Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria
Abstract
1. Introduction
2. Rapid AST Methods through Metabolic Profiling of Bacteria
2.1. Adenosine Triphosphate (ATP) Bioluminescence-Based Methods
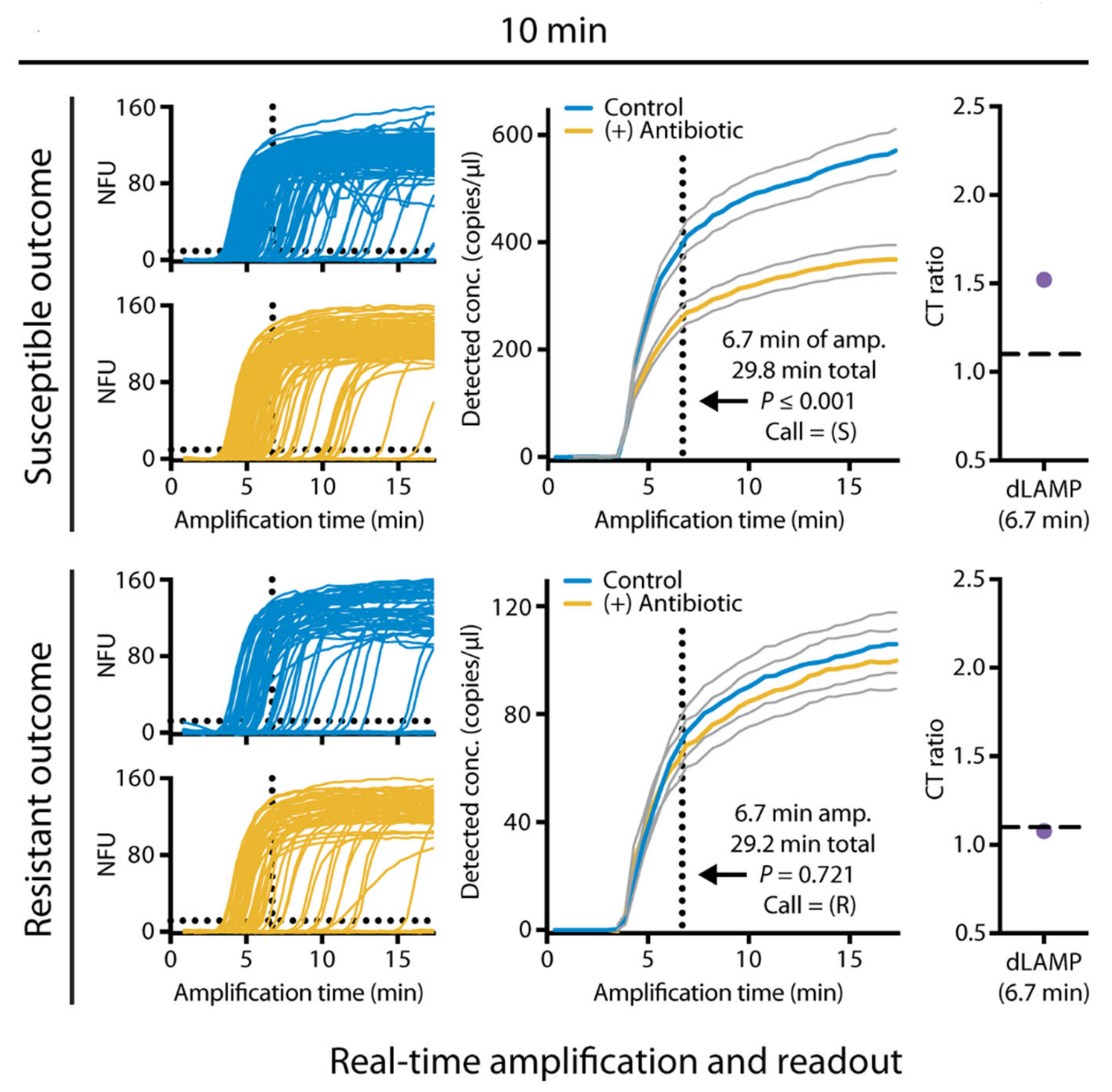
2.2. Nucleic Acid-Based Biochemical Methods
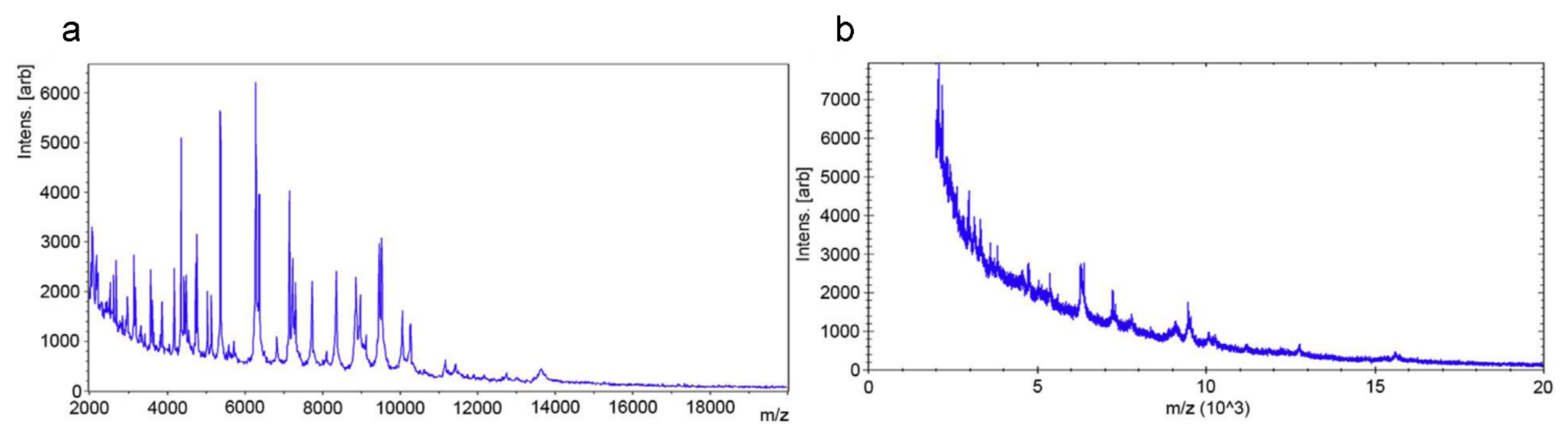
2.3. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Based Methods
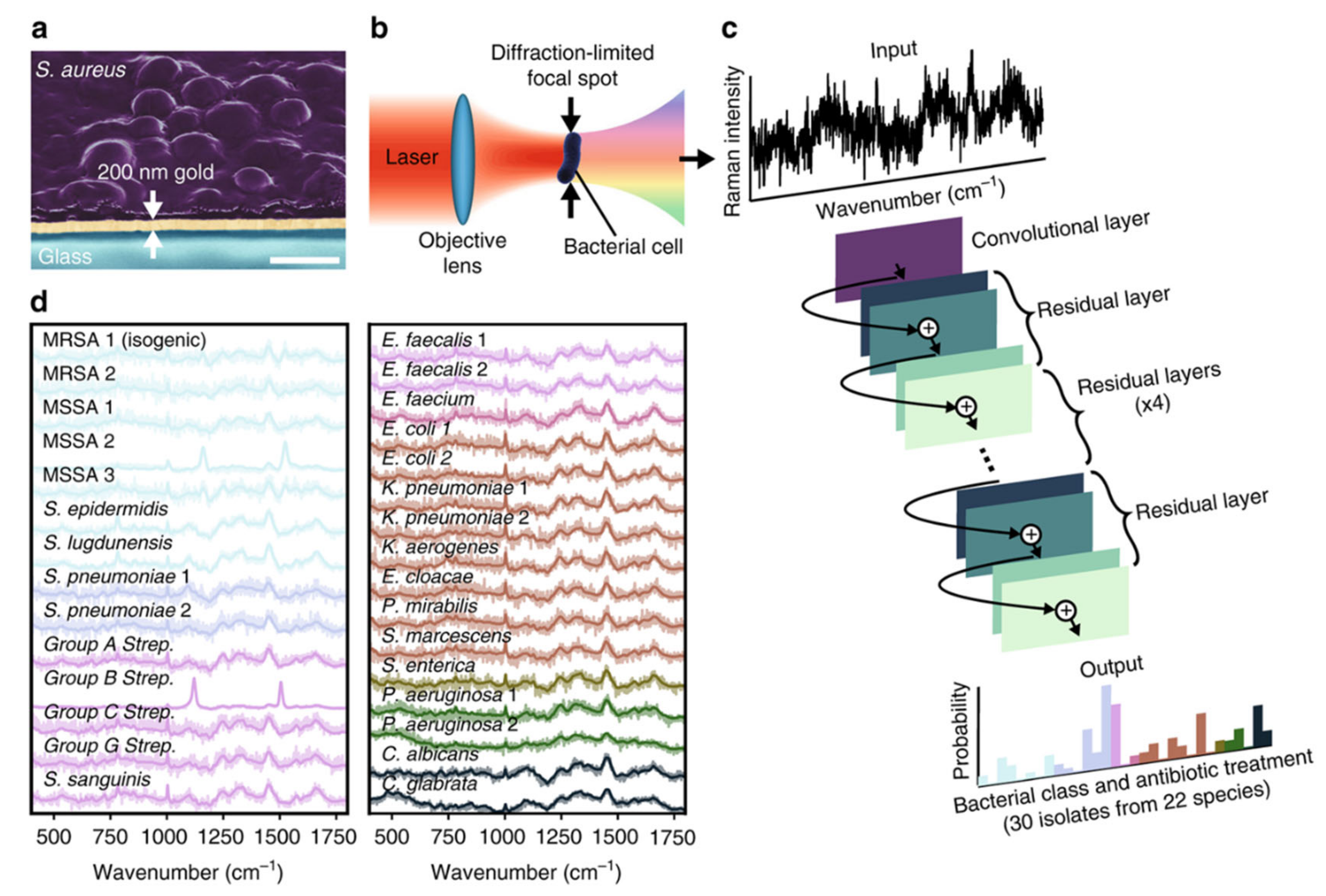
2.4. Raman Technology-Based Methods
2.4.1. Raman Spectroscopy
2.4.2. Stimulated Raman Scattering (SRS) Imaging
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, G.S. Sepsis, severe sepsis and septic shock: Changes in incidence, pathogens and outcomes. Expert. Rev. Anti. Infect. Ther. 2012, 10, 701–706. [Google Scholar] [CrossRef]
- Gal-Mor, O. Persistent infection and long-term carriage of typhoidal and nontyphoidal salmonellae. Clin. Microbiol. Rev. 2018, 32, e00088-18. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Katsumata, R. Antibiotic resistance in bacteria and its future for novel antibiotic development. Biosci. Biotechnol. Biochem. 2006, 70, 1060–1075. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of candida to azoles and echinocandins worldwide. Clin. Microbio. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Jim, O.N. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet. Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Vasala, A.; Hytonen, V.; Laitinen, O. Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 341, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.; Micek, S.; Hampton, N.; Doherty, J.A.; Kumar, A. Septic shock attributed to candida infection: Importance of empiric therapy and source control. Clin. Infect. Dis. 2012, 54, 1739–1746. [Google Scholar] [CrossRef]
- Morrell, M.; Fraser, V.; Kollef, M. Delaying the empiric treatment of candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef]
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: A multi-institutional study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Avesar, J.; Rosenfeld, D.; Truman-Rosentsvit, M.; Ben-Arye, T.; Geffen, Y.; Bercovici, M.; Levenberg, S. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C. Identification and antibiotic-susceptibility profiling of infectious bacterial agents: A review of current and future trends. J. Biotechnol. 2019, 14, 1700750. [Google Scholar] [CrossRef]
