Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing
Abstract
1. Introduction
2. Current Antimicrobial Susceptibility Testing (AST) Methods
3. The Atomic Force Microscope (AFM) and the Cantilever as a Mass Sensor
4. Nanomotion Detection
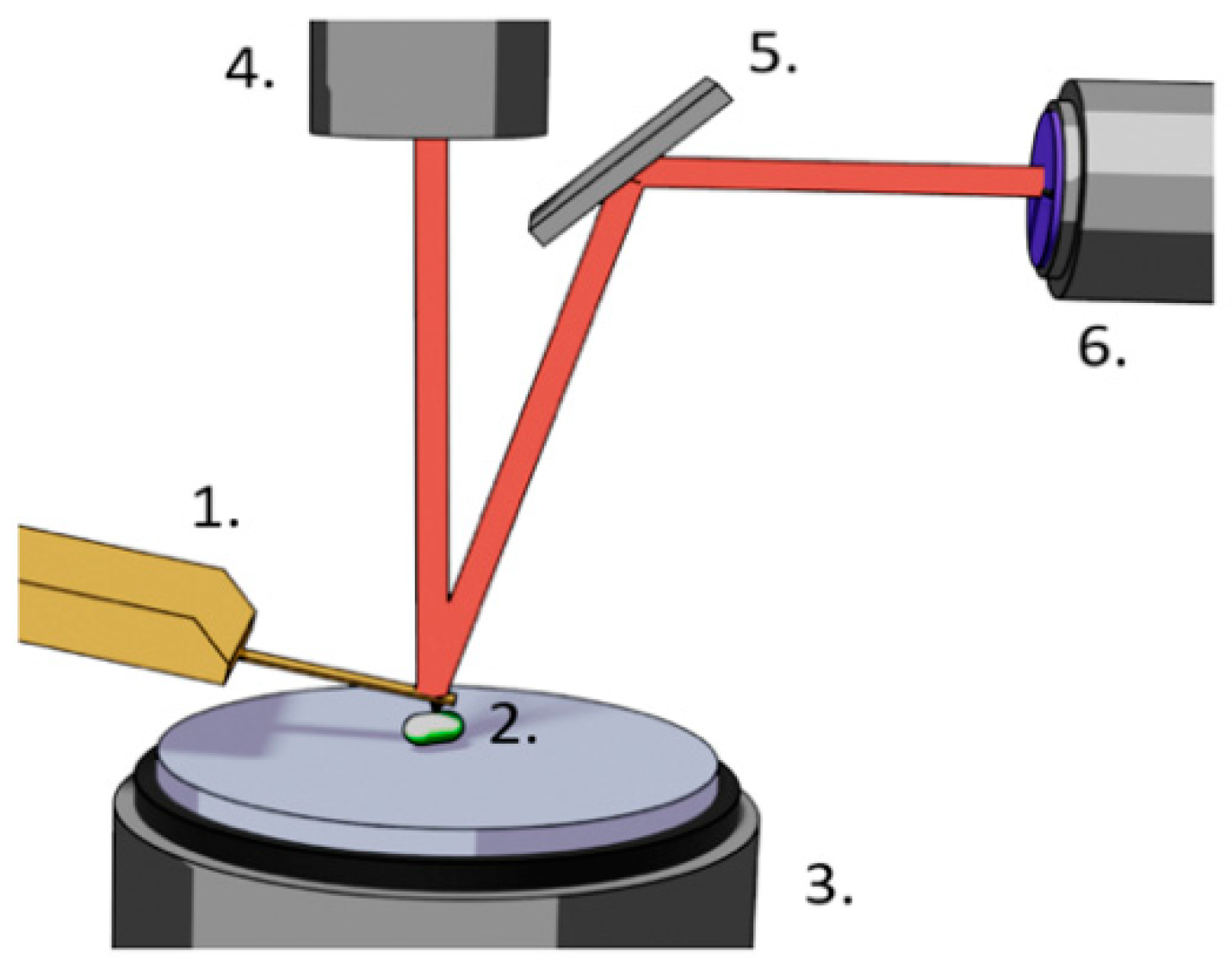
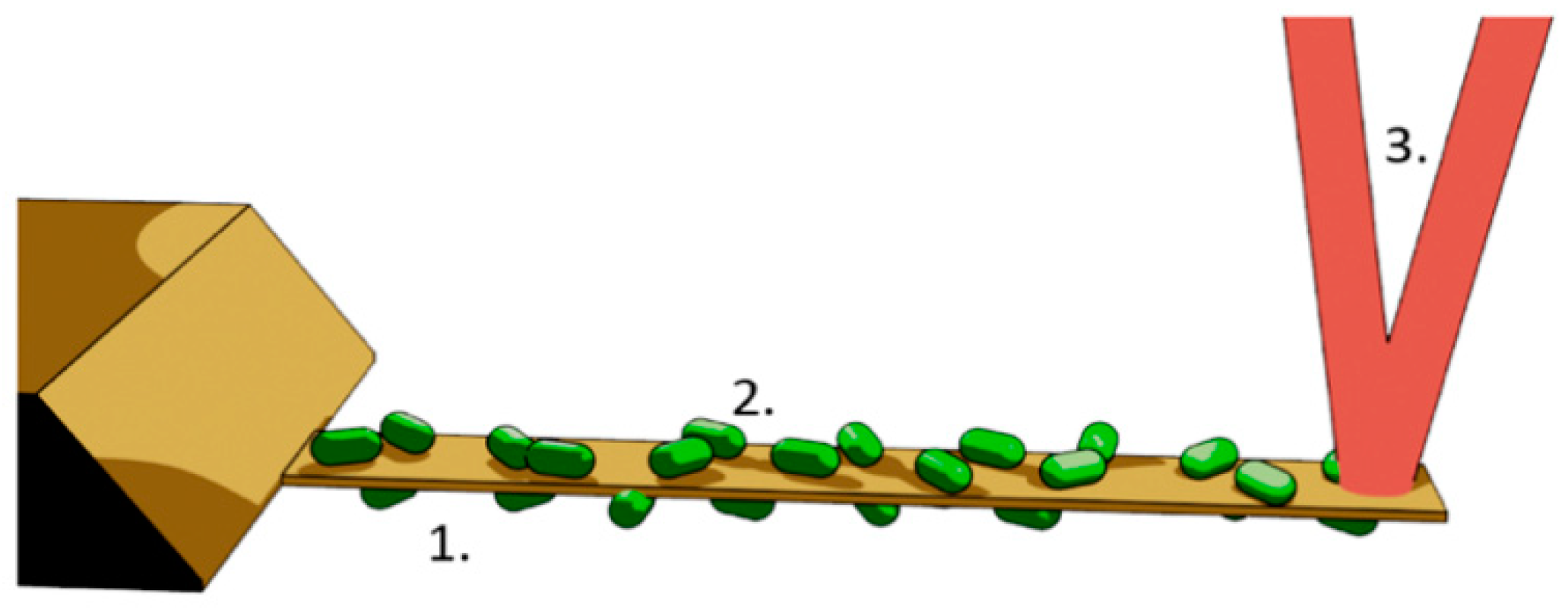
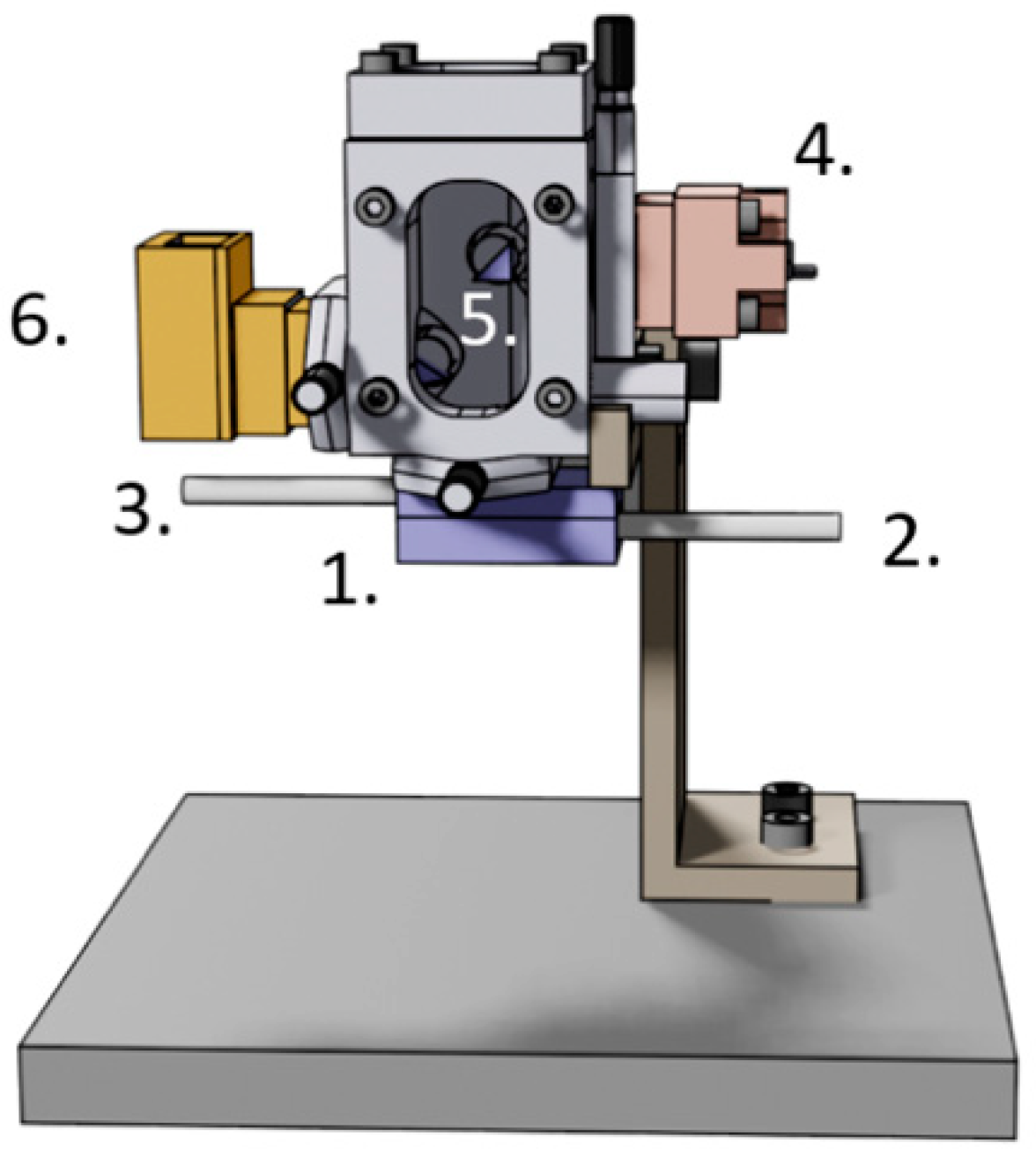
5. AFM Nanomotion Setup and Measurement
6. AFM Nanomotion Data Processing
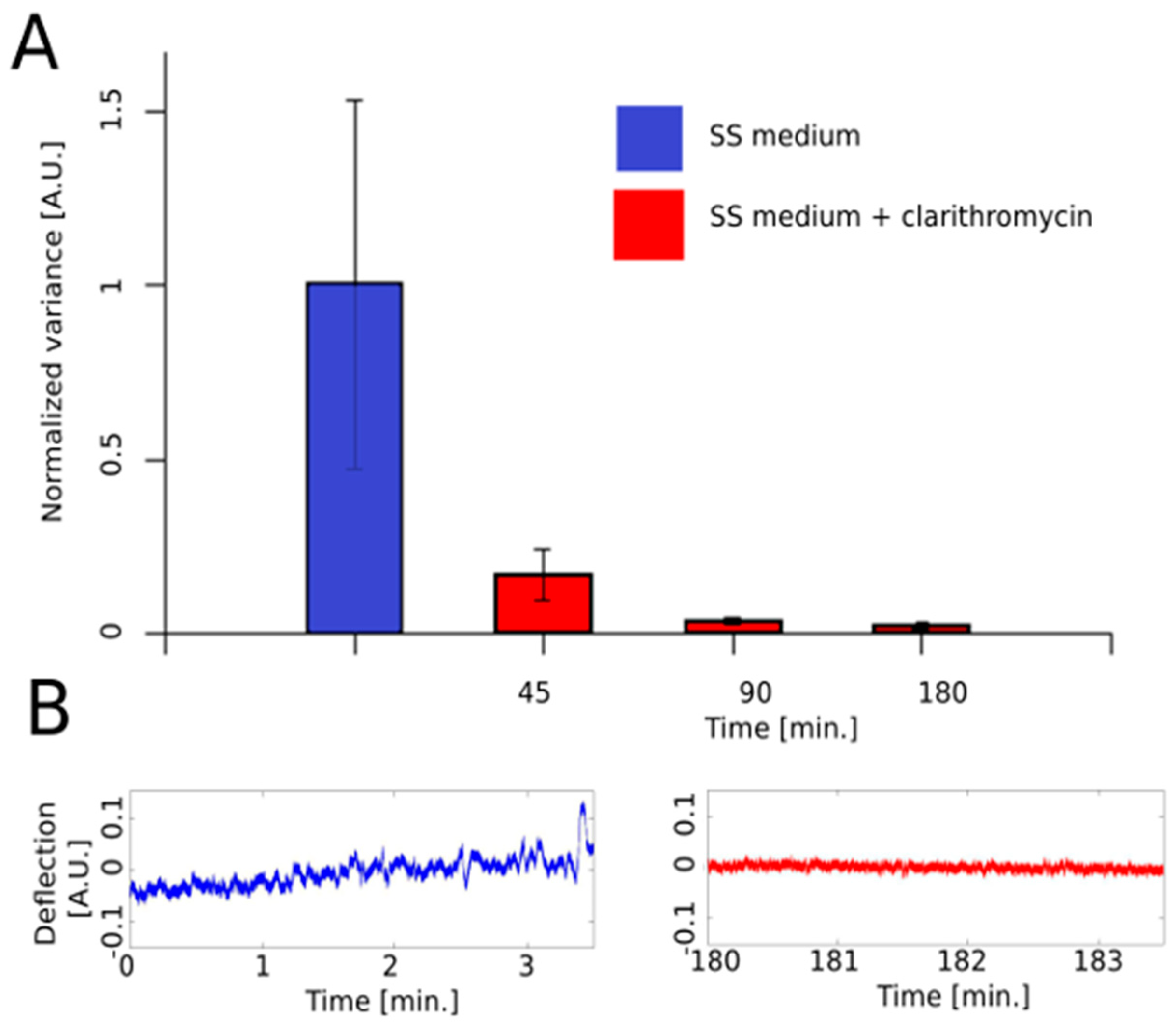
7. Application Example
8. Advantages and Drawbacks of the AFM Nanomotion AST Technique.
9. Future Developments
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- WHO. Antimicrobial Resistance: Global Report on Surveillance 2014; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Heath 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Van Boeckel, T.; Laxminarayan, R. Correction to global antibiotic consumption data. Lancet Infect. Dis. 2017, 17, 476–477. [Google Scholar] [CrossRef]
- Humphries, R.M.; Hindler, J.A. Emerging Resistance, new antimicrobial agents … but no tests! the challenge of antimicrobial susceptibility testing in the current us regulatory landscape. Clin. Infect. Dis. 2016, 63, 83–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K.; et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2019, 17, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Horvat, R.T. Review of Antibiogram Preparation and Susceptibility Testing Systems. Hosp. Pharm. 2010, 45, 6–9. [Google Scholar] [CrossRef]
- Kronvall, G.; Giske, C.G.; Kahlmeter, G. Setting interpretive breakpoints for antimicrobial susceptibility testing using disk diffusion. Int. J. Antimicrob. Agents 2011, 38, 281–290. [Google Scholar] [CrossRef]
- Matuschek, E.; Brown, D.F.J.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef]
- Fournier, P.E.; Drancourt, M.; Colson, P.; Rolain, J.M.; La Scola, B.; Raoult, D. Modern clinical microbiology: New challenges and solutions. Nat. Rev. Microbiol. 2013, 11, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Dinarelli, S.; Girasole, M.; Kasas, S.; Longo, G. Nanotools and molecular techniques to rapidly identify and fight bacterial infections. J. Microbiol. Methods 2017, 138, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Bowden, R.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012, 13, 601–612. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Lavallée, C.; Rouleau, D.; Gaudreau, C.; Roger, M.; Tsimiklis, C.; Locas, M.C.; Gagnon, S.; Delorme, J.; Labbé, A.C. Performance of an agar dilution method and a Vitek 2 card for detection of inducible clindamycin resistance in Staphylococcus spp. J. Clin. Microbiol. 2010, 48, 1354–1357. [Google Scholar] [CrossRef]
