Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm
Abstract
1. Introduction
2. Results
2.1. Virulence Profiling
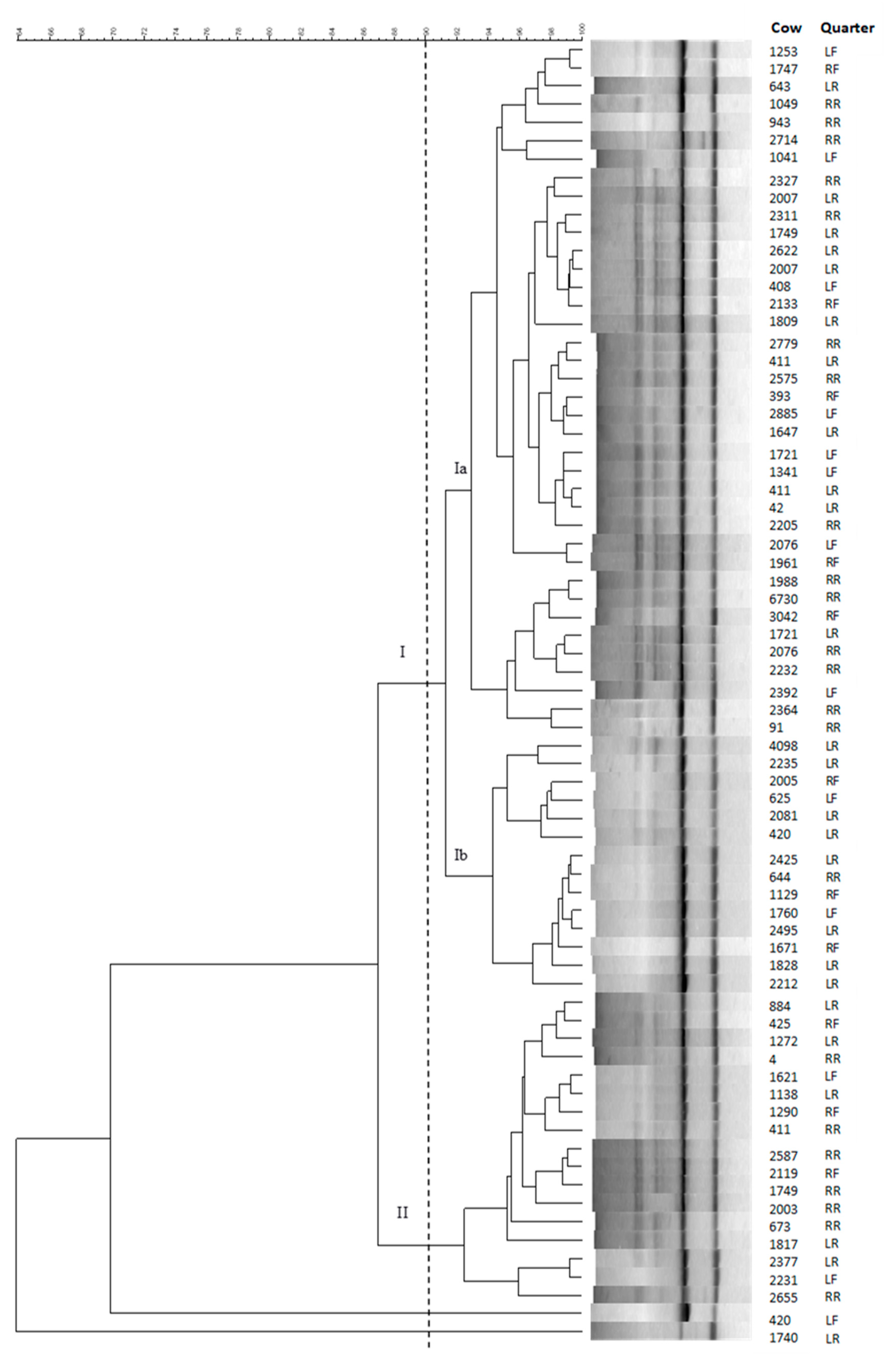
2.2. Genotyping
2.3. Antimicrobial Profiling
2.4. Distribution of Antimicrobial Resistance between Genotypic Clusters
3. Discussion
4. Materials and Methods
4.1. Herd
4.2. Sample Collection
4.3. Bacteriological Culture and MALDI-TOF Confirmation
4.4. Molecular Characterization
4.5. Cluster Analysis
4.6. Antimicrobial Susceptibility Testing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bradley, A.J.; Leach, K.A.; Breen, J.E.; Green, L.E.; Green, M.J. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 2007, 160, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Botrel, M.A.; Haenni, M.; Morignat, E.; Sulpice, P.; Madec, J.Y.; Calavas, D. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog. Dis. 2010, 7, 479–487. [Google Scholar] [CrossRef]
- Verbeke, J.; Piepers, S.; Supre, K.; De Vliegher, S. Pathogen-specific incidence rate of clinical mastitis in Flemish dairy herds, severity, and association with herd hygiene. J. Dairy Sci. 2014, 97, 6926–6934. [Google Scholar] [CrossRef] [PubMed]
- Poutrel, B.; Bareille, S.; Lequeux, G.; Leboeuf, F. Prevalence of mastitis pathogens in France: Antimicrobial susceptibility of Staphylococcus aureus, Streptococcus uberis and Escherichia coli. J. Vet. Sci. Technol. 2018, 9, 522. [Google Scholar] [CrossRef]
- Zadoks, R.N.; Tikofsky, L.L.; Boor, K.J. Ribotyping of Streptococcus uberis from a dairy’s environment, bovine feces and milk. Vet. Microbiol. 2005, 109, 257–265. [Google Scholar] [CrossRef]
- Lopez-Benavides, M.G.; Williamson, J.H.; Pullinger, G.D.; Lacy-Hulbert, S.J.; Cursons, R.T.; Leigh, J.A. Field observations on the variation of Streptococcus uberis populations in a pasture-based dairy farm. J. Dairy Sci. 2007, 90, 5558–5566. [Google Scholar] [CrossRef]
- Ericsson Unnerstad, H.; Lindberg, A.; Persson Waller, K.; Ekman, T.; Artursson, K.; Nilsson-Ost, M.; Bengtsson, B. Microbial aetiology of acute clinical mastitis and agent-specific risk factors. Vet. Microbiol. 2009, 137, 90–97. [Google Scholar] [CrossRef]
- Wente, N.; Klocke, D.; Paduch, J.H.; Zhang, Y.; Seeth, M.T.; Zoche-Golob, V.; Reinecke, F.; Mohr, E.; Krömker, V. Associations between Streptococcus uberis strains from the animal environment and clinical bovine mastitis cases. J. Dairy Sci. 2019, 102, 9360–9369. [Google Scholar] [CrossRef]