- Martínez, J.L.; Rojo, F. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.; Caliendo, A.; Mylonakis, E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Yanle, W.; Liu, Y.; Tan, F.; Chen, L.; Luo, Z.; Wu, Y. Effects of exogenous glucose on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. Microbiol. Open 2019, 8, e933. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.; Haydel, S.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Tannert, A.; Grohs, R.; Popp, J.; Neugebauer, U. Phenotypic antibiotic susceptibility testing of pathogenic bacteria using photonic readout methods: Recent achievements and impact. Appl. Microbiol. Biotechnol. 2019, 103, 549–566. [Google Scholar] [CrossRef]
- Leonard, H.; Colodner, R.; Halachmi, S.; Segal, E. Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sensors 2018, 3, 2202–2217. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Becker, K. How to accelerate antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2019, 25, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Burnham, C.A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Kouda, M.; Ouchi, Y.; Takasaki, Y.; Maeda, T.; Matsuzaki, H.; Nakaya, R. Bioluminescent assay as a potential method of rapid susceptibility testing. Microbiol. Immunol. 1985, 29, 309–315. [Google Scholar] [CrossRef] [PubMed]
- McWalter, P.W. Determination of susceptibility of Staphylococcus aureus to methicillin by luciferim-luciferase assay of bacterial adenosine triphosphate. J. Appl. Bacteriol. 1984, 56, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wheat, P.; Hastings, J.; Spencer, R. Rapid antibiotic susceptibility tests on Enterobacteriaceae by ATP bioluminescence. J. Med. Microbiol. 1988, 25, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wheat, P.; Spencer, R.; Hastings, J. A novel luminometer for rapid antimicrobial susceptibility tests on gram-positive cocci by ATP bioluminescence. J. Med. Microbiol. 1989, 29, 277–282. [Google Scholar] [CrossRef]
- Ivancic, V.; Mastali, M.; Percy, N.; Gornbein, J.; Babbitt, J.; Li, Y.; Landaw, E.; Bruckner, D.; Churchill, B.; Haake, D. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J. Clin. Microbiol. 2008, 46, 1213–1219. [Google Scholar] [CrossRef]
- Mempin, R.; Tran, H.; Chen, C.; Gong, H.; Ho, K.; Lu, S. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 2013, 13, 301. [Google Scholar] [CrossRef]
- Heller, A.; Spence, D. A rapid method for post-antibiotic bacterial susceptibility testing. PLoS ONE 2019, 14, e0210534. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Niimi, H.; Uchiho, Y.; Kawabe, S.; Noda, H.; Kitajima, I. A rapid ATP Bioluminescence-based test for detecting levofloxacin resistance starting from positive blood culture bottles. Sci. Rep. 2019, 9, 13565. [Google Scholar] [CrossRef]
