods, molecular analysis and DNA sequencing [5,6]. Moreover, patients with MDR-TB require intensive treatment for at least nine months (up to 20 months), supported by constant pharmacovigilance to reduce adverse events [7,8].
Streptomycin (Str) was the first antibiotic used for the therapeutic control of TB [9,10]. However, drug-associated side effects and the emergence of Str-resistant (StrR) strains prompted its removal from the group of first-line anti-TB drugs [11,12,13]. Despite this, WHO recommends using it in patients infected with a confirmed Str-susceptible strain [8,14,15]. Str is active against growing bacteria by inhibiting protein synthesis. It acts through irreversible binding to S12 protein and 16S rRNA, two molecular constituents of the small subunit of bacterial ribosomes [13,16].
Mutations in the M. tuberculosis rpsL, rrs and gidB genes, respectively, encoding the ribosomal protein S12, 16S rRNA and glucose-inhibited division protein B, have been associated with Str resistance [13,17,18,19,20,21]. The non-synonymous substitutions K43R and K88R in rpsL and nucleotide variations in the «530 loop» and «912 region» of 16S rRNA are the most frequent StrR-linked mutations. Remarkably, non-synonymous substitutions in gidB are usually associated with low-resistance levels [19,20,21,22,23,24,25].
The gidB gene product is an S-adenosylmethionine (SAM)-dependent 7-methylguanosine methyltransferase specific for 16S rRNA [26]. In particular, the GidB-catalyzed methylation occurs at nucleoside G527 of the E. coli 16S rRNA, which corresponds to G518 in the M. tuberculosis counterpart [27]. Thus, given that Str interacts with such a nucleoside, an impaired GidB function would affect the G518 methylation status of M. tuberculosis 16S rRNA, interfering with drug binding and, consequently, producing the observed StrR phenotype [21,25,26,27,28].
Herein, we performed a molecular analysis of the rpsL, rrs and gidB genes in clinical isolates of M. tuberculosis that showed a noticeable StrR phenotype to determine the current prevalence of known or novel mutations in our region. The combined outcome of DNA sequencing and gene comparisons allowed us to identify point mutations associated with such drug resistance. Furthermore, a biocomputational analysis was used as a theoretical approach to study the apparent genotype-phenotype association observed in two StrR isolates containing the 301c>t mutation in gidB, which produces the non-synonymous substitution L101F in the gene product.
2. Results and Discussion
2.1. Molecular Analysis of the M. tuberculosis rpsL, rrs and gidB Genes
Patients suspected of having active TB disease who attended the GHT’s TB Clinic provided the sputum samples. From a collection period of one year, 11 clinical isolates met the inclusion criteria for this study: positive Ziehl-Nielsen (ZN) staining and streptomycin resistance (minimum inhibitory concentration, MIC > 0.8 µg/mL). Using standard molecular methods for M. tuberculosis gDNA extraction and PCR amplification, the expected gene fragments for molecular analysis: 628 bp (rpsL), 645 bp (rrs) and 719 bp (gidB), were obtained. Once purified, the amplicons were sequenced and analyzed with various bioinformatics tools: NCBI’s BLAST, SequentiX’s DNA Dragon, SnapGene® Viewer and EBI’s Clustal Omega. Table 1 summarizes the data obtained from dsDNA sequencing and gene analysis.

Table 1.
Overall results of the molecular analysis of rpsL, rrs and gidB genes from 11 clinical isolates of M. tuberculosis showing resistance to streptomycin.
As initial analysis of the molecular results, none of the StrR isolates showed mutations in rpsL, i.e., fully identical to the reference gene sequence (Mycobrowser ID: Rv0682). Furthermore, most of them (10 out of 11) showed 100% identity within the 365–978 nucleotide segment of the reference rrs gene (Mycobrowser ID: MTB000019). The exception isolate (03R) showed the 491c>t mutation, which was also detected in Str-susceptible isolates, confirming a lack of genotype-phenotype association. Moreover, it represents an epidemiological biomarker assigned to the M. tuberculosis Latin American and Mediterranean sublineage 3 (LAM3) [28].
In contrast, the gidB analysis produced mixed results. Five isolates, 03R-07R, showed sequences 100% identical to the reference (Mycobrowser ID: Rv3919c). Another three: 02R, 09R and 11R, showed the 236t>c mutation (causing the L79S substitution). This variant is associated with a low-level StrR phenotype when detected as a sole mutation. However, when concurring with mutations in other genes (e.g., rrs 517c>t or rpsL K43R), isolates exhibit significant resistance to Str [24,29,30,31]. Two others (R01 and R10) showed the 301t>c mutation (causing the L101F substitution). Knowledge about this gene variant and its contribution to the phenotype is limited. However, a common feature of StrR isolates containing such a variation is the absence of mutations in their rrs and rpsL genes, suggesting a genuine association with the observed phenotype [29]. Lastly, isolate 08R showed two mutations, 37g>c and 47t>g, causing the G13R and L16R substitutions. While G13R seems to be a novel mutation, L16R is a natural polymorphism associated with the LAM lineage [20,24,29,31,32].
