Abstract
Mortality in neonates with Gram-negative bloodstream infections has remained unacceptably high. Very few data are available on the impact of resistance profiles, virulence factors, appropriateness of empirical treatment and clinical characteristics on patients’ mortality. A survival analysis to investigate 28-day mortality probability and predictors was performed including (I) infants <90 days (II) with an available Enterobacterales blood isolate with (III) clinical, treatment and 28-day outcome data. Eighty-seven patients were included. Overall, 299 virulence genes were identified among all the pathogens. Escherichia coli had significantly more virulence genes identified compared with other species. A strong positive correlation between the number of resistance and virulence genes carried by each isolate was found. The cumulative probability of death obtained by the Kaplan-Meier survival analysis was 19.5%. In the descriptive analysis, early age at onset, gestational age at onset, culture positive for E. coli and number of classes of virulence genes carried by each isolate were significantly associated with mortality. By Cox multivariate regression, none of the investigated variables was significant. This pilot study has demonstrated the feasibility of investigating the association between neonatal sepsis mortality and the causative Enterobacterales isolates virulome. This relationship needs further exploration in larger studies, ideally including host immunopathological response, in order to develop a tailor-made therapeutic strategy.
Keywords:
infant; newborn; bacteremia; Gram-negative bacteria; drug resistance; microbial; virulence factors; mortality 1. Introduction
Mortality in neonates with Gram-negative bloodstream infections (GN-BSIs) has remained unacceptably high. Appropriate empirical treatment is considered crucially important in reducing mortality. However, despite the improvement in neonatal care, the fatality rate in babies with GN-BSIs remains around 15–20%, also during the emergence of antimicrobial resistance (AMR) [1,2,3,4,5,6].
Very few data are available on the impact of different treatment regimens on clinical outcome in neonates with GN-BSIs. Previous studies conducted in both adults and children showed conflicting results on the impact of resistance profiles, appropriateness of empirical treatment and clinical characteristics on patients’ mortality [7,8,9,10,11,12,13,14,15]. In the last decade, the implementation of modern bioinformatics to assist next-generation sequencing data analysis greatly improved the knowledge on the genetic characterization of pathogenic strains that may serve as target for new therapies [16]. There has been a growing body of evidence about the role of virulence factors (VFs) in the pathogenesis of invasive infections. Enterobacterales employ many strategies to enhance invasiveness, overcome host defenses and cause infections. Different strains can use alternative VFs with similar functions during the infection process, with this plasticity leading to unique combinations of such factors [17]. Some VFs are disease-specific whereas others seem to play different roles in different types of infection [18]. This plasticity enables pathogenic strains to colonize and infect different tissues and hosts. Some major classes of VFs, such as capsule, siderophores and fimbriae, have been characterized well [18]. However, several other factors were recently identified and have yet to be defined to fully understand their mechanisms of action and clinical significance (summarized in Table 1). To achieve this goal, a genomic approach can be used to identify genes encoding specific virulence determinants. In adults, several genes present in the great majority of bacteremic strains and involved in virulence have been identified [19,20,21]. These are presumably essential for the infection process. However, the specific VFs that are relevant in causing neonatal GN-BSIs are not well defined yet, partly due the variability among the few studies available so far [2]. These have mostly been conducted on neonates and children with Escherichia coli bacteremia, with virtually no data available on other Enterobacterales [22,23,24,25]. Also, in the recent years, several data have been published investigating a potential role of the bacterial virulome, defined as the set of genes contributing to the bacterial virulence, in determining the outcome of patients with both Gram-positive and GN-BSIs. Again, these studies demonstrate a mixed picture reporting a significant correlation between virulence factors and mortality in both children and adults [2,19,26,27,28,29].

Table 1.
Main virulence factors in Enterobacterales.
Clarifying the role of the main determinants leading to adverse outcomes could help to define targeted interventions to decrease mortality. With this study, we aimed to investigate potential associations between patient characteristics, pathogen characteristics and antibiotic treatment regimen on the clinical outcome of neonates/infants affected by culture-proven GN-BSIs.
2. Results
2.1. Demographic and Clinical Data
Overall, 87 infants from six European countries (the United Kingdom: 49, Estonia: 21, Greece: 7, Italy: 7, Lithuania: 2, Spain: 1) between 2010–2015 fulfilled our inclusion criteria and were included in the study. Forty patients were retrieved from the neonatal infection surveillance network (NeonIN) study [30], 38 from NeoMero [31], and 9 from the Collaborations for Leadership in Applied Health Research and Care (CLAHRC) study [32]. Patients, pathogens and treatment characteristics of the included episodes are summarised in Table 2. At the BSI onset, the median age of the selected neonates was 15.2 days (interquartile range (IQR) 6.7–31), with a median gestational age (GA) of 33 weeks (IQR 28–37). Forty-nine out of 87 babies (56%) had a central line in situ at the episode onset. A total of 37 different antibiotic regimens have been reported among the 87 patients in the first 48 h of treatment.