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl. Med. 2017, 9, eaal3693. [Google Scholar] [CrossRef]
- Kostić, T.; Ellis, M.; Williams, M.R.; Stedtfeld, T.M.; Kaneene, J.B.; Stedtfeld, R.D.; Hashsham, S.A. Thirty-minute screening of antibiotic resistance genes in bacterial isolates with minimal sample preparation in static self-dispensing 64 and 384 assay cards. Appl. Microbiol. Biotechnol. 2015, 99, 7711–7722. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, T.; Seki, M. PCR-based method for rapid and minimized electrochemical detection of mecA gene of methicillin-resistant Staphylococcus aureus and methicillinresistant Staphylococcus epidermidis. General Med. Open Access 2015. [Google Scholar] [CrossRef]
- Zboromyrska, Y.; Vergara, A.; Cosgaya, C.; Verger, G.; Mosqueda, N.; Almela, M.; Pitart, C.; Roca, I.; Marco, F.; Vila, J. Rapid detection of b-lactamases directly from positive blood cultures using a loop-mediated isothermal amplification (LAMP)-based assay. Int. J. Antimicrob. Agents 2015, 46, 355–356. [Google Scholar] [CrossRef] [PubMed]
- Baltekin, Ö.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef]
- Tuite, N.; Reddington, K.; Barry, T.; Zumla, A.; Enne, V. Rapid nucleic acid diagnostics for the detection of antimicrobial resistance in Gram-negative bacteria: Is it time for a paradigm shift? J. Antimicrob. Chemother. 2014, 69, 1729–1733. [Google Scholar] [CrossRef]
- Pulido, M.R.; García-Quintanilla, M.; Martín-Peña, R.; Cisneros, J.M.; McConnell, M.J. Progress on the development of rapid methods for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013, 68, 2710–2717. [Google Scholar] [CrossRef]
- Mezger, A.; Wistrand-Yuen, E.; Göransson, J.; Zorzet, A.; Herthnek, D.; Tano, E.; Nilsson, M.; Andersson, D. A general method for rapid determination of antibiotic susceptibility and species in bacterial infections. J. Clin. Microbiol. 2015, 53, 425–432. [Google Scholar] [CrossRef]
- Rolain, J.M.; Mallet, M.N.; Fournier, P.E.; Raoult, D. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 2004, 54, 538–541. [Google Scholar] [CrossRef]
- Steinberger-Levy, I.; Shifman, O.; Zvi, A.; Ariel, N.; Beth-Din, A.; Israeli, O.; Gur, D.; Aftalion, M.; Maoz, S.; Ber, R. A rapid molecular test for determining Yersinia pestis susceptibility to ciprofloxacin by the quantification of differentially expressed marker genes. Front. Microbiol. 2016, 7, 763. [Google Scholar] [CrossRef]
- Barczak, A.; Gomez, J.; Kaufmann, B.; Hinson, E.; Cosimi, L.; Borowsky, M.; Onderdonk, A.; Stanley, S.; Kaur, D.; Bryant, K.; et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc. Natl. Acad. Sci. USA 2012, 109, 6217–6222. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, G.A.; Brabant, W.H. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 1997, 179, 4457–4463. [Google Scholar] [CrossRef] [PubMed]
