Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
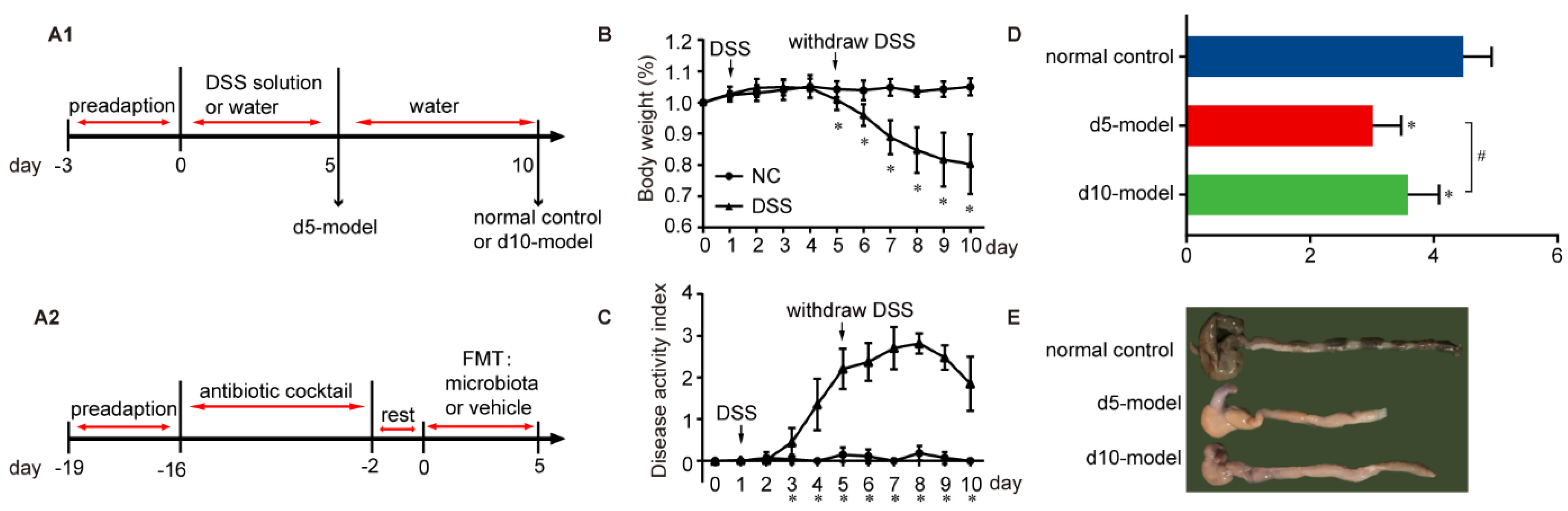
2.2. Mouse Models
2.3. Microbiota Transplantation
2.4. ELISA
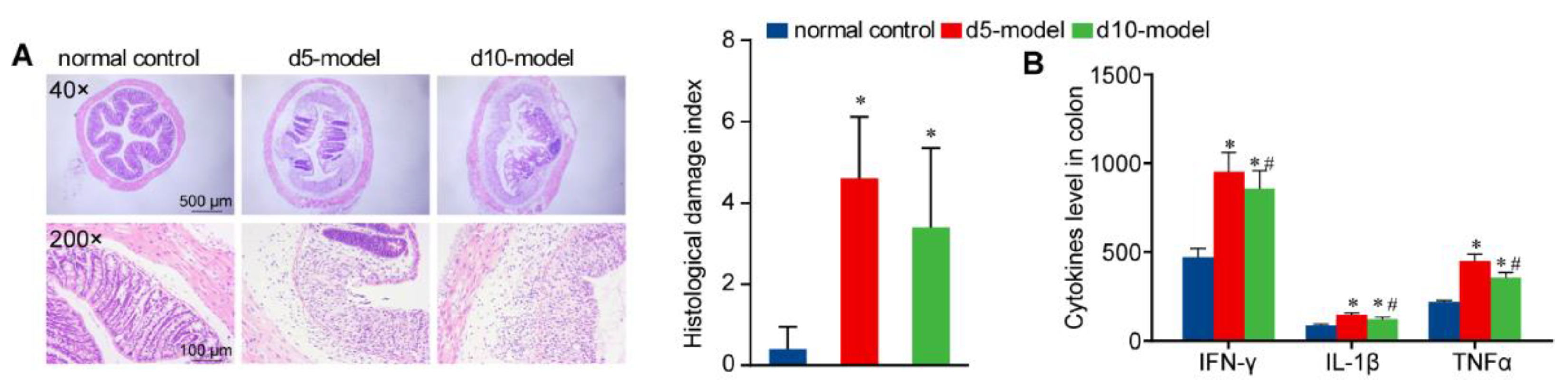
2.5. Histology
2.6. Microbiota Composition by 16S rRNA Sequencing Analysis
2.7. Statistical Analysis
3. Results
3.1. Clinical Signs
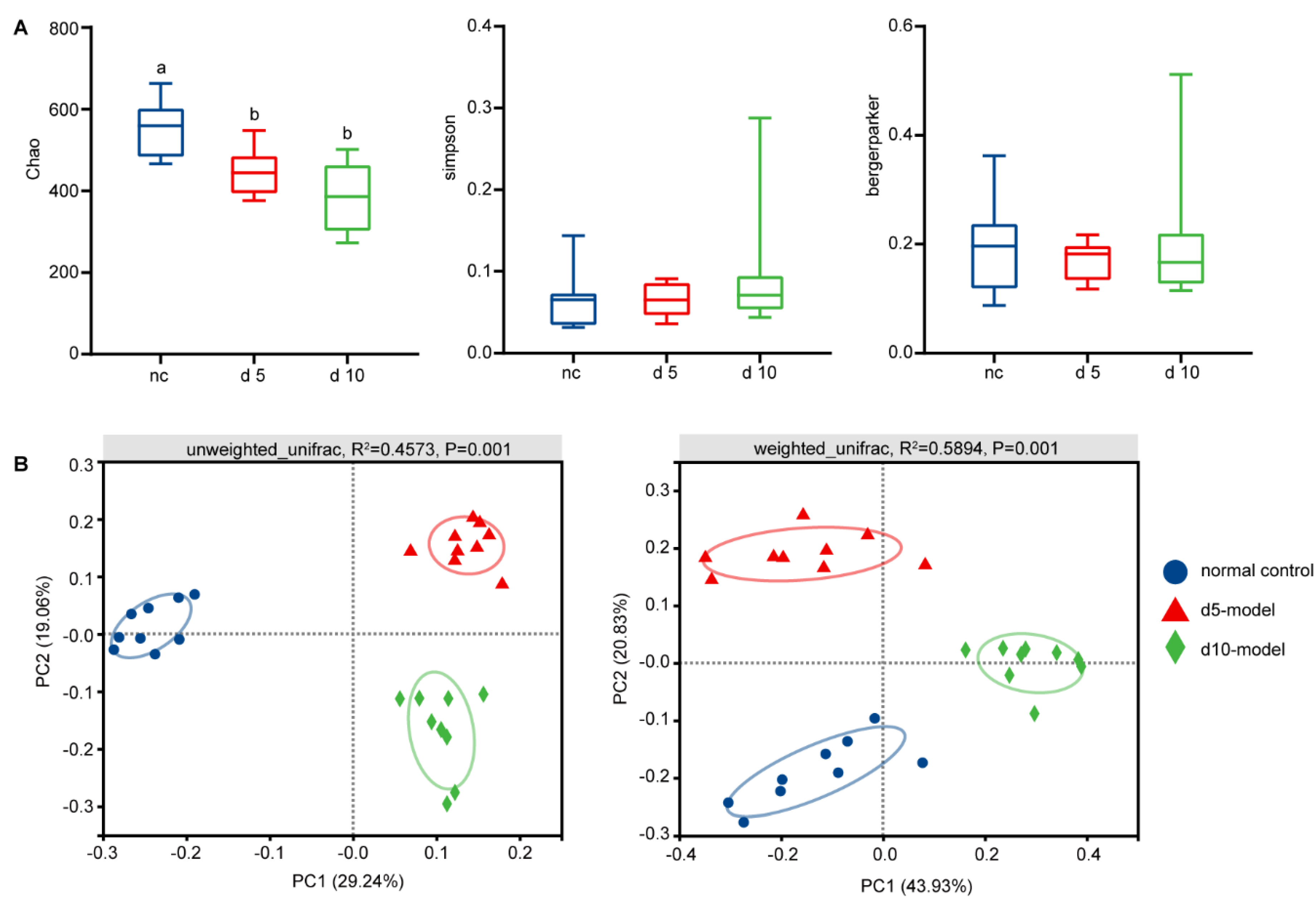
3.2. Gut Microbiota Altered in Colitis Mouse Model
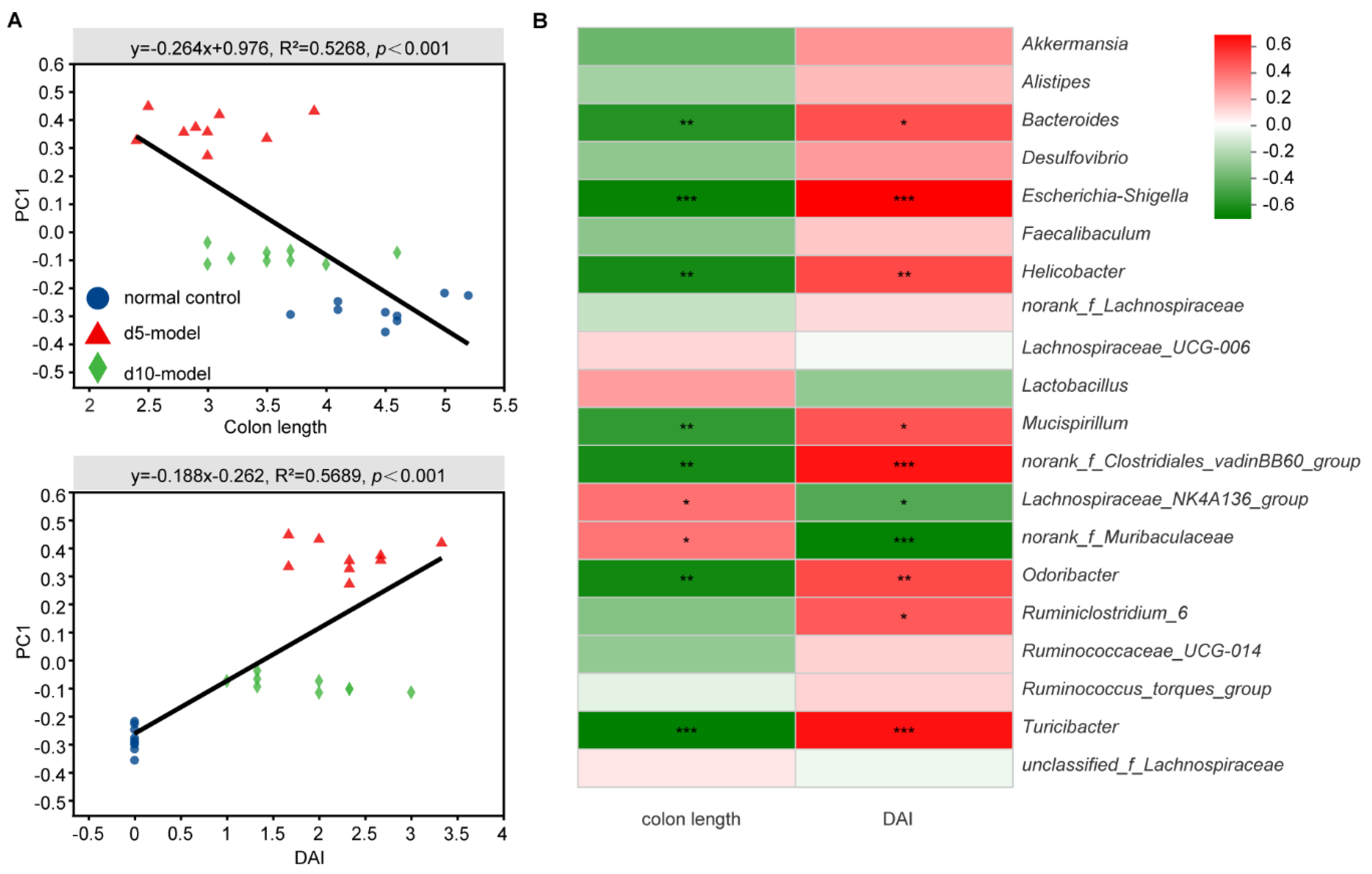
3.3. Correlation Analysis between Specific Microbiota and Disease Indicator
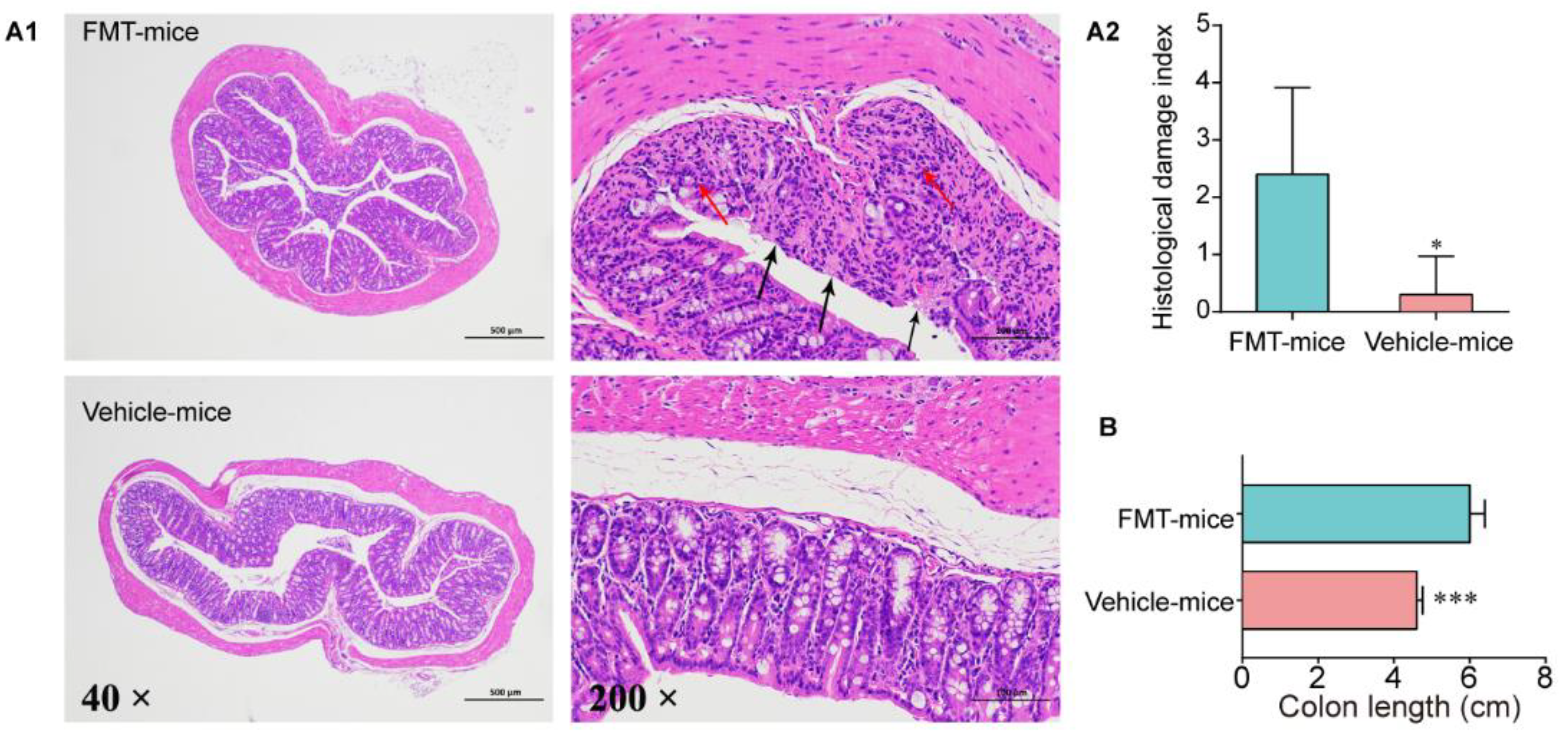
3.4. Colitis Mouse Model Colonic Commensal Microbiota Contributed to Colitis
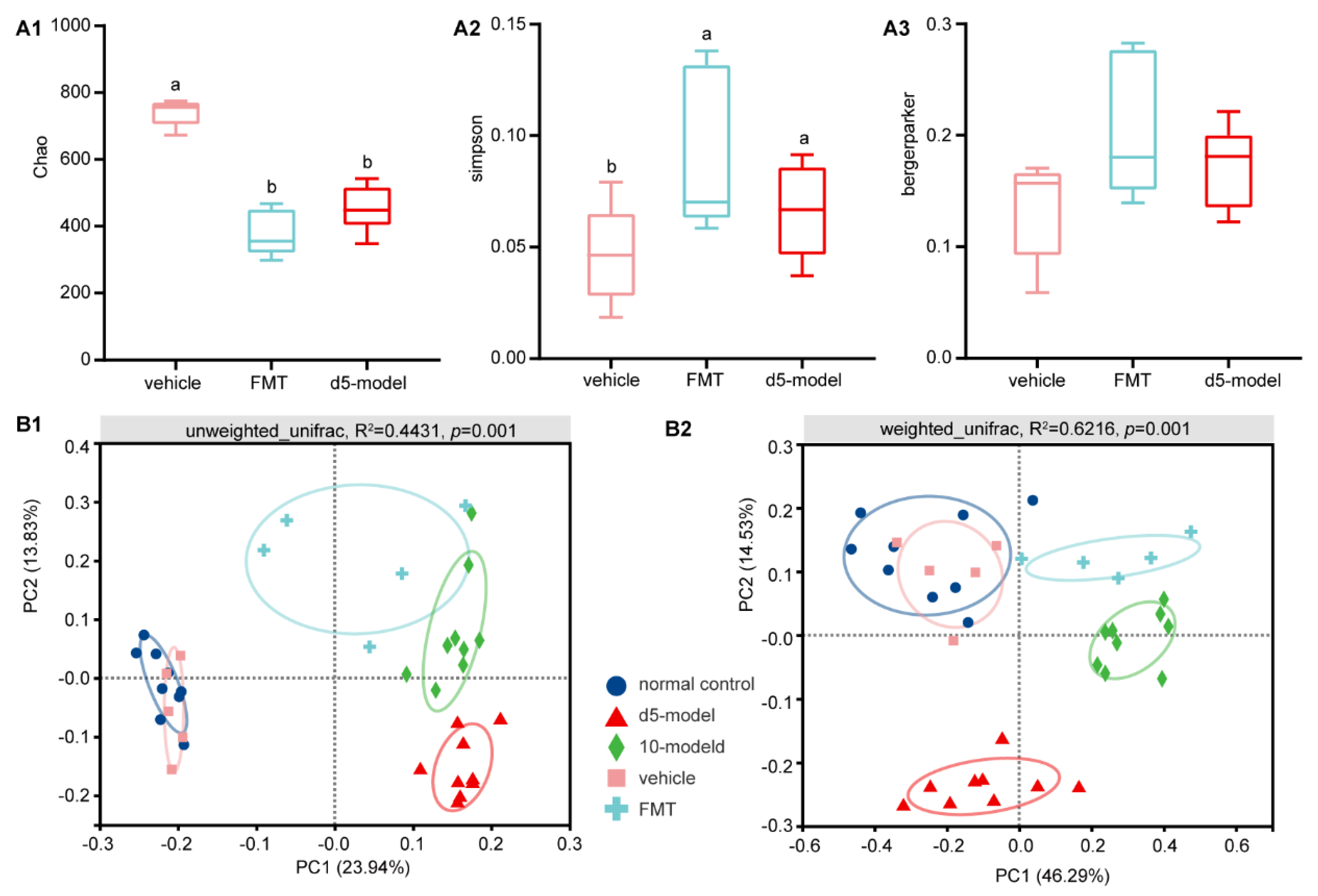
3.5. Microbial Diversity Changes after FMT
3.6. Key Microorganisms Associated with Colitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Ethics Approval
Abbreviations
| IBD | Inflammatory bowel disease |
| OTUs | Operational taxonomic units |
| CD | Crohn’s disease |
| UC | Ulcerative colitis |
| FMT | Fecal microbiota transplantation |
| SCFAs | Short-chain fatty acids |
References