Table 2.
Demographic and clinical characteristics of included patients.
2.2. Microbiological Data
The species IDs identified on matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry were all confirmed by sequencing. The isolate-specific accession numbers are indicated in Supplementary Table S1. The most frequently isolated pathogen was E. coli followed by Enterobacter spp. and Klebsiella spp. (Table 2). The percentage of multidrug-resistance (MDR) isolates was 30% (26/87). Based on interpretation of the in vitro susceptibility profile, 16/87 (18%) were suspected of producing an extended-spectrum beta lactamase (ESBL) enzyme, and only one isolate (Klebsiella pneumoniae) was resistant to carbapenems. Susceptibilities of single species to the investigated antibiotics are presented in Table 3.

Table 3.
Percentages of susceptibility for the isolated pathogens.
A total of 50 different sequence types (STs) were found, with ST131 and ST90 as the most frequent in E. coli and Enterobacter cloacae, respectively. The median number of classes of resistance genes carried per isolate was 5 (IQR 4–5) whereas the median number of classes of virulence genes was 7 (IQR 7–9). Twenty-five isolates harbored blaTEM-type genes, two non-Klebsiella spp. strains the blaSHV-type determinant, and two E. coli strains the blaCTX-M-type genes. One K. pneumoniae ST17 carried blaVIM-12 gene, and two Enterobacter asburiae (ST484) the mcr-9 determinant.
Overall, the genome sequencing identified 299 different virulence genes among all the pathogens. There was a strong positive correlation between the number of resistance and virulence genes carried by each isolate (Rho = 0.79; p = 0.001) (Figure 1).
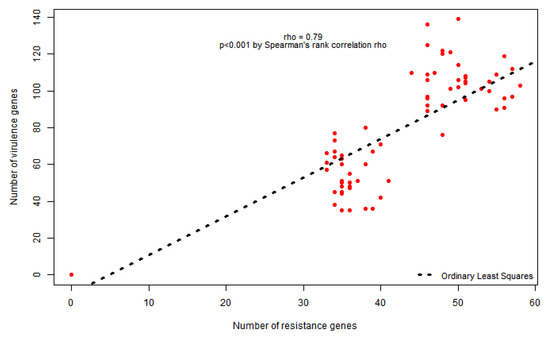
Figure 1.
Spearman’s rank correlation between number of resistance and virulence genes.
E. coli strains showed the highest mean number of virulence genes (105 vs. <65 in the other species overall), mainly those involved in fimbriae production (p < 0.0001) (Table 4). The genes that were more frequently carried by the isolates are summarized in Table 5. Among the most represented strains (E. coli, E. cloacae, K. pneumoniae, K. oxytoca) the following virulence genes were carried by all the isolates: gal, gnd, rcs (capsule); ompA (cell invasion); ent, fep (iron metabolism); acr (pumps). Some genes were shown to be strain-specific. Among them, the adhesion mrk gene cluster was sequenced in all Klebsiella spp. isolates as well as the secretion system’s exe and impA.tss genes. Conversely, genes encoding for motility and chemotaxis proteins (che, flg and fli) were only carried by E. coli and Enterobacter spp. The clb, esp and hly genes encoding for toxin proteins were sequenced only in E. coli strains. One hypervirulent K. pneumoniae with a hypermucoviscous phenotype harboring the rmpA and wca genes was found [33].

Table 4.
Median number of classes of virulence genes carried per bacterial strain.

Table 5.
Number and percentage of the most represented virulence genes.
2.3. Determinants of 28-Day Case-Fatality
The cumulative probability of death obtained by the Kaplan-Meier survival analysis was 19.5% with the greater percentage of deaths happening in the first week. In the descriptive analysis, early age at onset (p = 0.002), culture positive for E. coli (p = 0.029), number of classes of virulence genes carried per isolate (p = 0.022) and GA (weeks) at the onset (p = 0.003) were significantly associated with mortality (Table 6). By Cox multivariate regression, none of the investigated variables was significant (Table 7).

Table 6.
Descriptive analysis of potential association between variables and mortality.

Table 7.
Multivariate regression analysis on the 28-day mortality predictors.