2.2. Significance of the Molecular Analysis
So far, the observed results allow us to separate the analyzed StrR isolates into two groups (I and II, Table 1). Group I comprises of those lacking mutations in the commonly associated genes (i.e., rpsL, rrs and gidB) and those with mutations previously identified as polymorphisms (e.g., 491c>t in rrs). Interestingly, the identification of this group implies the existence of additional genes associated with the phenotype, as suggested by proteomic analyses [33,34,35]. On the other hand, group II includes those containing mutations in gidB and lacking known genetic variations in rpsL and rrs. Furthermore, while a subset of this group involves those carrying a mutation associated with a low level of resistance (i.e., L79S), another subset contains those revealing either G13R or L101F as a novel non-synonymous substitution.
Regarding the latter, G13R is within the N-terminal domain, and L101F is in the SAM-dependent methyltransferase (SAM-MTase) domain, within the SAM-interacting region [25,36,37]. As there is no prior evidence regarding the G13R genotype contribution to the StrR phenotype, the effect of such a mutation on MtGidB function will not be analyzed further. However, to gain additional knowledge on the molecular basis of Str resistance in M. tuberculosis, we performed a biocomputational analysis of the MtGidB L101F variant to predict its functional consequences (given the conserved structure-function relationship among SAM-MTase domains).
2.3. Biocomputational Analysis of the M. tuberculosis GidB L101F Variant
2.3.1. Structural and Functional Analysis
A primary structure-based biocomputational analysis of MtGidB provided the initial information about the effect of the L101F substitution on protein function (Table 2). The combined results of three bioinformatics tools predicted a negative effect of such a mutation on MtGidB function.

Table 2.
Effect of L101F substitution on MtGidB function.
Template-based modeling resulted in a reliable 3D structure to test whether L101F affects the functional conformation of MtGidB, as suggested before. The predicted 3D model showed a high confidence score (Figure 1A): 1.17 (in the range −5 to 2, a higher value means high confidence). After refinement, the quality assessment result validated its structural accuracy: a global score of 0.7045 (values ≥ 0.4 are good scores). Furthermore, the Ramachandran plot showed that 89.1% of the non-Gly/Pro residues are in the most favored regions plus an additional 8.7% in allowed regions (Figure 1B). In addition, the estimated Z-score for overall quality, −6.44, is within the range of values typically found for proteins of similar size (Figure 1C).
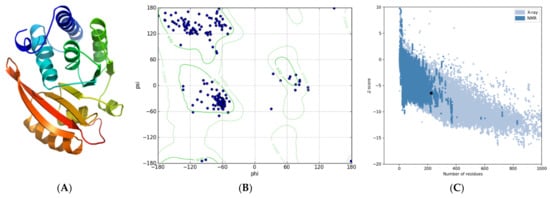
Figure 1.
Predicted tertiary structure for MtGidB. (A) Best 3D model (ribbon representation, rainbow-colored using default settings). (B) Ramachandran plot. (C) ProSA analysis (Z-score plot). A black dot denotes the estimated Z-Score.
Before further structure-function analysis, the predicted model served as a framework to identify the presumed SAM-interacting residues: G69, S70, G71, L74, E92, P93, L94, R97, G117, R118, A119, E120, R137 and A138. A tertiary structure-based bioinformatic analysis of the MtGidB 3D model provided additional knowledge about the effect of the L101F substitution on protein stability. Overall, four biocomputational methods, along with a thermodynamic assumption: destabilizing (ΔΔG < −1.0 Kcal/mol), neutral (−1.0 ≤ ΔΔG ≤ 1.0 Kcal/mol) and stabilizing (ΔΔG > 1.0 Kcal/mol) [38], confirmed structural destabilization (Table 3).

Table 3.
Effect of L101F substitution on MtGidB stability.
2.3.2. Polymorphic Site Interaction Analysis
The analysis of interatomic contacts at the polymorphic site provided further insights into the local interactions and changes derived from residue substitution (Figure 2). In this regard, a comparative examination of the respective networks of non-covalent bonds revealed that the mutant residue (F101) establishes new interactions and loses others, in contrast to the wild-type (L101). A supplementary analysis of interatomic contacts of structural units confirmed this observation (Table 4). Furthermore, as L101 is involved in a residue contact network that includes SAM-interacting residues (i.e., G71, E92 and R97), it seems reasonable to suggest that the L101F substitution affects the MtGidB function by altering the ligand-binding site of the SAM-MTase domain.