- Wald, R.; Baumgartner, M.; Gutschireiter, J.; Bazzanella, B.; Lichtmannsperger, K.; Wagner, M.; Wittek, T.; Stessl, B. Comparison of the population structure of Streptococcus uberis mastitis isolates from Austrian small-scale dairy farms and a Slovakian large-scale farm. J. Dairy Sci. 2020, 103, 1820–1830. [Google Scholar] [CrossRef]
- Gilbert, F.B.; Fromageau, A.; Lamoureux, J.; Poutrel, B. Evaluation of tandem repeats for MLVA typing of Streptococcus uberis isolated from bovine mastitis. BMC Vet. Res. 2006, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.; Rodriguez-Lecompte, J.C.; Blanchard, A.; McClure, J.T.; Sánchez, J. Molecular variability of Streptococcus uberis isolates from intramammary infections in Canadian dairy farms from the Maritime region. Can. J. Vet. Res. 2019, 83, 168–176. [Google Scholar] [PubMed]
- Leelahapongsathon, K.; Schukken, Y.H.; Srithanasuwan, A.; Suriyasathaporn, W. Molecular epidemiology of Streptococcus uberis intramammary infections: Persistent and transient patterns of infection in a dairy herd. J. Dairy Sci. 2020, 103, 3565–3576. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.L.; Leigh, J.A.; Bradley, A.J.; Archer, S.C.; Emes, R.D.; Green, M.J. Molecular Epidemiology of Streptococcus uberis Clinical Mastitis in Dairy Herds: Strain Heterogeneity and Transmission. J. Clin. Microbiol. 2016, 54, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, R.N.; Gillespie, B.E.; Barkema, H.W.; Sampimon, O.C.; Oliver, S.P.; Schukken, Y.H. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol. Infect. 2003, 130, 335–349. [Google Scholar] [CrossRef]
- Tomazi, T.; Freu, G.; Alves, B.G.; de Souza Filho, A.F.; Heinemann, M.B.; Veiga Dos Santos, M. Genotyping and antimicrobial resistance of Streptococcus uberis isolated from bovine clinical mastitis. PLoS ONE. 2019, 14, e0223719. [Google Scholar] [CrossRef]
- Gilchrist, T.L.; Smith, D.G.; Fitzpatrick, J.L.; Zadoks, R.N.; Fontaine, M.C. Comparative molecular analysis of ovine and bovine Streptococcus uberis isolates. J. Dairy Sci. 2013, 96, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.A.; Dego, O.K.; Headrick, S.I.; Lewis, M.J.; Oliver, S.P. Role of Streptococcus uberis adhesion molecule in the pathogenesis of Streptococcus uberis mastitis. Vet. Microbiol. 2015, 179, 332–335. [Google Scholar] [CrossRef]