3. Discussion
This study included 87 European neonates and infants younger than 90 days with GN-BSIs due to Enterobacterales. Overall, 299 virulence genes were identified in these pathogens. Among the different organisms, E. coli had significantly more virulence genes identified compared with other species. Gal, gnd, rcs, ompA, ent, fep, and acr virulence genes were identified from all the pathogens, likely being essential for the infection process. Conversely, some other genes were shown to be strain-specific. A strong positive correlation between the number of resistance and virulence genes carried by each isolate was found. By survival analysis, the 28-day probability of death was 19.5%. In the descriptive analysis, early age at onset, GA at the onset, culture positive for E. coli and number of classes of virulence genes carried by each isolate were significantly associated with mortality whereas discordant therapy was not related to mortality. By Cox multivariate regression, none of the investigated variables was significant.
Many studies have been conducted in both adults and neonates trying to define the main determinants of mortality in patients with GN-BSIs. AMR has been broadly investigated in the adult population, with the majority of studies reporting a significant association between multiple resistance to antibiotics and patients mortality [7,34,35,36]. Some large cohorts have not found a clear correlation between AMR and adverse outcome [8,12,13]. Data from this small study did not confirm a significant impact of resistance profile on neonatal mortality [5].
In recent years, an increasing number of studies are being conducted trying to investigate the impact of virulence genes on the outcome of patients with GN-BSIs. Among them, E. coli was the most frequently investigated pathogen followed by K. pneumoniae. Independent risk factors associated with 30-day mortality among adult patients with ESBL-producing E. coli bacteremia included siderophores iroN and iss positivity [21], the siderophore fyuA gene, and the presence of the afimbrial adhesin afa gene [19,37]. In a large prospective study investigating the main determinants for adverse outcome in patients with K. pneumoniae BSIs, the cytotoxicity pks gene cluster carriage by causative strains was an independent risk factor for 30-day mortality when accompanied by MDR [38]. Lastly, the siderophore-related iutA gene was found to be an independent predictor of the 30-day mortality in K. pneumoniae bacteremia [39]. However, almost all of these studies were conducted with pre-selected virulence genes searched by polymerase chain reaction (PCR) rather than sequencing the entire bacterial virulome. This led to a wide heterogeneity, with each group analyzing different genes and hampering the comparison of the results. Very little data have been published on the relationship between bacterial virulence factors and BSI mortality in children, and almost all in patients with E. coli bacteremia. In a prospective cohort of 43 septic neonates, the adhesin hek/hra gene was found to be significantly more frequent in isolates from newborns who died than in isolates from survivors [2]. On the other hand, in a cohort of children 0–17 years old (median age 2.4 months), none of the 20 virulence factors tested by PCR was found to correlate with sepsis severity [26]. The Burden of Antibiotic Resistance in Neonates from Developing Societies (BARNARDS) study was conducted to assess the burden of AMR in neonates in seven low-middle income countries [6]. In this study, Gram-negative (GN) pathogens from neonatal sepsis were isolated and characterized through whole genome sequencing (WGS) for resistance and virulence genes. The number of virulence genes carried by each isolate through a virulence score was used. The results obtained suggested that yersiniabactin and/or aerobactin/salmochelin virulence genes may be involved in a more rapid onset and mortality. However, the inability to follow up all neonates and additional local factors likely to contribute to patient’s death hampered the authors’ capacity to attribute mortality singularly to the presence/absence of genomic traits.
Our results showed a strong positive correlation between the number of resistance and virulence genes carried by each isolate. Many studies have been conducted on either AMR or virulence. However, the biological effect and connection between these two factors are of particular importance [40,41]. Indeed, a negative or positive relationship can be found among them. Enhanced virulence or AMR frequently has been reported to have a fitness cost on bacteria but their relationship changes according to different bacterial species, the resistance and virulence genes involved and the host’s immune system [42,43]. Some antibiotics, such as ceftazidime, cefotaxime and quinolones, have been reported to enhance the increase of the deletion and transposition of DNA regions that are specific for VFs [42]. By contrast, a positive correlation has been shown between AMR and virulence with the use of other antibiotics. In particular, uropathogenic strains of E. coli carrying the blaCTX-M-15 resistance gene also harbored more colV, colE2-E9, colIa-Ib, hlyA, and csgA genes as well as the blaOXA-2 beta lactamase was correlated with increased expression of colM, colB, colE, and crl genes [44]. Prophages are another mechanism that has been shown to be involved in both virulence and resistance diffusion through the spread of toxins to other resistant strains [45]. Porins and biofilm also play an important role in the relationship between virulence and resistance, with the first acting as a channel controlling the entrance of both antibiotics and VFs into the pathogens (i.e., the OmpC gene) and the second favoring antimicrobial treatment tolerance and infection persistence at the same time also increasing the transfer of resistance and virulence genes among the cells [46]. Lastly, an enhancement in both resistance and virulence characteristics of the pathogens can occur through mobile genetic elements like plasmids. These self-replicating extra-chromosomal elements are capable of transferring among different bacteria while carrying virulence and resistance genes [47]. This mechanism is independent of any antibiotic pressure.