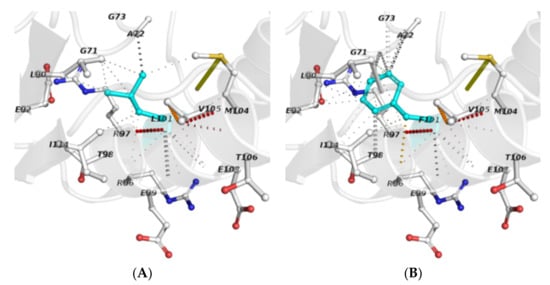
Figure 2.
Interatomic contacts among residues that occur at the polymorphic site: (A) L101 (wild-type) and (B) F101 (mutant). Polymorphic residues (sticks) are depicted in cyan, while the others are displayed using the element-coloring settings. Non-covalent interactions are presented in default colors, as stated in the Arpeggio server.

Table 4.
Residues in contact with L101 (wild-type) or F101 (mutant) in the MtGidB 3D structure. Classification of non-covalent interactions as detected by the LPC/CSU software program.
2.3.3. Protein Dynamics Analysis
Molecular dynamics (MD) simulations are commonly applied to study protein mobility and flexibility [39,40]. Using coarse-grained (CG) models as reduced representations of protein residues, this theoretical approach provided additional knowledge about the conformational structure of MtGidB and its changes due to the L101F substitution in a 2000 ps time frame. Interestingly, both systems (wild-type and mutant) depicted a short phase of continuous decrease in the UNRES (united residue) potential energy followed by an apparent steady-state, which remained until the simulation end (Figure 3A,B). However, the radius of gyration plots showed that the mutant system exhibits a higher degree of structural mobility than the wild-type system (Figure 3C,D), supporting the hypothesis that implies changes in local flexibility are the consequence of the L101F mutation on the MtGidB conformation.
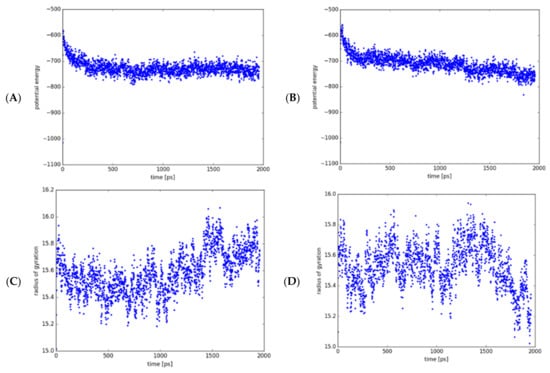
Figure 3.
MD analysis of the wild-type (WT, L101) and mutant (MU, F101) systems (MtGidB) in a 2000-ps simulation. Plots of potential energy (Kcal/mol) for WT (A) and MU (B). Plots of the radius of gyration (Å) for WT (C) and MU (D).
Supplementary analysis of atomic fluctuations completed the knowledge on residue-based flexibility. Even though both retained the secondary structure, the mutant system showed increased overall flexibility than the wild-type system (Figure 4A,B), with significant deviations in residues and regions of the SAM-MTase domain (Figure 4C). This structural behavior suggests that the L101F substitution indirectly affects the flexibility of other residues and, thus, the global MtGidB structural stability.
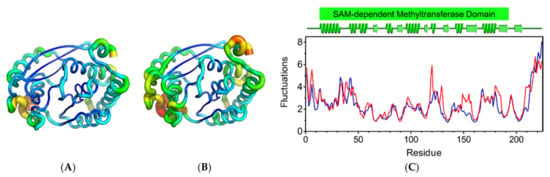
Figure 4.
Analysis of atomic fluctuations. Cartoon putty representations of MtGidB structures: (A) wild-type (WT, L101) and (B) mutant (MU, F101). Blue represents the lowest value for B-factor and red the highest. The size of the tube reflects the value of the B-factor (i.e., the larger the B-factor, the thicker the tube). (C) A plot of the residue-based fluctuations (Å) estimated for WT (blue) and MU (red). The predicted secondary structure (α-helices and β-strands) for MtGidB and the stretch corresponding to its SAM-MTase domain are colored green (PDB representation, top panel).
2.4. Significance of the Biocomputational Analysis
Overall, the MD results suggest that the L101F mutation affects the flexibility and stability of the MtGidB structure, probably due to local and global intramolecular perturbations. Furthermore, as the L101 residue is involved in a contact network that includes SAM-interacting residues (i.e., G71, E92 and R97), it seems reasonable to suggest that the Leu>Phe substitution at position 101 affects the MtGidB function by altering the ligand-binding site of the SAM-MTase domain. However, an experimental approach is required to test the latter hypothesis and accurately establish the genotype-phenotype association observed in the clinical isolates of M. tuberculosis.