- Halford, C.; Gonzalez, R.; Campuzano, S.; Hu, B.; Babbitt, J.T.; Liu, J.; Wang, J.; Churchill, B.M.; Haake, D.A. Rapid antimicrobial susceptibility testing by sensitive detection of precursor rRNA using a novel electrochemical biosensing platform. Antimicrob. Agents Chemother. 2013, 57, 936–943. [Google Scholar] [CrossRef]
- Waldeisen, J.R.; Wang, T.; Mitra, D.; Lee, L.P. A real-time PCR antibiogram for drug-resistant sepsis. PLoS ONE 2011, 6, e28528. [Google Scholar] [CrossRef]
- Schoepp, N.G.; Khorosheva, E.M.; Schlappi, T.S.; Curtis, M.S.; Humphries, R.M.; Hindler, J.A.; Ismagilov, R.F. Digital quantification of DNA replication and chromosome segregation enables determination of antimicrobial susceptibility after only 15 min of antibiotic exposure. Angew. Chem. Int. Ed. 2016, 55, 9557–9561. [Google Scholar] [CrossRef]
- Whale, A.S.; Huggett, J.F.; Cowen, S.; Speirs, V.; Shaw, J.; Ellison, S.; Foy, C.A.; Scott, D.J. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012, 40, e82. [Google Scholar] [CrossRef]
- Selck, D.A.; Karymov, M.A.; Sun, B.; Ismagilov, R.F. Increased robustness of single-molecule counting with microfluidics, digital isothermal amplification, and a mobile phone versus real-time kinetic measurements. Anal. Chem. 2013, 85, 11129–11136. [Google Scholar] [CrossRef] [PubMed]
- Oviaño, M.; Bou, G. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin. Microbiol. Rev. 2018, 32, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-offlight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.; Virdi, J. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yang, Q.; Kudinha, T.; Sun, L.; Zhang, R.; Liu, C.; Yu, S.; Xiao, M.; Kong, F.; Zhao, Y.; et al. An improved in-house MALDI-TOF MS protocol for direct cost-effective identification of pathogens from blood cultures. Front. Microbiol. 2017, 8, 1824. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, B.; Sánchez-Carrillo, C.; Ruiz, A.; Marín, M.; Cercenado, E.; Rodríguez-Créixems, M.; Bouza, E. Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 2014, 20, O421–O427. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 h. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef] [PubMed]
- Oviaño, M.; Gómara, M.; Barba, M.; Revillo, M.; Barbeyto, L.; Bou, G. Towards the early detection of β-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J. Antimicrob. Chemother. 2017, 72, 2259–2262. [Google Scholar] [CrossRef] [PubMed]