There are several limitations in the study design, due to both confounding factors and heterogeneity of our sample. Firstly, the study is not powered to demonstrate a significant correlation between the presence of single resistance/virulence genes and neonates’ clinical outcome; this required us to analyze resistome and virulome by grouping genes according to the class. Moreover, we are well aware of the rapid dynamics of bacterial genetics, and the selection of a single time point isolate can cause bias and affect the results. At the same time, considering the possible implication of heteroresistance, the selection of certain colonies in the first place could have potentially missed relevant isolates. We found a significantly higher number of virulence genes in E. coli isolates compared with other species; this could be due to an over representation of E. coli genes in the virulence database (VFDB). Different pathogens can have different impacts on neonates with sepsis, and pooling data on multiple strains could alter the results. Lastly, neonates and infants represent a broadly heterogeneous population, being characterized by different gestational ages, birth weight, underlying conditions, and risk factors. Despite these limitations, this was the first study to correlate virulence factors of non-E. coli Enterobacterales with mortality in septic neonates. Although a number of studies have tried to correlate single virulence genes with Gram-negative sepsis outcome, virtually none of them have investigated the role of the entire virulome in causing mortality. Considering the huge number of virulence genes involved and the limited sample size, we presented a potential way to use the entire body of information gained from the WGS by grouping genes as the number of classes of virulence genes involved. Having access to a very complete dataset including patient-level treatment, outcome data, and isolates available for a comprehensive genetic characterization, we believe that our data provide new and relevant information on the molecular picture of GN pathogens causing neonatal BSIs.
4. Materials and Methods
4.1. Study Design and Data Source
A case series of neonates with clinical sepsis and microbiologically confirmed GN-BSIs was constructed from three separate studies: the NeonIN [30], the NeoMero clinical trial [31], and the CLAHRC [32]. The NeonIN is a multinational network which prospectively collect data on neonatal infections from neonatal units. The detailed data procedures have been previously described [48]. NeoMero was a European-based randomized controlled trial to compare the efficacy of meropenem with standard of care (ampicillin + gentamicin or cefotaxime + gentamicin) in the treatment of late onset neonatal sepsis. CLAHRC is a United Kingdom-based prospective cohort study to collect data for patients with GN-BSIs in three hospitals in South London which aims to characterize clinical management of patients with GN-BSIs in all ages and identify potential risk factors associated with 28-day treatment outcomes.
4.2. Selection Criteria, Available Data and Definitions
Patients with a microbiologically-confirmed diagnosis of GN-BSIs due to Enterobacterales were selected from the above studies among European Neonatal Intensive Care Units between 2010–2015. Inclusion criteria were (I) age between 0–90 days, (II) Enterobacterales blood isolate available for the sequencing, (III) patient-level clinical and treatment data, and (IV) 28-day clinical outcome data.
Data available for the analysis included demographic, risk factors, clinical characteristics, antibiotic treatment, susceptibility results, resistome and virulome of the isolated bacteria, and 28-day treatment outcome.
The date of the blood culture was considered as the day of the sepsis onset. The MDR of the isolates was defined according to Magiorakos A.-P. et al. as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [49]. The first 48-h antibiotic treatment was defined as concordant or discordant based on the inclusion of at least an active drug against the blood isolate. The evaluated outcome was defined as 28-day case-fatality (patient alive/dead).
4.3. Microbiological Methods
Bacterial isolates from blood were cultured from frozen stocks (−80 °C) on blood agar plates. Species were identified by MALDI-TOF mass spectrometry (Bruker, Karlsruhe, Germany). Antibiotic susceptibility profiles were obtained according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2019 Clinical Breakpoints with disk diffusion tests for the following antibiotics: amikacin, ampicillin, amoxicillin-clavulanate, ceftriaxone, ceftazidime, aztreonam, ciprofloxacin, gentamicin, meropenem, piperacillin-tazobactam, and trimethoprim-sulphametoxazole. To facilitate the analyses, isolates that were defined as having increased exposure were classified as non-susceptible. The isolates included in the study were subjected to WGS using the Illumina MiSeq platform (Illumina, San Diego, CA, USA), with paired-end runs of 2 × 300 bp, after Nextera XT library preparation. The obtained reads were assembled using SPAdes [50]. For each genome, we determined the ST using an in-house script (available upon request) and the Multilocus sequence typing (MLST) schemes and gene alleles sequences available on PubMLST (pubmlst.org). The isolates were further characterized at the genomic level with the identification of resistant and virulence genes using ABRicate (Seemann T, Abricate, Github https://github.com/tseemann/abricate, accessed on: 11 November 2019) and the following databases: The Comprehensive Antibiotic Resistance Database (CARD) [51] and Resfinder [52] for the resistance genes and VFDB [53] for the virulence genes.