- Gardiner, B.J.; Grayson, M.L.; Wood, G.M. Inducible resistance to clindamycin in Staphylococcus aureus: Validation of Vitek-2 against CLSI D-test. Pathology 2013, 45, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.E.; Ng, L.S.Y.; Tan, T.Y. Evaluation of Enterococcus faecalis clinical isolates with “penicillin-resistant, ampicillin-susceptible” phenotype as reported by Vitek-2 Compact system. Pathology 2014, 46, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Won, E.J.; Shin, J.H.; Kim, M.N.; Choi, M.J.; Joo, M.Y.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Evaluation of the BD Phoenix system for identification of a wide spectrum of clinically important yeast species: A comparison with Vitek 2-YST. Diagn. Microbiol. Infect. Dis. 2014, 79, 477–480. [Google Scholar] [CrossRef]
- McGregor, A.; Schio, F.; Beaton, S.; Boulton, V.; Perman, M.; Gilbert, G. The microscan walkaway diagnostic microbiology system—An evaluation. Pathology 1995, 27, 172–176. [Google Scholar] [CrossRef]
- Winstanley, T.; Courvalin, P. Expert systems in clinical microbiology. Clin. Microbiol. Rev. 2011, 24, 515–556. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.W.; Munier, G.K.; Johnson, C.L. Direct comparison of the BD phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 2008, 46, 2327–2333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mittman, S.A.; Huard, R.C.; Della-Latta, P.; Whittier, S. Comparison of BD Phoenix to Vitek 2, MicroScan MICroSTREP, and Etest for antimicrobial susceptibility testing of Streptococcus pneumoniae. J. Clin. Microbiol. 2009, 47, 3557–3561. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Pérez-Vázquez, M.; Oliver, A.; Sánchez Del Saz, B.; Gutiérrez, M.O.; Martínez-Ferrer, M.; Baquero, F. Evaluation of the wider system, a new computer-assisted image-processing device for bacterial identification and susceptibility testing. J. Clin. Microbiol. 2000, 38, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Swenson, J.M.; Anderson, K.F.; Lonsway, D.R.; Thompson, A.; McAllister, S.K.; Limbago, B.M.; Carey, R.B.; Tenover, F.C.; Patel, J.B. Accuracy of commercial and reference susceptibility testing methods for detecting vancomycin-intermediate Staphylococcus aureus. J. Clin. Microbiol. 2009, 47, 2013–2017. [Google Scholar] [CrossRef] [PubMed]
- Junkins, A.D.; Lockhart, S.R.; Heilmann, K.P.; Dohrn, C.L.; Von Stein, D.L.; Winokur, P.L.; Doern, G.V.; Richter, S.S. BD Phoenix and Vitek 2 detection of mecA-mediated resistance in Staphylococcus aureus with cefoxitin. J. Clin. Microbiol. 2009, 47, 2879–2882. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Sheehan, D.J.; Rex, J.H. Determination of Fungicidal Activities against Yeasts and Molds: Lessons Learned from Bactericidal Testing and the Need for Standardization. Clin. Microbiol. Rev. 2004, 17, 268–280. [Google Scholar] [CrossRef]