- Boonyayatra, S.; Tharavichitkul, P.; Oliver, S.P. Virulence-associated genes and molecular typing of Streptococcus uberis associated with bovine mastitis in northern Thailand. Turk. J. Vet. Anim. Sci. 2018, 42, 73–81. [Google Scholar] [CrossRef]
- Calonzi, D.; Romano, A.; Monistero, V.; Moroni, P.; Luini, M.V.; Biscarini, F.; Castiglioni, B.; Cremonesi, P. Technical note: Development of multiplex PCR assays for the molecular characterization of Streptococcus uberis strains isolated from bovine mastitis. J. Dairy Sci. 2020, 103, 915–921. [Google Scholar] [CrossRef]
- Fang, W.; Oliver, S.P. Identification of lactoferrin-binding proteins in bovine mastitis-causing Streptococcus uberis. FEMS Microbiol. Lett. 1999, 176, 91–96. [Google Scholar] [CrossRef]
- Pol, M.; Ruegg, P.L. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J. Dairy Sci. 2007, 90, 249–261. [Google Scholar] [CrossRef]
- Saini, V.; McClure, J.T.; Léger, D.; Dufour, S.; Sheldon, A.G.; Scholl, D.T.; Barkema, H.W. Antimicrobial use on Canadian dairy farms. J. Dairy Sci. 2012, 95, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; Garch, F.E.; Simjee, S.; Moyaert, H.; Rose, M.; Youala, M.; Siegwart, E.; VetPath Study Group. Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet. Microbiol. 2018, 213, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Käppeli, N.; Morach, M.; Zurfluh, K.; Corti, S.; Nüesch-Inderbinen, M.; Stephan, R. Sequence Types and Antimicrobial Resistance Profiles of Streptococcus uberis Isolated From ovine Mastitis. Front. Vet. Sci. 2019, 6, 234. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Feβler, A.T.; Kaspar, H.; Lindeman, C.J.; Stegemann, M.R.; Peters, T.; Mankertz, J.; Watts, J.L.; Schwarz, S. A proposal of interpretive criteria for cefoperazone applicable to bovine mastitis pathogens. Vet. Microbiol. 2012, 157, 226–231. [Google Scholar] [CrossRef]
- Lang, I.; Rose, M.; Thomas, E.; Zschiesche, E. A field study of cefquinome for the treatment of pigs with respiratory disease. Rev. Méd. Vét. 2002, 153, 575–580. [Google Scholar]
- Comité de l’antibiogramme de la société française de microbiologie. Groupe de Travail Antibiogramme Vétérinaire. In Recommandations Vétérinaires 2019; SFM: Paris, France, 2019. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 1 July 2020).