4.4. Statistical Analysis
Qualitative variables were summarized by absolute frequencies and percentages, and quantitative variables by median and IQR. A descriptive analysis was conducted with the potential association between variables and outcome of interest evaluated by chi-squared or Fisher’s exact test as more appropriate for qualitative variables, and Mann–Whitney or t-test as appropriate for quantitative variables. A Spearman’s rank correlation was calculated to evaluate the correlation between the number of resistance genes and virulence genes carried by each isolate. We performed a survival analysis to investigate the 28-day mortality predictors using the Cox regression model (primary endpoint), after evaluated the proportional hazard (PH) assumption. In case of non-PH assumption, the weighted Cox regression model was performed [54]. A bivariate analysis (univariate) was carried out and the variables for which the p-value was <0.10 in univariate analysis were included in the multivariate model. All variables entered as covariates were evaluated at the baseline. As secondary endpoint, a survival analysis was conducted using the Kaplan-Meier curves to assess the probability of death at 28 days. To facilitate the analysis, the resistome and virulome data obtained in sequencing were categorized as number of classes of virulence and resistance genes carried by each isolate. Because of the huge heterogeneity of treatment regimens among the included patients, antibiotics were coded according to the WHO ATC/DDD Index 2020 at the 4th level [55]. p values <0.05 were considered as statistically significant.
All statistical analyses were performed using R Statistical Software (version 4.0.2) [56].
4.5. Ethics
The source studies were approved by the Ethical Committees of the participating institutions, and all enrolled patients’ legal guardians provided informed consent. Given the retrospective nature of the present study, ethical approval for this analysis was not necessary.
5. Conclusions
To conclude, this pilot study demonstrated the feasibility of investigating the association between neonatal sepsis mortality and the causative Enterobacterales isolates virulome. The limited sample size of our cohort did not allow us to determine the role of single virulence genes in neonatal GN-BSIs but grouping genes as the number of classes involved allowed us to investigate the impact of the entire virulome in neonatal sepsis outcome. This knowledge may be useful for predicting clinical outcomes, detecting virulent strains, and helping with vaccine development. Expanding research on anti-virulence molecules together with the development of new antibiotics could be crucial to improving the management of these fragile patients. Further research would be advisable to elucidate the correlation with the timing of sepsis onset to personalize the clinical approach. These findings need further exploration in larger global studies, ideally including host immunopathological response, in order to develop a tailor-made therapeutic strategy.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10060706/s1, Table S1: Isolate-specific accession numbers.
Author Contributions
Conceptualization, L.F., M.S.; methodology, D.D.C., F.C., A.P., Y.H.; validation, D.D.C., F.C., A.A.W., I.B., Y.H., K.L., I.M., J.B., A.A., G.V.Z., T.P., P.T.H., M.S.; formal analysis, L.F., D.D.C., F.C., A.P., A.A.W., K.L., I.M.; investigation, L.F., K.L., I.M.; resources, L.F., M.S.; data curation, L.F., Y.H.; writing—original draft preparation, L.F.; writing—review and editing, D.D.C., F.C., A.P., A.A.W., I.B., Y.H., K.L., I.M., J.B., A.A., G.V.Z., T.P., P.T.H., M.S.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study and the APC were funded by Penta Foundation.
Institutional Review Board Statement
The source studies were approved by the Ethical Committees of the participating institutions. Given the retrospective nature of the present study, ethical approval for this analysis was not necessary.
Informed Consent Statement
All enrolled patients’ legal guardians provided informed consent.
Data Availability Statement
The corresponding author confirms that he had full access to all the data in the study and had final responsibility for the decision to submit for publication. Whole-genome shotgun projects have been deposited in GenBank (project code PRJEB44870). The isolate-specific accession numbers are indicated in Supplementary Table S1.
Acknowledgments
We acknowledge “The NeoMero Consortium” for providing data regarding patients enrolled in the NeoMero 1 clinical trial for the present study.
Conflicts of Interest
The authors declare no conflict of interest related to this work.