- Pardo, C.A.; Tan, R.N.; Hennequin, C.; Beyrouthy, R.; Bonnet, R.; Robin, F. Rapid detection of AAC(6′)-lb-cr production using a MALDI-TOF MS strategy. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 2047–2051. [Google Scholar] [CrossRef] [PubMed]
- Oviaño, M.; Gómara, M.; Barba, M.; Sparbier, K.; Pascual, Á.; Bou, G. Quantitative and automated MALDI-TOF MS-based detection of the plasmid-mediated quinolone resistance determinant AAC(6′)-lb-cr in Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2952–2954. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B. New approaches for antifungal susceptibility testing. Clin. Microbio. Infect. 2017, 23, 931–934. [Google Scholar] [CrossRef]
- Florio, W.; Tavanti, A.; Ghelardi, E.; Lupetti, A. MALDI-TOF MS applications to the detection of antifungal resistance: State of the art and future perspectives. Front. Microbiol. 2018, 9, 2577. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Schubert, S.; Jung, J.; Kostrzewa, M.; Sparbier, K. Quantitative matrixassisted laser desorption ionization-time of flight mass spectrometry for rapid resistance detection. J. Clin. Microbiol. 2014, 52, 4155e62. [Google Scholar] [CrossRef]
- Jung, J.S.; Hamacher, C.; Gross, B.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S. Evaluation of a semiquantitative matrix-assisted laser desorption ionization–time of flight mass spectrometry method for rapid antimicrobial susceptibility testing of positive blood cultures. J. Clin. Microbiol. 2016, 54, 2820–2824. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018, 24, 738e43. [Google Scholar] [CrossRef] [PubMed]
- Demirev, P.; Hagan, N.; Antoine, M.; Lin, J.; Feldman, A. Establishing drug resistance in microorganisms by mass spectrometry. J. Am. Soc. Mass Spectrom. 2013, 24, 1194–1201. [Google Scholar] [CrossRef] [PubMed]
- Sparbier, K.; Lange, C.; Jung, J.; Wieser, A.; Schubert, S.; Kostrzewa, M. MALDI biotyper-based rapid resistance detection by stable-isotope labeling. J. Clin. Microbiol. 2013, 51, 3741–3748. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Eberl, T.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S.; Wieser, A. Rapid detection of antibiotic resistance based on mass spectrometry and stable isotopes. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H. Endoscopic Raman spectroscopy for molecular fingerprinting of castric cancer: Principle to implementation. Biomed. Res. Int. 2015, 2015, 670121. [Google Scholar]
- Raman, C.V.; Krishnan, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Cheng, J.-X.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350, aaa8870. [Google Scholar] [CrossRef]