- Hossain, M.; Egan, S.A.; Coffey, T.; Ward, P.N.; Wilson, R.; Leigh, J.A.; Emes, R.D. Virulence related sequences; Insights provided by comparative genomics of Streptococcus uberis of differing virulence. BMC Genom. 2015, 16, 334. [Google Scholar] [CrossRef]
- Fessia, A.S.; Diesera, S.A.; Raspantia, C.G.; Odierno, L.M. Genotyping and study of adherence-related genes of Streptococcus uberis isolates from bovine mastitis. Microb. Pathog. 2019, 130, 295–301. [Google Scholar] [CrossRef]
- Moshynskyy, I.; Jiang, M.; Fontaine, M.C.; Perez-Casal, J.; Babiuk, L.A.; Potter, A.A. Characterization of a bovine lactoferrin binding protein of Streptococcus uberis. Microb. Pathog. 2003, 35, 203–215. [Google Scholar] [CrossRef]
- Ward, P.N.; Field, T.R.; Rapier, C.D.; Leigh, J.A. The activation of bovine plasminogen by PauA is not required for virulence of Streptococcus uberis. Infect. Immun. 2003, 71, 7193–7196. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Kitt, A.J.; Ward, P.N.; Leigh, J.A. Isolation and characterization of a mutant strain of Streptococcus uberis, which fails to utilize a plasmin derived beta-casein peptide for the acquisition of methionine. J. Appl. Microbiol. 2002, 93, 631–639. [Google Scholar] [CrossRef]
- Parin, U.; Kirkan, S.; Cicek, E.; Yuksel, H.T. Detection of virulence genes in Streptococcus uberis isolated from bovine mastitis in Aydin province by multiplex polymerase chain reaction. F.Ü. Sağ. Bil. Vet. Derg. 2017, 31, 213–219. [Google Scholar]
- Ward, P.N.; Holden, M.T.; Leigh, J.A.; Lennard, N.; Bignell, A.; Barron, A.; Clark, L.; Quail, M.A.; Woodward, J.; Barrell, B.G.; et al. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genom. 2009, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Kromker, V.; Reinecke, F.; Paduch, J.H.; Grabowski, N. Bovine Streptococcus uberis Intramammary Infections and Mastitis. Clin. Microbial. 2014, 3, 157. [Google Scholar] [CrossRef]
- Phuektes, P.; Mansell, P.D.; Dyson, R.S.; Hooper, N.D.; Dick, J.S.; Browning, G.F. Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J. Clin. Microbiol. 2001, 39, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.Y.; Zheng, N.; Han, R.W.; Ho, H.; Wang, J.; Wang, Y.T.; Wang, S.Q.; Li, H.G.; Liu, H.W.; Yu, Z.N. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019, 131, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Gajda, A.; Posyniak, A.; Żmudzki, J.; Rożańska, H. Occurrence of tetracyclines in tissues and food of animal origin: Causes and consequences. Med. Weter. 2012, 68, 650–655. [Google Scholar]
- Rato, M.G.; Bexiga, R.; Florindo, C.; Cavaco, L.M.; Vilela, C.L.; Santos-Sanches, I. Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet. Microbiol. 2013, 161, 286–294. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Bertin, S.; Leblond, P.; Madec, J.Y.; Payot, S. Diversity and Mobility of Integrative and Conjugative Elements in Bovine Isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl. Environ. Microbiol. 2010, 76, 7957–7965. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Chaussière, S.; Treilles, M.; Madec, J.Y. ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet. J. 2011, 189, 356–358. [Google Scholar] [CrossRef]