References
- Ballot, D.E.; Bandini, R.; Nana, T.; Bosman, N.; Thomas, T.; Davies, V.A.; Cooper, P.A.; Mer, M.; Lipman, J. A review of -multidrug-resistant Enterobacteriaceae in a neonatal unit in Johannesburg, South Africa. BMC Pediatr. 2019, 19, 320. [Google Scholar] [CrossRef]
- Cole, B.K.; Ilikj, M.; McCloskey, C.B.; Chavez-Bueno, S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS ONE 2019, 14, e0219352. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Chen, F.; Cai, K.; Tan, J.; Xie, W.; Qian, R.; Liu, X.; Zhang, W.; Du, H.; et al. A risk score based on pediatric sequential organ failure assessment predicts 90-day mortality in children with Klebsiella pneumoniae bloodstream infection. BMC Infect. Dis. 2020, 20, 916. [Google Scholar] [CrossRef]
- Nour, I.; Eldegla, H.E.; Nasef, N.; Shouman, B.; Abdel-Hady, H.; Shabaan, A.E. Risk factors and clinical outcomes for carbapenem-resistant Gram-negative late-onset sepsis in a neonatal intensive care unit. J. Hosp. Infect. 2017, 97, 52–58. [Google Scholar] [CrossRef]
- Thatrimontrichai, A.; Premprat, N.; Janjindamai, W.; Dissaneevate, S.; Maneenil, G. Risk Factors for 30-Day Mortality in Neonatal Gram-Negative Bacilli Sepsis. Am. J. Perinatol. 2020, 37, 689–694. [Google Scholar] [CrossRef]
- Sands, K.; Carvalho, M.J.; Portal, E.; Thomson, K.; Dyer, C.; Akpulu, C.; Andrews, R.; Ferreira, A.; Gillespie, D.; Hender, T.; et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 2021, 6, 512–523. [Google Scholar] [CrossRef]
- Budhram, D.R.; Mac, S.; Bielecki, J.M.; Patel, S.N.; Sander, B. Health outcomes attributable to carbapenemase-producing Enterobacteriaceae infections: A systematic review and meta-analysis. Infect. Control. Hosp. Epidemiol. 2020, 41, 37–43. [Google Scholar] [CrossRef]
- Burnham, J.P.; Lane, M.A.; Kollef, M.H. Impact of Sepsis Classification and Multidrug-Resistance Status on Outcome among Patients Treated with Appropriate Therapy. Crit. Care Med. 2015, 43, 1580–1586. [Google Scholar] [CrossRef] [PubMed]
- Folgori, L.; Bernaschi, P.; Piga, S.; Carletti, M.; Cunha, F.P.; Lara, P.H.; de Castro Peixoto, N.C.; Alves Guimarães, B.G.; Sharland, M.; Araujo da Silva, A.R.; et al. Healthcare-Associated Infections in Pediatric and Neonatal Intensive Care Units: Impact of Underlying Risk Factors and Antimicrobial Resistance on 30-Day Case-Fatality in Italy and Brazil. Infect. Control. Hosp. Epidemiol. 2016, 37, 1302–1309. [Google Scholar] [CrossRef]
- Gutiérrez-Gutiérrez, B.; Salamanca, E.; de Cueto, M.; Hsueh, P.R.; Viale, P.; Paño-Pardo, J.R.; Venditti, M.; Tumbarello, M.; Daikos, G.; Cantón, R.; et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): A retrospective cohort study. Lancet Infect. Dis. 2017, 17, 726–734. [Google Scholar] [CrossRef]
- Kyo, M.; Ohshimo, S.; Kosaka, T.; Fujita, N.; Shime, N. Impact of inappropriate empiric antimicrobial therapy on mortality in pediatric patients with bloodstream infection: A retrospective observational study. J. Chemother. 2019, 31, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Sakellariou, C.; Gürntke, S.; Steinmetz, I.; Kohler, C.; Pfeifer, Y.; Gastmeier, P.; Schwab, F.; Kola, A.; Deja, M.; Leistner, R. Sepsis Caused by Extended-Spectrum Beta-Lactamase (ESBL)-Positive, K. pneumoniae and E. coli: Comparison of Severity of Sepsis, Delay of Anti-Infective Therapy and ESBL Genotype. PLoS ONE 2016, 11, e0158039. [Google Scholar] [CrossRef]
- Sianipar, O.; Asmara, W.; Dwiprahasto, I.; Mulyono, B. Mortality risk of bloodstream infection caused by either Escherichia coli or Klebsiella pneumoniae producing extended-spectrum β-lactamase: A prospective cohort study. BMC Res. Notes 2019, 12, 719. [Google Scholar] [CrossRef]
- Tanır Basaranoglu, S.; Ozsurekci, Y.; Aykac, K.; Karadag Oncel, E.; Bıcakcigil, A.; Sancak, B.; Cengiz, A.B.; Kara, A.; Ceyhan, M. A comparison of blood stream infections with extended spectrum beta-lactamase-producing and non-producing Klebsiella pneumoniae in pediatric patients. Ital. J. Pediatr. 2017, 43, 79. [Google Scholar] [CrossRef]