- Choi, C.R.; Al Bakir, I.; Ding, N.J.; Lee, G.; Askari, A.; Warusavitarne, J.; Moorghen, M.; Humphries, A.; Ignjatovic-Wilson, A.; Thomas-Gibson, S.; et al. Cumulative burden of inflammation predicts colorectal neoplasia risk in ulcerative colitis: A large single-centre study. Gut 2019, 68, 414–422. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Nagalingam, N.A.; Kao, J.Y.; Young, V.B. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 2011, 17, 917–926. [Google Scholar] [CrossRef]
- Heimesaat, M.M.; Fischer, A.; Siegmund, B.; Kupz, A.; Niebergall, J.; Fuchs, D.; Jahn, H.K.; Freudenberg, M.; Loddenkemper, C.; Batra, A.; et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS ONE 2007, 2, e662. [Google Scholar] [CrossRef]
- Berry, D.; Schwab, C.; Milinovich, G.; Reichert, J.; Ben, M.K.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091–2106. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; Allison, L.S.A.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef]
- Steinhart, H. The Current Use of Antibiotic Therapies for IBD. Gastroenterol. Hepatol. 2006, 2, 406–407. [Google Scholar]
- Sambuelli, A.; Boerr, L.; Negreira, S.; Gil, A.; Camartino, G.; Huernos, S.; Kogan, Z.; Cabanne, A.; Graziano, A.; Peredo, H.; et al. Budesonide enema in pouchitis-A double-blind, double-dummy, controlled trial. Aliment. Pharmacol. Ther. 2002, 16, 27–34. [Google Scholar] [CrossRef]