- Ruegg, P.L.; Oliveira, L.; Jin, W.; Okwumabua, O. Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J. Dairy Sci. 2015, 98, 4521–4534. [Google Scholar] [CrossRef]
- Kaczorek, E.; Małaczewska, J.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J. Dairy Sci. 2017, 100, 6442–6453. [Google Scholar] [CrossRef]
- Thomas, V.; de Jong, A.; Moyaert, H.; Simjee, S.; El Garch, F.; Morrissey, I.; Marion, H.; Vallé, M. Antimicrobial susceptibility monitoring of mastitis pathogens isolated from acute cases of clinical mastitis in dairy cows across Europe: VetPath results. Int. J. Antimicrob. Agents. 2015, 46, 13–20. [Google Scholar] [CrossRef]
- Entorf, M.; Feßler, A.T.; Kaspar, H.; Kadlec, K.; Peters, T.; Schwarz, S. Comparative erythromycin and tylosin susceptibility testing of Streptococci from bovine mastitis. Vet. Microbiol. 2016, 194, 36–42. [Google Scholar] [CrossRef]
- McDougall, S.; Hussein, H.; Petrovski, K. Antimicrobial resistance in Staphylococcus aureus, Streptococcus uberis and Streptococcus dysgalactiae from dairy cows with mastitis. N. Z. Vet. J. 2014, 62, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Galofaro, L.; Ythier, M.; Giddey, M.; Majcherczyk, P.; Moreillon, P.; Madec, J.Y. Penicillin-Binding Protein Gene Alterations in Streptococcus uberis Isolates Presenting Decreased Susceptibility to Penicillin. Antimicrob. Agents Chemother. 2010, 54, 1140–1145. [Google Scholar] [CrossRef]
- McDougall, S.; Clausen, L.; Ha, H.J.; Gibson, I.; Bryan, M.; Hadjirin, N.; Lay, E.; Raisen, C.; Ba, X.; Restif, O.; et al. Mechanisms of β-lactam resistance of Streptococcus uberis isolated from bovine mastitis cases. Vet. Microbiol. 2020, 242, 108592. [Google Scholar] [CrossRef] [PubMed]
- Wenz, J.R.; Barrington, G.M.; Garry, F.B.; Dinsmore, R.P.; Callan, R.J. Use of systemic disease signs to assess disease severity in dairy cows with acute coliform mastitis. J. Am. Vet. Med. Assoc. 2001, 218, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. Managing mastitis and producing quality milk. In Dairy Production Medicine, 1st ed.; Risco, C.A., Melendez, P.R., Eds.; Wiley-Blackwell: West Sussex, UK, 2011; pp. 207–232. [Google Scholar]
- National Mastitis Council. Laboratory Handbook on Bovine Mastitis, 3rd ed.; NMC: New Prague, MN, USA, 2017. [Google Scholar]
- Cremonesi, P.; Castiglioni, B.; Malferrari, G.; Biunno, I.; Vimercati, C.; Moroni, P.; Morandi, S.; Luzzana, M. Technical Note: Improved Method for Rapid DNA Extraction of Mastitis Pathogens Directly from Milk. J. Dairy Sci. 2006, 89, 163–169. [Google Scholar] [CrossRef]
- Schmitt-Van de Leemput, E.; Zadoks, R.N. Genotypic and phenotypic detection of macrolide and lincosamide resistance in Streptococcus uberis. J. Dairy Sci. 2007, 90, 5089–5096. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; Standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
| Antimicrobials | Range (µg/mL) | Breakpoints (µg/mL) and Susceptibility | MIC50 (μg/mL) | MIC90 (μg/mL) | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S 1 | [%] | I 2 | [%] | R 3 | [%] | |||||
| AMC 4 | 0.064/0.032–64/32 | ≤0.25/0.125 | 98.6 | 0.5/0.25 | 0 | >1/0.5 | 1.4 | 0.25/0.125 | 0.25/0.125 | [25] |
| AMP 5 | 0.016–16 | ≤0.25 | 98.6 | 0.5 | 0 | >2 | 1.4 | 0.25 | 0.25 | [25] |