- Zarkotou, O.; Pournaras, S.; Tselioti, P.; Dragoumanos, V.; Pitiriga, V.; Ranellou, K.; Prekates, A.; Themeli-Digalaki, K.; Tsakris, A. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin. Microbiol. Infect. 2011, 17, 1798–1803. [Google Scholar] [CrossRef]
- Lisboa, T.; Waterer, G.; Rello, J. We should be measuring genomic bacterial load and virulence factors. Crit. Care Med. 2010, 38 (Suppl. S10), S656–S662. [Google Scholar] [CrossRef]
- Ron, E.Z. Distribution and evolution of virulence factors in septicemic Escherichia coli. Int. J. Med. Microbiol. 2010, 300, 367–370. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Daga, A.P.; Koga, V.L.; Soncini, J.G.M.; de Matos, C.M.; Peruginim, M.R.E.; Pelisson, M.; Kobayashi, R.K.T.; Vespero, E.C. Escherichia coli Bloodstream Infections in Patients at a University Hospital: Virulence Factors and Clinical Characteristics. Front. Cell Infect. Microbiol. 2019, 9, 191. [Google Scholar] [CrossRef]
- Du, F.; Wei, D.D.; Wan, L.G.; Cao, X.W.; Zhang, W.; Liu, Y. Evaluation of ompK36 allele groups on clinical characteristics and virulence features of Klebsiella pneumoniae from bacteremia. J. Microbiol. Immunol. Infect. 2019, 52, 779–787. [Google Scholar] [CrossRef]
- Hung, W.T.; Cheng, M.F.; Tseng, F.C.; Chen, Y.S.; Shin-Jung Lee, S.; Chang, T.H.; Lin, H.H.; Hung, C.H.; Wang, J.L. Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: The role of virulence genes. J. Microbiol. Immunol. Infect. 2019, 52, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Tsau, Y.K.; Kuo, C.Y.; Su, L.H.; Lin, T.Y. Comparison of extended virulence genotypes for bacteria isolated from pediatric patients with urosepsis, acute pyelonephritis, and acute lobar nephronia. Pediatr. Infect. Dis. J. 2010, 29, 736–740. [Google Scholar] [CrossRef]
- Palma, N.; Gomes, C.; Riveros, M.; García, W.; Martínez-Puchol, S.; Ruiz-Roldán, L.; Mateu, J.; García, C.; Jacobs, J.; Ochoa, T.J.; et al. Virulence factors profiles and ESBL production in Escherichia coli causing bacteremia in Peruvian children. Diagn. Microbiol. Infect. Dis. 2016, 86, 70–75. [Google Scholar] [CrossRef]
- Roy, S.; Datta, S.; DAS, P.; Gaind, R.; Pal, T.; Tapader, R.; Mukherjee, S.; Basu, S. Insight into neonatal septicaemic Escherichia coli from India with respect to phylogroups, serotypes, virulence, extended-spectrum-β-lactamases and association of ST131 clonal group. Epidemiol. Infect. 2015, 143, 3266–3276. [Google Scholar] [CrossRef] [PubMed]
- Tapader, R.; Chatterjee, S.; Singh, A.K.; Dayma, P.; Haldar, S.; Pal, A.; Basu, S. The high prevalence of serine protease autotransporters of Enterobacteriaceae (SPATEs) in Escherichia coli causing neonatal septicemia. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2015–2024. [Google Scholar] [CrossRef]
- Burdet, C.; Clermont, O.; Bonacorsi, S.; Laouénan, C.; Bingen, E.; Aujard, Y.; Mentré, F.; Lefort, A.; Denamur, E. Escherichia coli bacteremia in children: Age and portal of entry are the main predictors of severity. Pediatr. Infect. Dis. J. 2014, 33, 872–879. [Google Scholar] [CrossRef][Green Version]
- Horváth, A.; Dobay, O.; Sahin-Tóth, J.; Juhász, E.; Pongrácz, J.; Iván, M.; Fazakas, E.; Kristóf, K. Characterisation of antibiotic resistance, virulence, clonality and mortality in MRSA and MSSA bloodstream infections at a tertiary-level hospital in Hungary: A 6-year retrospective study. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Zürn, K.; Lander, F.; Hufnagel, M.; Monecke, S.; Berner, R. Microarray Analysis of Group B Streptococci Causing Invasive Neonatal Early- and Late-onset Infection. Pediatr. Infect. Dis. J. 2020, 39, 449–453. [Google Scholar] [CrossRef]
- Xu, M.; Fu, Y.; Kong, H.; Chen, X.; Chen, Y.; Li, L.; Yang, Q. Bloodstream infections caused by Klebsiella pneumoniae: Prevalence of bla(KPC), virulence factors and their impacts on clinical outcome. BMC Infect. Dis. 2018, 18, 358. [Google Scholar] [CrossRef]
- Neonatal Infection Surveillance Database. Available online: https://www.neonin.org.uk/ (accessed on 15 April 2021).