3. Materials and Methods
3.1. Sample Collection and Mycobacteriological Analysis
Sputum samples from patients suspected of having active TB were collected at the TB Clinic of the General Hospital of Tijuana (GHT) by qualified personnel. All samples were digested-decontaminated using a BBL MycoPrep™ System (Becton Dickinson). The mycobacteriological analyses and drug-susceptibility assays were performed at the TB Diagnostic Unit (GHT), using standard protocols. Out of 157 independent samples from a one-year collection period, 11 tested positive for two inclusion criteria: acid-fast bacilli by Ziehl-Nielsen (ZN) staining and Str-resistant (MIC > 0.8 µg/mL) by MGIT analysis (BACTEC 960 System, Becton Dickinson). In this case, 15 ZN-positive Str-sensitive samples, selected at random, were used as controls. Sample handling and subsequent procedures were according to standard protocols approved by the GHT’s Ethics Committee.
3.2. Molecular Methods
3.2.1. Mycobacterial DNA Extraction
Genomic DNA was isolated using the DNAzol reagent (Becton Dickinson) and the protocol recommended by the manufacturer. Briefly, 0.2 mL of MycoPrep’s sediment and 0.5 mL of 1X Dulbecco’s PBS solution were mixed, and bacterial lysis was completed by heating at 80 °C (10 min). After cooling for 1 min on an ice bath, 1 mL of DNAzol reagent was added, mixed thoroughly, and centrifuged at 10,000 rpm for 10 min (to remove cell debris). The supernatant was mixed with 0.5 mL of cold ethanol and then centrifuged for 10 min at 14,500 rpm. The precipitated DNA was air-dried for 10 min and then dissolved in 30 µL of 8 mL NaOH.
3.2.2. PCR Amplification of Gene Fragments
Typical PCR reactions (20 μL) contained 10 picomoles of each primer (i.e., Fw/Rv) and 1 µL of template DNA in 1X Taq Master Mix (New England Biolabs). Table 5 lists the synthetic oligonucleotides used as primers for PCR amplification of the M. tuberculosis rpsL, rrs, gidB gene fragments. Reactions were completed in a C1000 Touch™ Thermal Cycler (Biorad) using the following settings: an initial denaturation step (2 min at 94 °C), 45 cycles of exponential amplification (20 s at 94 °C, 20 s at 55 °C, 20 s at 72 °C) and a final extension step (7 min at 72 °C). A reaction lacking template DNA was used as a negative control, while another containing M. tuberculosis H37Rv genomic DNA (1 ng) was positive (and reference DNA for comparative purposes).

Table 5.
Gene-specific primers for PCR amplification or DNA sequencing.
3.2.3. Analysis and Purification of Amplicons
The amplification products were analyzed by 2.0% agarose gel electrophoresis, using EtBr as a fluorescent dye (0.5 µg/mL, final), and visualized/documented with a GelDoc™ EZ Imager (Biorad). The 100-bp DNA Ladder and λ-DNA/HindIII Digest (New England Biolabs) were the DNA markers used to assess molecular weights. The amplicons: 628 bp for rpsL, 645 bp for rrs and 719 bp for gidB, were purified using a QIAquick PCR Purification Kit, as recommended by the manufacturer (Qiagen).
3.2.4. DNA Sequencing and Data Analysis
All Amplicons were sequenced by the Sanger method using gene-specific primers (Table 5) and a SeqStudio™ Genetic Analyzer (ThermoFisher Scientific). The NCBI BLAST engine (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 September 2020) [41] was the computational tool used to perform DNA sequence comparisons against M. tuberculosis H37Rv (as a reference). The double-stranded DNA sequences were analyzed using two different biocomputational packages, the SequentiX’s DNA Dragon—Sequence Contig Assembler (https://www.dna-dragon.com/, accessed on 15 September 2020) and SnapGene® Viewer (https://www.snapgene.com/snapgene-viewer/, accessed on 15 September 2020). The multi-sequence alignments were generated by Clustal Omega [42,43], using the EMBL-EBI server (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 15 September 2020).