| CEZ 6 | 0.125–32 | ≤2 | 98.6 | 4 | 0 | >8 | 1.4 | 0.5 | 0.5 | [25] |
| CPZ 7 | 0.125–16 | ≤2 | 97.2 | 4 | 1.4 | >8 | 1.4 | 1 | 2 | [26] |
| CEQ 8 | 0.125–8 | ≤2 | 98.6 | >4 | 1.4 | ≤0.125 | 0.25 | [27] | ||
| CEF 9 | 0.125–32 | ≤2 | 98.6 | 4 | 0 | >8 | 1.4 | 0.5 | 1 | [25] |
| ENRO 10 | 0.016–8 | ≤0.5 | 91.6 | 1–2 | 7 | >4 | 1.4 | 0.5 | 0.5 | [25] |
| ERY 11 | 0.125–8 | ≤0.25 | 91.6 | 0.5 | 2.8 | >1 | 5.6 | ≤0.125 | ≤0.125 | [25] |
| FLL 12 | 0.064–64 | ≤2 | 94.4 | 4 | 5.6 | >8 | 0 | 2 | 2 | [25] |
| LIN 13 | 1–8 | ≤2 | 7 | 4–8 | 11.3 | >18 | 81.7 | >8 | >8 | [28] |
| OXA 14 | 0.125–4 | ≤2 | 95.8 | >4 | 4.2 | 2 | 2 | [28] | ||
| PEN 15 | 0.0625–16 | ≤0.125 | 84.5 | 0.25–2 | 12.7 | >4 | 2.8 | 0.125 | 0.25 | [25] |
| TET 16 | 0.032–16 | ≤2 | 14.1 | 4 | 2.8 | >8 | 83.1 | >16 | >16 | [25] |
| T/S 17 | 0.016/0.304–32/608 | ≤1/19 | 98.6 | >2/38 | 1.4 | 0.062/1.18 | 0.125/2.37 | [29] | ||
| Antimicrobial Agents | Cluster I | Cluster II | ||||||
|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | MIC50 | MIC90 | |||||
| (μg/mL) | [%] | (μg/mL) | [%] | (μg/mL) | [%] | (μg/mL) | [%] | |
| AMC 1 | 0.25/0.125 | 98 | 0.25/0.125 | 98 | 0.125/0.064 | 53 | 0.25/0.125 | 100 |
| AMP 2 | 0.25 | 98 | 0.25 | 98 | 0.125 | 53 | 0.25 | 100 |
| CEZ 3 | 0.5 | 92 | 0.5 | 92 | 0.5 | 88 | 1 | 100 |
| CPZ 4 | 1 | 73 | 2 | 96 | 1 | 65 | 2 | 100 |
| CEQ 5 | 0.25 | 96 | 0.25 | 96 | ≤0.125 | 71 | 0.25 | 94 |
| CEF 6 | 0.5 | 69 | 1 | 96 | 0.5 | 94 | 0.5 | 94 |
| ENRO 7 | 0.5 | 88 | 1 | 94 | 0.5 | 100 | 0.5 | 100 |
| ERY 8 | ≤0.125 | 90 | ≤0.125 | 90 | ≤0.125 | 94 | ≤0.125 | 94 |
| FLL 9 | 2 | 94 | 2 | 94 | 2 | 94 | 2 | 94 |
| LIN 10 | >8 | 100 | >8 | 100 | >8 | 100 | >8 | 100 |
| OXA 11 | 2 | 96 | 2 | 96 | 2 | 94 | 2 | 94 |
| PEN 12 | 0.125 | 81 | 0.25 | 96 | 0.125 | 94 | 0.125 | 94 |
| TET 13 | >16 | 100 | >16 | 100 | >16 | 100 | >16 | 100 |
| T/S 14 | 0.062/1.18 | 83 | 0.125/2.37 | 96 | 0.062/1.18 | 94 | 0.062/1.18 | 94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monistero, V.; Barberio, A.; Cremonesi, P.; Castiglioni, B.; Morandi, S.; Lassen, D.C.K.; Astrup, L.B.; Locatelli, C.; Piccinini, R.; Addis, M.F.; et al. Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm. Antibiotics 2021, 10, 644. https://doi.org/10.3390/antibiotics10060644
Monistero V, Barberio A, Cremonesi P, Castiglioni B, Morandi S, Lassen DCK, Astrup LB, Locatelli C, Piccinini R, Addis MF, et al. Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm. Antibiotics. 2021; 10(6):644. https://doi.org/10.3390/antibiotics10060644
Chicago/Turabian StyleMonistero, Valentina, Antonio Barberio, Paola Cremonesi, Bianca Castiglioni, Stefano Morandi, Desiree C. K. Lassen, Lærke B. Astrup, Clara Locatelli, Renata Piccinini, M. Filippa Addis, and et al. 2021. "Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm" Antibiotics 10, no. 6: 644. https://doi.org/10.3390/antibiotics10060644
APA StyleMonistero, V., Barberio, A., Cremonesi, P., Castiglioni, B., Morandi, S., Lassen, D. C. K., Astrup, L. B., Locatelli, C., Piccinini, R., Addis, M. F., Bronzo, V., & Moroni, P. (2021). Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm. Antibiotics, 10(6), 644. https://doi.org/10.3390/antibiotics10060644