- Lutsar, I.; Chazallon, C.; Trafojer, U.; de Cabre, V.M.; Auriti, C.; Bertaina, C.; Calo Carducci, F.I.; Canpolat, F.E.; Esposito, S.; Fournier, I.; et al. Meropenem vs standard of care for treatment of neonatal late onset sepsis (NeoMero1): A randomised controlled trial. PLoS ONE 2020, 15, e0229380. [Google Scholar] [CrossRef]
- CLAHRC-Infection Theme-Gram-Negative Project—Bloodstream Infection. Available online: http://www.clahrc-southlondon.nihr.ac.uk/infection/investigating-causes-and-care-septicaemia-caused-gram-negative-bacteria (accessed on 15 April 2021).
- Derakhshan, S.; Najar Peerayeh, S.; Bakhshi, B. Association between Presence of Virulence Genes and Antibiotic Resistance in Clinical Klebsiella Pneumoniae Isolates. Lab. Med. 2016, 47, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Pop-Vicas, A.; Opal, S.M. The clinical impact of multidrug-resistant gram-negative bacilli in the management of septic shock. Virulence 2014, 5, 206–212. [Google Scholar] [CrossRef]
- Viale, P.; Giannella, M.; Lewis, R.; Trecarichi, E.M.; Petrosillo, N.; Tumbarello, M. Predictors of mortality in multidrug-resistant Klebsiella pneumoniae bloodstream infections. Expert Rev. Anti Infect. Ther. 2013, 11, 1053–1063. [Google Scholar] [CrossRef]
- Ray, S.; Anandm, D.; Purwar, S.; Samanta, A.; Upadhye, K.V.; Gupta, P.; Dhar, D. Association of high mortality with extended-spectrum β-lactamase (ESBL) positive cultures in community acquired infections. J. Crit. Care 2018, 44, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Mora-Rillo, M.; Fernández-Romero, N.; Navarro-San Francisco, C.; Díez-Sebastián, J.; Romero-Gómez, M.P.; Fernández, F.A.; López, J.R.A.; Mingorance, J. Impact of virulence genes on sepsis severity and survival in Escherichia coli bacteremia. Virulence 2015, 6, 93–100. [Google Scholar] [CrossRef]
- Kim, D.; Park, B.Y.; Choi, M.H.; Yoon, E.J.; Lee, H.; Lee, K.J.; Park, Y.S.; Shin, J.H.; Uh, Y.; Shin, K.S.; et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J. Antimicrob. Chemother. 2019, 74, 190–199. [Google Scholar] [CrossRef]
- Namikawa, H.; Niki, M.; Niki, M.; Yamada, K.; Nakaie, K.; Sakiyama, A.; Oinuma, K.I.; Tsubouchi, T.; Tochino, Y.; Takemoto, Y.; et al. Clinical and virulence factors related to the 30-day mortality of Klebsiella pneumoniae bacteremia at a tertiary hospital: A case-control study. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2291–2297. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Interplay between Antibiotic Resistance and Virulence during Disease Promoted by Multidrug-Resistant Bacteria. J. Infect. Dis. 2017, 215 (Suppl. S1), S9–S17. [Google Scholar] [CrossRef]
- Gharrah, M.M.; El-Mahdy, A.M.; Barwa, R.F. Association between Virulence Factors and Extended Spectrum Beta-Lactamase Producing Klebsiella pneumoniae Compared to Nonproducing Isolates. Interdiscip. Perspect. Infect. Dis. 2017, 2017, 7279830. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Uhlemann, A.C. Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella pneumoniae. J. Infect. Dis. 2017, 215 (Suppl. S1), S18–S27. [Google Scholar] [CrossRef]
- Abd El-Baky, R.M.; Ibrahim, R.A.; Mohamed, D.S.; Ahmed, E.F.; Hashem, Z.S. Prevalence of Virulence Genes and Their Association with Antimicrobial Resistance Among Pathogenic, E. coli Isolated from Egyptian Patients with Different Clinical Infections. Infect. Drug Resist. 2020, 13, 1221–1236. [Google Scholar] [CrossRef]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef]
- Vergnano, S.; Menson, E.; Kennea, N.; Embleton, N.; Russell, A.B.; Watts, T.; Robinson, M.J.; Collinson, A. Heath PT. Neonatal infections in England: The NeonIN surveillance network. Arch. Dis. Child. Fetal. Neonatal. Ed. 2011, 96, F9–F14. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Schemper, M.; Wakounig, S.; Heinze, G. The estimation of average hazard ratios by weighted Cox regression. Stat. Med. 2009, 28, 2473–2489. [Google Scholar] [CrossRef] [PubMed]
- ATC/DDD Index 2020. Available online: https://www.whocc.no/atc_ddd_index/ (accessed on 23 November 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 15 April 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).