- Konaté, K.; Mavoungou, J.F.; Lepengué, A.N.; Aworet-Samseny, R.R.R.; Hilou, A.; Souza, A.; Dicko, M.H.; M’Batchi, B. Antibacterial activity against β- lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: Fractional Inhibitory Concentration Index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- White, R.L.; Burgess, D.S.; Manduru, M.; Bosso, J.A. Comparison of three different in vitro methods of detecting synergy: Time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 1996, 40, 1914–1918. [Google Scholar] [CrossRef]
- Clancy, C.J.; Huang, H.; Cheng, S.; Derendorf, H.; Nguyen, M.H. Characterizing the effects of caspofungin on Candida albicans, Candida parapsilosis, and Candida glabrata isolates by simultaneous time-kill and postantifungal-effect experiments. Antimicrob. Agents Chemother. 2006, 50, 2569–2572. [Google Scholar] [CrossRef]
- Klepser, M.E.; Ernst, E.J.; Lewis, R.E.; Ernst, M.E.; Pfaller, M.A. Influence of test conditions on antifungal time-kill curve results: Proposal for standardized methods. Antimicrob. Agents Chemother. 1998, 42, 1207–1212. [Google Scholar] [CrossRef]
- Chantell, C. Multiplexed Automated Digital Microscopy for Rapid Identification and Antimicrobial Susceptibility Testing of Bacteria and Yeast Directly from Clinical Samples. Clin. Microbiol. Newsl. 2015, 37, 161–167. [Google Scholar] [CrossRef]
- Price, C.S.; Kon, S.E.; Metzger, S. Rapid antibiotic susceptibility phenotypic characterization of Staphylococcus aureus using automated microscopy of small numbers of cells. J. Microbiol. Methods 2014, 98, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Douglas, I.S.; Price, C.S.; Overdier, K.H.; Wolken, R.F.; Metzger, S.W.; Hance, K.R.; Howson, D.C. Rapid automated microscopy for microbiological surveillance of ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 2015, 191, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Fredborg, M.; Andersen, K.R.; Jørgensen, E.; Droce, A.; Olesen, T.; Jensen, B.B.; Rosenvinge, F.S.; Sondergaard, T.E. Real-time optical antimicrobial susceptibility testing. J. Clin. Microbiol. 2013, 51, 2047–2053. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, Y.G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.F.J.; Brown, L. Evaluation of the e test, a novel method of quantifying antimicrobial activity. J. Antimicrob. Chemother. 1991, 27, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Jiang, J.; McBride, M.; Yang, Y.; Mo, M.; Iriya, R.; Peterman, J.; Jing, W.; Grys, T.; Haydel, S.E.; et al. Direct Antimicrobial Susceptibility Testing on Clinical Urine Samples by Optical Tracking of Single Cell Division Events. Small 2020, 16, 2004148. [Google Scholar] [CrossRef]