- Madden, M.V.; McIntyre, A.S.; Nicholls, R.J. Double-blind crossover trial of metronidazole versus placebo in chronic unremitting pouchitis. Dig. Dis. Sci. 1994, 39, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Achkar, J.P.; Lashner, B.A.; Ormsby, A.H.; Remzi, F.H.; Brzezinski, A.; Bevins, C.L.; Bambrick, M.L.; Seidner, D.L.; Fazio, V.W. A randomized clinical trial of ciprofloxacin and metronidazole to treat acute pouchitis. Inflamm. Bowel Dis. 2001, 7, 301–305. [Google Scholar] [CrossRef]
- Sokol, H. Probiotics and antibiotics in IBD. Dig. Dis. 2014, 32, 10–17. [Google Scholar] [CrossRef]
- Ohkusa, T.; Nomura, T.; Terai, T.; Miwa, H.; Kobayashi, O.; Hojo, M.; Takei, Y.; Ogihara, T.; Hirai, S.; Okayasu, I.; et al. Effectiveness of antibiotic combination therapy in patients with active ulcerative colitis: A randomized, controlled pilot trial with long-term follow-up. Scand. J. Gastroenterol. 2005, 40, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Perencevich, M.; Burakoff, R. Use of antibiotics in the treatment of inflammatory bowel disease. Inflamm. Bowel Dis. 2006, 12, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, S.; Vitali, B.; Klinder, A.; Kolida, S.; Ndagijimana, M.; Laghi, L.; Calanni, F.; Brigidi, P.; Gibson, G.R.; Costabile, A. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: An in vitro approach using a continuous culture colonic model system. J. Antimicrob. Chemother. 2010, 65, 2556–2565. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gillilland, M.R.; Owyang, C. Rifaximin, Gut microbes and mucosal inflammation: Unraveling a complex relationship. Gut Microbes 2014, 5, 571–575. [Google Scholar] [CrossRef]
- Rafii, F.; Ruseler-Van, E.J.; van Lieshout, L.M. Changes in bacterial enzymes and PCR profiles of fecal bacteria from a patient with ulcerative colitis before and after antimicrobial treatments. Dig. Dis. Sci. 1999, 44, 637–642. [Google Scholar] [CrossRef]
- Ganji-Arjenaki, M.; Rafieian-Kopaei, M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta analysis and systematic review. J. Cell. Physiol. 2018, 233, 2091–2103. [Google Scholar] [CrossRef]
- Ledder, O.; Turner, D. Antibiotics in IBD: Still a Role in the Biological Era? Inflamm. Bowel Dis. 2018, 24, 1676–1688. [Google Scholar] [CrossRef]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 8, 1. [Google Scholar] [CrossRef]
- Tang, C.; Kakuta, S.; Shimizu, K.; Kadoki, M.; Kamiya, T.; Shimazu, T.; Kubo, S.; Saijo, S.; Ishigame, H.; Nakae, S.; et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat. Immunol. 2018, 19, 755–765. [Google Scholar] [CrossRef]