3.3. Biocomputational Methods
3.3.1. Sequence-Based Function Predictions
Three bioinformatic predictors determined the effect of L101F substitution on the function of MtGidB: PolyPhen-2, PROVEAN and SIFT, with default settings. PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/, accessed on 1 October 2020) is an algorithm that combines sequence and structure-based attributes and uses a naive Bayesian classifier to identify missense mutations with an impact on the phenotype. Output levels of probably (0.85–1.0) and possibly (0.15–0.84) damaging are significant [44,45,46]. PROVEAN (http://provean.jcvi.org/, accessed on 1 October 2020) is an alignment-based method that estimates the influence of amino acid substitutions on protein function. The final score designates the mutation as deleterious or neutral, according to a predefined threshold. Protein variants with a score equal to or less than −2.5 are deleterious [47,48]. SIFT (https://sift.bii.a-star.edu.sg/, accessed on 1 October 2020) is a sequence homology-based tool that classifies amino acid substitutions as tolerant (neutral) or intolerant (deleterious) mutations. Protein variants with a normalized probability value equal to or less than 0.05 are deleterious [49].
3.3.2. Template-Based Protein Modeling
The three-dimensional (3D) structure of MtGidB was generated by template-based modeling using the I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/, accessed on 1 November 2020), a unified platform that uses a hierarchical approach for automated 3D structure prediction [50,51,52]. C-score was used to measure the modeling confidence [53,54]. The top-ranked 3D structure was improved using ReFOLD (http://www.reading.ac.uk/bioinf/ReFOLD/, accessed on 10 November 2020), a computational tool for model refinement guided by accurate quality estimates [55,56], and the structural precision was estimated using ModFold (https://www.reading.ac.uk/bioinf/ModFOLD/, accessed on 20 November 2020), a server for global and local quality assessment [57]. The 3D model was further analyzed using Procheck’s Ramachandran plot [58] and ProSA’s Z-score plot [59,60]. PyMol (Schrödinger, LLC.) and UCSF Chimera [61] were the molecular graphics systems used to visualize protein structures.
3.3.3. Structure-Based Stability Predictions
Five biocomputational methods predicted the effect of L101F substitution on the stability of MtGidB: DeepDDG, DUET, mCSM and SDM, with default settings. DeepDDG (http://protein.org.cn/ddg.html, accessed on 5 December 2020) employs a well-trained, neural network-based method to predict changes in protein stability due to point mutations [62]. DUET (http://biosig.unimelb.edu.au/duet, accessed on 5 December 2020) predicts the effects of missense mutations on protein stability by combining two complementary approaches in a consensus prediction [63,64]. mCSM (http://biosig.unimelb.edu.au/mcsm, accessed on 5 December 2020) uses graph-based signatures to predict the impact of missense mutations on protein stability, encoding distance patterns between atoms [65]. SDM (http://marid.bioc.cam.ac.uk/sdm2, accessed on 5 December 2020) applies conformationally constrained environment-specific substitution tables to predict the effect of a missense mutation and calculate the change in protein stability [66,67].
3.3.4. Examination of the Interatomic Contacts
The interatomic contacts at the polymorphic site were estimated using Arpeggio (http://biosig.unimelb.edu.au/arpeggioweb/, accessed on 15 December 2020), a web service for calculating the interatomic interactions in protein structures [68]. The MtGidB model was the wild-type structure, and its L101F variant the mutant. The local network of non-covalent interactions was analyzed using the PyMol system. The specific contacts were detected using the LPC/CSU software (http://oca.weizmann.ac.il/oca-bin/lpccsu, accessed on 15 December 2020) by automatic analysis of interatomic contacts of structural units (CSU) [69].
3.3.5. Coarse-Grained Molecular Dynamics Simulations
Coarse-grained (CG) models for (MD) simulations are an effective biocomputational approach for adequate sampling of the conformational space while maintaining physical rigor [70]. The CG-MD simulations were performed online by the UNRES web server (https://unres.pl/, accessed on 10 January 2021), using the MtGidB 3D model as a wild-type structure and its L101F variant as the mutant structure with default settings for standard protein dynamics. The CG united residue (i.e., UNRES) model is a highly-reduced physics-based representation of proteins, in which only two interaction sites per residue (united side chains and united peptide groups) are present [71,72,73]. The automatic output data, such as plots of potential energy and radius of gyration, were downloaded and analyzed as generated by the server. The fluctuations results were analyzed using the Pymol system.
Author Contributions
Conceptualization and supervision, R.E.M.-A. and M.A.R.-I.; methodology, R.E.M.-A., P.L.A.M.-M., S.G.M.-L., A.F.L.-N. and M.A.R.-I.; validation, R.E.M.-A., A.F.L.-N. and M.A.R.-I.; formal analysis, all authors; investigation, Á.R.-G., S.R.-R., R.A.G.-S. and J.B.-S.; writing—original draft preparation, Á.R.-G., R.E.M.-A. and M.A.R.-I.; writing—review and editing, R.E.M.-A., P.L.A.M.-M., S.G.M.-L., A.F.L.-N. and M.A.R.-I. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded in part by grants from Mexico’s National Council for Science and Technology (CONACyT; CB-2019/01-170715) and the Autonomous University of Baja California (UABC; CPI/300/735/E, CPI/300/2344 and CPI/300/2596).