- Tawil, N.; Mouawad, F.; Lévesque, S.; Sacher, E.; Mandeville, R.; Meunier, M. The differential detection of methicillin-resistant, methicillin-susceptible and borderline oxacillin-resistant Staphylococcus aureus by surface plasmon resonance. Biosens. Bioelectron. 2013, 49, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gupta, K.; Ekinci, K.L. All-electrical monitoring of bacterial antibiotic susceptibility in a microfluidic device. Proc. Natl. Acad. Sci. USA 2020, 117, 10639–10644. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.C.; Paton, T.F.; Mulroney, K.T.; Inglis, T.J.J.; Sutton, J.M.; Morgan, H. A fast impedance-based antimicrobial susceptibility test. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Robello, E.; Battaglini, F.; Mikkelsen, S.R. Rapid antibiotic susceptibility testing via electrochemical measurement of ferricyanide reduction by Escherichia coli and Clostridium sporogenes. Anal. Chem. 2000, 72, 4957–4964. [Google Scholar] [CrossRef] [PubMed]
- Mann, T.S.; Mikkelsen, S.R. Antibiotic susceptibility testing at a screen-printed carbon electrode array. Anal. Chem. 2008, 80, 843–848. [Google Scholar] [CrossRef]
- Onishi, K.; Enomoto, J.; Araki, T.; Takagi, R.; Suzuki, H.; Fukuda, J. Electrochemical microdevices for rapid and on-site determination of the minimum inhibitory concentration of antibiotics. Analyst 2018, 143, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Lee, S.M.; Oh, J.; Park, I.H.; Song, J.H.; Han, M.; Yong, D.; Lim, K.J.; Shin, J.S.; Yoo, K.H. Electrical antimicrobial susceptibility testing based on aptamer-functionalized capacitance sensor array for clinical isolates. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Kinnunen, P.; McNaughton, B.H.; Albertson, T.; Sinn, I.; Mofakham, S.; Elbez, R.; Newton, D.W.; Hunt, A.; Kopelman, R. Self-assembled magnetic bead biosensor for measuring bacterial growth and antimicrobial susceptibility testing. Small 2012, 8, 2477–2482. [Google Scholar] [CrossRef] [PubMed]
- Frye, J.G.; Jesse, T.; Long, F.; Rondeau, G.; Porwollik, S.; McClelland, M.; Jackson, C.R.; Englen, M.; Fedorka-Cray, P.J. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Int. J. Antimicrob. Agents 2006, 27, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Frye, J.G.; Lindsey, R.L.; Rondeau, G.; Porwollik, S.; Long, F.; McClelland, M.; Jackson, C.R.; Englen, M.D.; Meinersmann, R.J.; Berrang, M.E.; et al. Development of a DNA microarray to detect antimicrobial resistance genes identified in the national center for biotechnology information database. Microb. Drug Resist. 2010, 16, 9–19. [Google Scholar] [CrossRef]