- Kirchhoff, J.; Glaser, U.; Bohnert, J.A.; Pletz, M.W.; Popp, J.; Neugebauer, U. Simple ciprofloxacin resistance test and determination of minimal inhibitory concentration within 2 h using Raman spectroscopy. Anal. Chem. 2018, 90, 1811–1818. [Google Scholar] [CrossRef]
- Schröder, U.C.; Beleites, C.; Assmann, C.; Glaser, U.; Hübner, U.; Pfister, W.; Neugebauer, U. Detection of vancomycin resistances in enterococci within 3 ½ hours. Sci. rep. 2015, 5, 8217. [Google Scholar] [CrossRef]
- Novelli-Rousseau, A.; Espagnon, I.; Filiputti, D.; Gal, O.; Douet, A.; Mallard, F.; Josso, Q. Culture-free antibiotic-susceptibility determination from single-bacterium Raman spectra. Sci. Rep. 2018, 8, 3957. [Google Scholar] [CrossRef]
- Schröder, U.-C.; Kirchhoff, J.; Hübner, U.; Mayer, G.; Glaser, U.; Henkel, T.; Pfister, W.; Fritzsche, W.; Popp, J.; Neugebauer, U. On-chip spectroscopic assessment of microbial susceptibility to antibiotics within 3.5 h. J. Biophotonics 2017, 10, 1547–1557. [Google Scholar] [CrossRef]
- Germond, A.; Ichimura, T.; Horinouchi, T.; Fujita, H.; Furusawa, C.; Watanabe, T.M. Raman spectral signature reflects transcriptomic features of antibiotic resistance in Escherichia coli. Commun. Biol. 2018, 1, 85. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman techniques: Fundamentals and frontiers. Nano Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, Y.; Huang, S.; Zhu, P.; Huang, W.E.; Ling, J.; Xu, J. Metabolic-Activity-Based assessment of antimicrobial effects by D2O-labeled single-cell Raman microspectroscopy. Anal. Chem. 2017, 89, 4108–4115. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Mader, E.; Lee, T.K.; Woebken, D.; Wang, Y.; Zhu, D.; Palatinszky, M.; Schintlmeister, A.; Schmid, M.C.; Hanson, B.T.; et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. USA 2015, 112, E194–E203. [Google Scholar] [CrossRef]
- Shi, L.; Zheng, C.; Shen, Y.; Chen, Z.; Silveira, E.S.; Zhang, L.; Wei, M.; Liu, C.; de Sena-Tomas, C.; Targoff, K.; et al. Optical imaging of metabolic dynamics in animals. Nat. Commun. 2018, 9, 2995. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Y.Z.; Tao, Y.F.; Muhamadali, H.; Goodacre, R.; Zhou, N.Y.; Preston, G.M.; Xu, J.; Huang, W.E. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal. Chem. 2016, 88, 9443–9450. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.-Z.; Zhu, X.; Su, J.-Q.; Ren, B.; Zhu, Y.-G.; Cui, L. Rapid antibiotic susceptibility testing of pathogenic bacteria using heavy-water-labeled single-cell Raman spectroscopy in clinical samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef] [PubMed]