- Emal, D.; Rampanelli, E.; Stroo, I.; Butter, L.M.; Teske, G.J.; Claessen, N.; Stokman, G.; Florquin, S.; Leemans, J.C.; Dessing, M.C. Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2017, 28, 1450–1461. [Google Scholar] [CrossRef]
- Josefsdottir, K.S.; Baldridge, M.T.; Kadmon, C.S.; King, K.Y. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 2017, 129, 729–739. [Google Scholar] [CrossRef]
- Hagerbrand, K.; Westlund, J.; Yrlid, U.; Agace, W.; Johansson-Lindbom, B. MyD88 signaling regulates steady-state migration of intestinal CD103+ dendritic cells independently of TNF-alpha and the gut microbiota. J. Immunol. 2015, 195, 2888–2899. [Google Scholar] [CrossRef] [PubMed]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, S.; Neurath, M.F. Mouse models of inflammatory bowel disease. Adv. Drug Deliv Rev. 2007, 59, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Erridge, C.; Duncan, S.H.; Bereswill, S.; Heimesaat, M.M. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS ONE 2010, 5, e9125. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Philip Schumm, L.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016–6029. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 2019, 19, 1–6. [Google Scholar] [CrossRef]
- Ma, N.; Guo, P.; Zhang, J.; He, T.; Kim, S.W.; Zhang, G.; Ma, X. Nutrients mediate intestinal bacteria-mucosal immune crosstalk. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Hague, A.; Butt, A.J.; Paraskeva, C. The role of butyrate in human colonic epithelial cells: An energy source or inducer of differentiation and apoptosis? Proc. Nutr. Soc. 1996, 55, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Balkwill, D.L.; Aldrich, H.C.; Drake, G.R.; Boone, D.R. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 1999, 49, 545–556. [Google Scholar] [CrossRef]
- Sun, C.Q.; O’Connor, C.J.; Turner, S.J.; Lewis, G.D.; Stanley, R.A.; Roberton, A.M. The effect of pH on the inhibition of bacterial growth by physiological concentrations of butyric acid: Implications for neonates fed on suckled milk. Chem. Biol. Interact. 1998, 113, 117–131. [Google Scholar] [CrossRef]
- Zeng, A.P.; Ross, A.; Biebl, H.; Tag, C.; Günzel, B.; Deckwer, W.D. Multiple product inhibition and growth modeling of clostridium butyricum and klebsiella pneumoniae in glycerol fermentation. Biotechnol. Bioeng. 1994, 44, 902–911. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, N.; Nam, R.H.; Park, J.H.; Choi, S.I.; Park, Y.; Kim, Y.; Seok, Y.; Shin, C.M.; Lee, D.H. Gut microbiota and butyrate level changes associated with the long-term administration of proton pump inhibitors to old rats. Sci. Rep. 2019, 9, 6626. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N.; Forbes, J.D. Gut microbiome in inflammatory bowel disease and other chronic immune-mediated inflammatory diseases. Inflamm. Intest. Dis. 2017, 2, 116–123. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, L.; Liu, H.; Yu, H.; Chen, M.; Yang, T.; Zeng, X.; Qiao, S. Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis. Antibiotics 2021, 10, 643. https://doi.org/10.3390/antibiotics10060643
Shang L, Liu H, Yu H, Chen M, Yang T, Zeng X, Qiao S. Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis. Antibiotics. 2021; 10(6):643. https://doi.org/10.3390/antibiotics10060643
Chicago/Turabian StyleShang, Lijun, Hongbin Liu, Haitao Yu, Meixia Chen, Tianren Yang, Xiangfang Zeng, and Shiyan Qiao. 2021. "Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis" Antibiotics 10, no. 6: 643. https://doi.org/10.3390/antibiotics10060643
APA StyleShang, L., Liu, H., Yu, H., Chen, M., Yang, T., Zeng, X., & Qiao, S. (2021). Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis. Antibiotics, 10(6), 643. https://doi.org/10.3390/antibiotics10060643