- Huletsky, A.; Giroux, R.; Rossbach, V.; Gagnon, M.; Vaillancourt, M.; Bernier, M.; Gagnon, F.; Truchon, K.; Bastien, M.; Picard, F.J.; et al. New Real-Time PCR Assay for Rapid Detection of Methicillin-Resistant Staphylococcus aureus Directly from Specimens Containing a Mixture of Staphylococci. J. Clin. Microbiol. 2004, 42, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid Molecular Detection of Tuberculosis and Rifampin Resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Mazza-Stalder, J.; Greub, G.; Jaton, K. The rapid molecular test Xpert MTB/RIF ultra: Towards improved tuberculosis diagnosis and rifampicin resistance detection. Clin. Microbiol. Infect. 2019, 25, 1370–1376. [Google Scholar] [CrossRef]
- Chakravorty, S.; Simmons, A.M.; Rowneki, M.; Parmar, H.; Cao, Y.; Ryan, J.; Banada, P.P.; Deshpande, S.; Shenai, S.; Gall, A.; et al. The new Xpert MTB/RIF ultra: Improving detection of Mycobacterium tuberculosis and resistance to Rifampin in an assay suitable for point-of-care testing. mBio 2017, 8, e00812-17. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef]
- Degand, N.; Carbonnelle, E.; Dauphin, B.; Beretti, J.L.; Le Bourgeois, M.; Sermet-Gaudelus, I.; Segonds, C.; Berche, P.; Nassif, X.; Ferroni, A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 2008, 46, 3361–3367. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Ledeboer, N.A.; Hodinka, R.L. Molecular detection of resistance determinants. J. Clin. Microbiol. 2011, 49, S20–S24. [Google Scholar] [CrossRef]
- Neville, S.A.; LeCordier, A.; Ziochos, H.; Chater, M.J.; Gosbell, I.B.; Maley, M.W.; Van Hal, S.J. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 2011, 49, 2980–2984. [Google Scholar] [CrossRef] [PubMed]
- Dubois, D.; Leyssene, D.; Chacornac, J.P.; Kostrzewa, M.; Schmit, P.O.; Talon, R.; Bonnet, R.; Delmas, J. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2010, 48, 941–945. [Google Scholar] [CrossRef]
- Saleeb, P.G.; Drake, S.K.; Murray, P.R.; Zelazny, A.M. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011, 49, 1790–1794. [Google Scholar] [CrossRef]
- Dhiman, N.; Hall, L.; Wohlfiel, S.L.; Buckwalter, S.P.; Wengenack, N.L. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 2011, 49, 1614–1616. [Google Scholar] [CrossRef]
- Croxatto, A.; Prod’hom, G.; Durussel, C.; Greub, G. Preparation of a blood culture pellet for rapid bacterial identification and antibiotic susceptibility testing. J. Vis. Exp. 2014, e51985. [Google Scholar] [CrossRef]
- Opota, O.; Croxatto, A.; Prod’hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Jaton, K.; Greub, G. Microbial diagnosis of bloodstream infection: Towards molecular diagnosis directly from blood. Clin. Microbiol. Infect. 2015, 21, 323–331. [Google Scholar] [CrossRef]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by vitek 2, CLSI broth microdilution, and etest method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar]