- Stöckel, S.; Kirchhoff, J.; Neugebauer, U.; Rösch, P.; Popp, J. The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman. Spectrosc. 2016, 47, 89–109. [Google Scholar] [CrossRef]
- Fu, D.; Yu, Y.; Folick, A.; Currie, E.; Farese, R.V.; Tsai, T.-H.; Xie, X.S.; Wang, M.C. In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated Raman scattering microscopy. J. Am. Chem. Soc. 2014, 136, 8820–8828. [Google Scholar] [CrossRef]
- Li, J.; Cheng, J.-X. Direct visualization of de novo lipogenesis in single living cells. Sci. Rep. 2014, 4, 6807. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, Y.; Suzuki, Y.; Iwata, O.; Nakashima, A.; Ito, T.; Hirose, M.; Domon, R.; Sugawara, M.; Tsumura, N.; Watarai, H.; et al. Probing the metabolic heterogeneity of live Euglena gracilis with stimulated Raman scattering microscopy. Nat. Microbiol. 2016, 1, 16124. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Chen, Z.; Zhang, L.; Shen, Y.; Wei, L.; Min, W. Vibrational imaging of glucose uptake activity in live cells and tissues by stimulated Raman scattering. Angew. Chem. Int. Ed. 2015, 54, 9821–9825. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, B.; Zhang, D.; Belew, M.Y.; Tissenbaum, H.A.; Cheng, J.-X. Imaging lipid metabolism in live Caenorhabditis elegans using fingerprint vibrations. Angew. Chem. 2014, 53, 11787–11792. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Karanja, C.W.; Abutaleb, N.S.; Younis, W.; Zhang, X.; Seleem, M.N.; Cheng, J.-X. Antibiotic susceptibility determination within one cell cycle at single-bacterium level by stimulated Raman metabolic imaging. Anal. Chem. 2018, 90, 3737–3743. [Google Scholar] [CrossRef] [PubMed]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hong, W.; Abutaleb, N.S.; Li, J.; Dong, P.-T.; Zong, C.; Wang, P.; Seleem, M.N.; Cheng, J.-X. Rapid determination of antimicrobial susceptibility by stimulated Raman scattering imaging of D2O metabolic incorporation in a single bacterium. Sci. Adv. 2020, 7, 2001452. [Google Scholar] [CrossRef] [PubMed]
- Athamanolap, P.; Hsieh, K.; O’Keefe, C.M.; Zhang, Y.; Yang, S.; Wang, T.-H. Nanoarray digital polymerase chain reaction with high-resolution melt for enabling broad bacteria identification and pheno–molecular antimicrobial susceptibility test. Anal. Chem. 2019, 91, 12784–12792. [Google Scholar] [CrossRef] [PubMed]