Institutional Review Board Statement
The study was conducted according to the Declaration-of-Helsinki’s guidelines and approved by the HGT’s Ethics Committee (CONBIOETICA-02-CEI-001-20170526, 30 October 2018). No informed consent was required. Sputum samples, DNA extracts and bacterial cultures were analyzed anonymously.
Data Availability Statement
All data presented in this study are available on request from the corresponding author, without undue reservation, to any qualified researcher.
Acknowledgments
The authors thank QFB. Concepción García-Castro (CLS) and QFB. Georgina Torres-Rodríguez (CLS) of the GHT’s Clinical Diagnostic Lab (TB Diagnostic Unit) for their exceptional technical assistance; and Rafael Laniado-Laborín (Head of the GHT’s TB Clinic) and QFB. Walther H. Suárez-Carrillo (Head of the GHT’s Clinical Diagnostic Lab) for their invaluable support during the study. Álvaro García-Rodriguez thanks GHT for both academic and administrative facilities granted to achieve his Ph.D. studies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mello, F.C.D.Q.; Silva, D.R.; Dalcolmo, M.P. Tuberculosis: Where are we? J. Bras. Pneumol. 2018, 44, 82. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2020; WHO: Geneva, Switzerland, 2020; ISBN 9789240013131. [Google Scholar]
- Kurz, S.G.; Furin, J.J.; Bark, C.M. Drug-Resistant Tuberculosis. Infect. Dis. Clin. N. Am. 2016, 30, 509–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seung, K.J.; Keshavjee, S.; Rich, M.L. Multidrug-Resistant Tuberculosis and Extensively Drug-Resistant Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a017863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.N.A.; Anton-Leberre, V.; Bañuls, A.-L.; Nguyen, T.V.A. Molecular Diagnosis of Drug-Resistant Tuberculosis; A Literature Review. Front. Microbiol. 2019, 10, 794. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Choi, H.J.; Kim, K.; Whang, J.; Sung, J.; Mitarai, S. Tuberculosis: A persistent unpleasant neighbour of humans. J. Infect. Public Health 2021, 14, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Trubnikov, A.; Hovhannesyan, A.; Akopyan, K.; Ciobanu, A.; Sadirova, D.; Kalandarova, L.; Parpieva, N.; Gadoev, J. Effectiveness and Safety of a Shorter Treatment Regimen in a Setting with a High Burden of Multidrug-Resistant Tuberculosis. Int. J. Environ. Res. Public. Health 2021, 18, 4121. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Kerantzas, C.A.; Jacobs, W.R. Origins of Combination Therapy for Tuberculosis: Lessons for Future Antimicrobial Development and Application. mBio 2017, 8, e01586-16. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.F.; Schraufnagel, D.; Hopewell, P.C. Treatment of Tuberculosis. A Historical Perspective. Ann. Am. Thorac. Soc. 2015, 12, 1749–1759. [Google Scholar] [CrossRef]
- Mase, S.R.; Chorba, T. Treatment of Drug-Resistant Tuberculosis. Clin. Chest Med. 2019, 40, 775–795. [Google Scholar] [CrossRef]
- Castro, E.A.T.; Mendes, M.; Freitas, S.; Roxo, P.C. Incidence and risk factors of major toxicity associated to first-line antituberculosis drugs for latent and active tuberculosis during a period of 10 years. Rev. Port. Pneumol. 2015, 21, 144–150. [Google Scholar] [CrossRef]
- Palomino, J.C.; Martin, A. Drug Resistance Mechanisms in Mycobacterium tuberculosis. Antibiotics 2014, 3, 317–340. [Google Scholar] [CrossRef] [Green Version]
- Nahid, P.; Mase, S.R.; Migliori, G.B.; Sotgiu, G.; Bothamley, G.H.; Brozek, J.L.; Cattamanchi, A.; Cegielski, J.P.; Chen, L.; Daley, C.L.; et al. Treatment of Drug-Resistant Tuberculosis. An Official ATS/CDC/ERS/IDSA Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2019, 200, e93–e142. [Google Scholar] [CrossRef]
- Cohen, K.A.; Stott, K.E.; Munsamy, V.; Manson, A.L.; Earl, A.M.; Pym, A.S. Evidence for Expanding the Role of Streptomycin in the Management of Drug-Resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2020, 64, e00860-20. [Google Scholar] [CrossRef]
- Dookie, N.; Rambaran, S.; Padayatchi, N.; Mahomed, S.; Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis: A review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 2018, 73, 1138–1151. [Google Scholar] [CrossRef] [Green Version]
- Musser, J.M. Antimicrobial agent resistance in mycobacteria: Molecular genetic insights. Clin. Microbiol. Rev. 1995, 8, 496–514. [Google Scholar] [CrossRef]