- Hazelton, B.; Thomas, L.C.; Olma, T.; Kok, J.; O′Sullivan, M.; Chen, S.C.A.; Iredell, J.R. Rapid and accurate direct antibiotic susceptibility testing of blood culture broths using MALDI sepsityper combined with the BD phoenix automated system. J. Med. Microbiol. 2014, 63, 1590–1594. [Google Scholar] [CrossRef] [PubMed]
- Morgenthaler, N.G.; Kostrzewa, M. Rapid identification of pathogens in positive blood culture of patients with sepsis: Review and meta-analysis of the performance of the Sepsityper kit. Int. J. Microbiol. 2015, 2015, 827416. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.S.; Eberl, T.; Sparbier, K.; Lange, C.; Kostrzewa, M.; Schubert, S.; Wieser, A. Rapid detection of antibiotic resistance based on mass spectrometry and stable isotopes. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 949–955. [Google Scholar] [CrossRef]
- Finger, S.; Wiegand, C.; Buschmann, H.J.; Hipler, U.C. Antibacterial properties of cyclodextrin-antiseptics-complexes determined by microplate laser nephelometry and ATP bioluminescence assay. Int. J. Pharm. 2013, 452, 188–193. [Google Scholar] [CrossRef]
- VOJTEK, L.; DOBES, P.; BUYUKGUZEL, E.; ATOSUO, J.; HYRSL, P. Bioluminescent assay for evaluating antimicrobial activity in insect haemolymph. Eur. J. Entomol. 2014, 111, 335–340. [Google Scholar] [CrossRef]
- Ivančić, V.; Mastali, M.; Percy, N.; Gornbein, J.; Babbitt, J.T.; Li, Y.; Landaw, E.M.; Bruckner, D.A.; Churchill, B.M.; Haake, D.A. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J. Clin. Microbiol. 2008, 46, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Andreu, N.; Fletcher, T.; Krishnan, N.; Wiles, S.; Robertson, B.D. Rapid measurement of antituberculosis drug activity in vitro and in macrophages using bioluminescence. J. Antimicrob. Chemother. 2012, 67, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; Lang, H.R.M.; Schimke, D.; Lammers, A. Evaluation of a bioluminescence assay for rapid antimicrobial susceptibility testing of mycobacteria. Eur. J. Clin. Microbiol. 1985, 4, 556–561. [Google Scholar] [CrossRef]
- Finger, S.; Wiegand, C.; Buschmann, H.J.; Hipler, U.C. Antimicrobial properties of cyclodextrin-antiseptics-complexes determined by microplate laser nephelometry and ATP bioluminescence assay. Int. J. Pharm. 2012, 436, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Galiger, C.; Brock, M.; Jouvion, G.; Savers, A.; Parlato, M.; Ibrahim-Granet, O. Assessment of efficacy of antifungals against Aspergillus fumigatus: Value of real-time bioluminescence imaging. Antimicrob. Agents Chemother. 2013, 57, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Mach, K.E.; Mohan, R.; Baron, E.J.; Shih, M.-C.; Gau, V.; Wong, P.K.; Liao, J.C. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J. Urol. 2011, 185, 148–153. [Google Scholar] [CrossRef]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef]