- Sauget, M.; Bertrand, X.; Hocquet, D. Rapid antibiotic susceptibility testing on blood cultures using MALDI-TOF MS. PLoS ONE 2018, 13, e0205603. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, C.; Rehnstam-Holm, A.-S.; Nilson, B. Rapid detection of antibiotic resistance in positive blood cultures by MALDI-TOF MS and an automated and optimized MBT-ASTRA protocol for Escherichia coli and Klebsiella pneumoniae. Infect. Dis. 2020, 52, 45–53. [Google Scholar] [CrossRef] [PubMed]

| AST Technology | Mechanism | Time of Sample Preparation | Time to Result | Direct on Clinical Samples | Provide MIC | References | Cost for Device | Cost for a Single Test |
|---|---|---|---|---|---|---|---|---|
| ATP bioluminescence | ATP concentration | 2 h from urine | Yes/urine | No | [27] | Low | Low | |
| 2 h for preculture | 20 min–1 h from cultured bacteria | No | No | [29] | ||||
| 15 min for centrifugation | At least 6 h from blood | Yes/ blood | Yes | [30] | ||||
| Digital PCR | Copy number of DNA | 30 min from urine | Yes/urine | No | [31] | High | High | |
| 14–18 h for preculture | 2 h from cultures in log phase | No | No | [45] | ||||
| 4 h from isolates or urine | Yes/urine | No | [89] | |||||
| MALDI-TOF MS | Change of mass spectrum | 2–3 h from cultured bacteria or blood | Yes/blood | No | [59] | High | Low | |
| 4 h from blood | Yes/blood | No | [60] | |||||
| 4–18 h from cultured bacteria | No | No | [61] | |||||
| 1 h for subculture | 4 h from blood | Yes/blood | No | [90] | ||||
| 20 min for centrifugation | 3.5 h from blood | Yes/blood | No | [91] | ||||
| Raman spectroscopy | Change of Raman spectrum | 2 h for preculture | 4 h from isolates | No | Yes | [70] | High | Low |
| Overnight-culture in D2O | At least 40 min from overnight cultures | No | Yes | [75] | ||||
| 15 min for filtration | 2.5 h from urine | Yes/urine | No | [79] | ||||
| SRS | Quantify deuterium incorporation | 2 h for preculture | At least 0.5 h from cultures in log phase | No | Yes | [86] | High | Low |
| 15 min for centrifugation and filtration | 2.5 h from isolates, urine, or blood | Yes/urine and blood | Yes | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Hong, W. Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria. Antibiotics 2021, 10, 311. https://doi.org/10.3390/antibiotics10030311
Chen C, Hong W. Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria. Antibiotics. 2021; 10(3):311. https://doi.org/10.3390/antibiotics10030311
Chicago/Turabian StyleChen, Chen, and Weili Hong. 2021. "Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria" Antibiotics 10, no. 3: 311. https://doi.org/10.3390/antibiotics10030311
APA StyleChen, C., & Hong, W. (2021). Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria. Antibiotics, 10(3), 311. https://doi.org/10.3390/antibiotics10030311