- Arjomandzadegan, M.; Gravand, S. Analysis of rpsL and rrs genes mutations related to streptomycin resistance in Mdr and Xdr clinical isolates of Mycobacterium tuberculosis. Tuberk. Toraks 2015, 63, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.-J.; Luo, J.-T.; Wong, S.-Y.; Lee, A.S.G. Analysis of rpsL and rrs mutations in Beijing and non-Beijing streptomycin-resistant Mycobacterium tuberculosis isolates from Singapore. Clin. Microbiol. Infect. 2010, 16, 287–289. [Google Scholar] [CrossRef] [Green Version]
- Jagielski, T.; Ignatowska, H.; Bakula, Z.; Dziewit, Ł.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Zwolska, Z.; Bielecki, J. Screening for Streptomycin Resistance-Conferring Mutations in Mycobacterium tuberculosis Clinical Isolates from Poland. PLoS ONE 2014, 9, e100078. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.Y.; Lee, J.S.; Kwak, H.K.; Via, L.; Boshoff, H.I.M.; Barry, C.E. Mutations ingidBConfer Low-Level Streptomycin Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 2515–2522. [Google Scholar] [CrossRef] [Green Version]
- Khosravi, A.D.; Etemad, N.; Hashemzadeh, M.; Dezfuli, S.K.; Goodarzi, H. Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J. Glob. Antimicrob. Resist. 2017, 9, 51–56. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, C.; Xiang, L.; Pi, R.; Guo, Z.; Zheng, C.; Li, S.; Zhao, Y.; Tang, K.; Luo, M.; et al. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Beijing-lineage and dual-mutation in gidB. Tuberculosis 2016, 96, 102–106. [Google Scholar] [CrossRef]
- Spies, F.S.; Ribeiro, A.W.; Ramos, D.; Ribeiro, M.O.; Martin, A.; Palomino, J.C.; Rossetti, M.L.R.; Da Silva, P.E.A.; Zaha, A. Streptomycin Resistance and Lineage-Specific Polymorphisms in Mycobacterium tuberculosis gidB Gene. J. Clin. Microbiol. 2011, 49, 2625–2630. [Google Scholar] [CrossRef] [Green Version]
- Verma, J.S.; Gupta, Y.; Nair, D.; Manzoor, N.; Rautela, R.S.; Rai, A.; Katoch, V.M. Evaluation of gidB alterations responsible for streptomycin resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2014, 69, 2935–2941. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- Wong, S.Y.; Javid, B.; Addepalli, B.; Piszczek, G.; Strader, M.B.; Limbach, P.A.; Barry, C.E. Functional Role of Methylation of G518 of the 16S rRNA 530 Loop by GidB in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2013, 57, 6311–6318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tudó, G.; Rey, E.; Borrell, S.; Alcaide, F.; Codina, G.; Coll, P.; Martín-Casabona, N.; Montemayor, M.; Moure, R.; Orcau, À.; et al. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis clinical isolates in the area of Barcelona. J. Antimicrob. Chemother. 2010, 65, 2341–2346. [Google Scholar] [CrossRef] [Green Version]
- Smittipat, N.; Juthayothin, T.; Billamas, P.; Jaitrong, S.; Rukseree, K.; Dokladda, K.; Chaiyasirinroje, B.; Disratthakit, A.; Chaiprasert, A.; Mahasirimongkol, S.; et al. Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J. Glob. Antimicrob. Resist. 2016, 4, 5–10. [Google Scholar] [CrossRef]
- Klopper, M.; Heupink, T.H.; Hill-Cawthorne, G.; Streicher, E.M.; Dippenaar, A.; De Vos, M.; Abdallah, A.M.; Limberis, J.; Merker, M.; Burns, S.; et al. A landscape of genomic alterations at the root of a near-untreatable tuberculosis epidemic. BMC Med. 2020, 18, 24. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Q.; Gao, H.; Zhang, Z.; Liu, Y.; Lu, J.; Dai, E. The roles of rpsL, rrs, and gidB mutations in predicting streptomycin-resistant drugs used on clinical Mycobacterium tuberculosis isolates from Hebei Province, China. Int. J. Clin. Exp. Pathol. 2019, 12, 2713–2721. [Google Scholar]
- Perdigão, J.; Macedo, R.; Machado, D.; Silva, C.; Jordao, L.; Couto, I.; Viveiros, M.; Portugal, I. GidB mutation as a phylogenetic marker for Q1 cluster Mycobacterium tuberculosis isolates and intermediate-level streptomycin resistance determinant in Lisbon, Portugal. Clin. Microbiol. Infect. 2014, 20, O278–O284. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Kumar, B.; Singhal, N.; Katoch, V.M.; Venkatesan, K.; Chauhan, D.S.; Bisht, D. Streptomycin induced protein expression analysis in Mycobacterium tuberculosis by two-dimensional gel electrophoresis & mass spectrometry. Indian J. Med. Res. 2010, 132, 400–408. [Google Scholar] [PubMed]