- Alexander, S.; Hellemans, L.; Marti, O.; Schneir, J.; Elings, V.; Hansma, P.K.; Longmire, M.; Gurley, J. An atomic-resolution atomic-force microscope implemented using an optical lever. J. Appl. Phys. 1989, 65, 164–167. [Google Scholar] [CrossRef]
- Kasas, S.; Fellay, B.; Cargnello, R. Observation of the action of penicillin onbacillus subtilis using atomic force microscopy: Technique for the preparation of bacteria. Surf. Interface Anal. 1994, 21, 400–401. [Google Scholar] [CrossRef]
- Braga, P.C.; Ricci, D. Atomic force microscopy: Application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Antimicrob. Agents Chemother. 1998, 42, 18–22. [Google Scholar] [CrossRef]
- Braga, P.C.; Ricci, D. Differences in the susceptibility of Streptococcus pyogenes to rokitamycin and erythromycin A revealed by morphostructural atomic force microscopy. J. Antimicrob. Chemother. 2002, 50, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Braga, P.C.; Ricci, D.; Dal Sasso, M. Daptomycin morphostructural damage in Bacillus cereus visualized by atomic force microscopy. J. Chemother. 2002, 14, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Soon, R.L.; Nation, R.L.; Hartley, P.G.; Larson, I.; Li, J. Atomic force microscopy investigation of the morphology and topography of colistin-heteroresistant Acinetobacter baumannii strains as a function of growth phase and in response to colistin treatment. Antimicrob. Agents Chemother. 2009, 53, 4979–4986. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, K.S.; Han, I.; Kim, M.H.; Jung, M.H.; Park, H.K. Quantitative and qualitative analysis of the antifungal activity of allicin alone and in combination with antifungal drugs. PLoS ONE 2012, 7, e38242. [Google Scholar] [CrossRef] [PubMed]
- Formosa, C.; Schiavone, M.; Martin-Yken, H.; François, J.M.; Duval, R.E.; Dague, E. Nanoscale effects of caspofungin against two yeast species, saccharomyces cerevisiae and candida albicans. Antimicrob. Agents Chemother. 2013, 57, 3498–3506. [Google Scholar] [CrossRef] [PubMed]
- Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B.M.; Roos, W.H.; Wuite, G.J.L.; Gaub, H.E.; Gerber, C.; Dufrêne, Y.F.; Müller, D.J. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 2019, 1, 41–57. [Google Scholar] [CrossRef]
- Demir, I.; Blockx, J.; Dague, E.; Guiraud, P.; Thielemans, W.; Muylaert, K.; Formosa-Dague, C. Nanoscale Evidence Unravels Microalgae Flocculation Mechanism Induced by Chitosan. ACS Appl. Bio Mater. 2020, 3, 8446–8459. [Google Scholar] [CrossRef]
- Kumar, A.; Ting, Y.P. Effect of sub-inhibitory antibacterial stress on bacterial surface properties and biofilm formation. Colloids Surf. B Biointerfaces 2013, 111, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Vadillo-Rodríguez, V.; Logan, B.E. Localized attraction correlates with bacterial adhesion to glass and metal oxide substrata. Environ. Sci. Technol. 2006, 40, 2983–2988. [Google Scholar] [CrossRef]