- Sharma, P.; Kumar, B.; Gupta, Y.; Singhal, N.; Katoch, V.M.; Venkatesan, K.; Bisht, D. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 2010, 8, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Bisht, D. Secretory Proteome Analysis of Streptomycin-Resistant Mycobacterium tuberculosis Clinical Isolates. SLAS Discov. Adv. Life Sci. R&D 2017, 22, 1229–1238. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.L. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, E.; Fariselli, P.; Rossi, I.; Casadio, R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinform. 2008, 9, S6. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [Green Version]
- Kmiecik, S.; Kouza, M.; Badaczewska-Dawid, A.E.; Kloczkowski, A.; Kolinski, A. Modeling of Protein Structural Flexibility and Large-Scale Dynamics: Coarse-Grained Simulations and Elastic Network Models. Int. J. Mol. Sci. 2018, 19, 3496. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A meth-od and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.; Jordan, D.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Cloete, R.; Akurugu, W.A.; Werely, C.J.; Van Helden, P.D.; Christoffels, A. Structural and functional effects of nucleotide variation on the human TB drug metabolizing enzyme arylamine N -acetyltransferase 1. J. Mol. Graph. Model. 2017, 75, 330–339. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [Green Version]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Xu, N.; Poisson, J.; Zhang, Y. A Protocol for Computer-Based Protein Structure and Function Prediction. J. Vis. Exp. 2011, 3259, e3259. [Google Scholar] [CrossRef] [Green Version]
- Shuid, A.N.; Kempster, R.; McGuffin, L.J. ReFOLD: A server for the refinement of 3D protein models guided by accurate quality estimates. Nucleic Acids Res. 2017, 45, W422–W428. [Google Scholar] [CrossRef] [Green Version]
- Adiyaman, R.; McGuffin, L.J. ReFOLD3: Refinement of 3D protein models with gradual restraints based on predicted local quality and residue contacts. Nucleic Acids Res. 2021, gkab300. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Aldowsari, F.M.F.; Alharbi, A.S.M.; Adiyaman, R. ModFOLD8: Accurate global and local quality estimates for 3D protein models. Nucleic Acids Res. 2021, gkab321. [Google Scholar] [CrossRef]
- Laskowski, R.; MacArthur, M.W.; Moss, D.S.; Thornton, J. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J. Chem. Inf. Model. 2019, 59, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Ascher, D.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Chen, J.; Blundell, T.L.; Ascher, D.B. In silico functional dissection of saturation mutagenesis: Interpreting the relationship between phenotypes and changes in protein stability, interactions and activity. Sci. Rep. 2016, 6, 19848. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.E.V.; Ascher, D.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [Green Version]
- Worth, C.; Preissner, R.; Blundell, T.L. SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011, 39, W215–W222. [Google Scholar] [CrossRef] [Green Version]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef] [Green Version]
- Jubb, H.C.; Higueruelo, A.; Ochoa-Montaño, B.; Pitt, W.; Ascher, D.B.; Blundell, T.L. Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef]
- Sobolev, V.; Sorokin, A.; Prilusky, J.; Abola, E.E.; Edelman, M. Automated analysis of interatomic contacts in proteins. Bioinformatics 1999, 15, 327–332. [Google Scholar] [CrossRef]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Badaczewska-Dawid, A.E.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef] [Green Version]
- Voth, G.A. (Ed.) Coarse-Graining of Condensed Phase and Biomolecular Systems; CRC Press: Boca Raton, FL, USA, 2009; ISBN 9781420059557. [Google Scholar]
- Liwo, A.; Baranowski, M.; Czaplewski, C.; Gołaś, E.; He, Y.; Jagieła, D.; Krupa, P.; Maciejczyk, M.; Makowski, M.; Mozolewska, M.A.; et al. A unified coarse-grained model of biological macromolecules based on mean-field multipole–multipole interactions. J. Mol. Model. 2014, 20, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Czaplewski, C.; Karczynska, A.; Sieradzan, A.K.; Liwo, A. UNRES server for physics-based coarse-grained simulations and prediction of protein structure, dynamics and thermodynamics. Nucleic Acids Res. 2018, 46, W304–W309. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).