- Fang, H.H.P.; Chan, K.Y.; Xu, L.C. Quantification of bacterial adhesion forces using atomic force microscopy (AFM). J. Microbiol. Methods 2000, 40, 89–97. [Google Scholar] [CrossRef]
- Dupres, V.; Menozzi, F.D.; Locht, C.; Clare, B.H.; Abbott, N.L.; Cuenot, S.; Bompard, C.; Raze, D.; Dufrêne, Y.F. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2005, 2, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, Y.; Deghorain, M.; Wang, L.; Xu, B.; Pollheimer, P.D.; Gruber, H.J.; Errington, J.; Hallet, B.; Haulot, X.; Verbelen, C.; et al. Single-molecule force spectroscopy and imaging of the vancomycin/D-Ala-D- Ala interaction. Nano Lett. 2007, 7, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.I.; Stupar, P.; Chomicki, W.; Bertacchi, M.; Dietler, G.; Arnal, L.; Vela, M.E.; Yantorno, O.; Kasas, S. Nanomotion Detection Method for Testing Antibiotic Resistance and Susceptibility of Slow-Growing Bacteria. Small 2018, 14, 1702671. [Google Scholar] [CrossRef]
- Vinckier, A.; Semenza, G. Measuring elasticity of biological materials by atomic force microscopy. FEBS Lett. 1998, 430, 12–16. [Google Scholar] [CrossRef]
- Hertz, H. Ueber die Berührung fester elastischer Körper. J. fur die Reine und Angew. Math. 1882, 1882, 156–171. [Google Scholar]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Tatara, Y. Extensive theory of force- approach relations of elastic spheres in compression and in impact. J. Eng. Mater. Technol. Trans. ASME 1989, 111, 163–168. [Google Scholar] [CrossRef]
- Cappella, B.; Dietler, G. Force-distance curves by atomic force microscopy. Surf. Sci. Rep. 1999, 34, 1–3. [Google Scholar] [CrossRef]
- Butt, H.J.; Cappella, B.; Kappl, M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surf. Sci. Rep. 2005, 59, 1–152. [Google Scholar]
- Xu, W.; Mulhern, P.J.; Blackford, B.L.; Jericho, M.H.; Firtel, M.; Beveridge, T.J. Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication for methane production. J. Bacteriol. 1996, 178, 3106–3112. [Google Scholar] [CrossRef][Green Version]
- Arnoldi, M.; Kacher, C.M.; Bäuerlein, E.; Radmacher, M.; Fritz, M. Elastic properties of the cell wall of Magnetospirillum gryphiswaldense investigated by atomic forcemicroscopy. Appl. Phys. A Mater. Sci. Process. 1998, 66, S613–S617. [Google Scholar] [CrossRef]
- Arce, F.T.; Carlson, R.; Monds, J.; Veeh, R.; Hu, F.Z.; Stewart, P.S.; Lal, R.; Ehrlich, G.D.; Avci, R. Nanoscale structural and mechanical properties of nontypeable haemophilus influenzae biofilms. J. Bacteriol. 2009, 191, 2512–2520. [Google Scholar] [CrossRef]
- Lau, P.C.Y.; Dutcher, J.R.; Beveridge, T.J.; Lam, J.S. Absolute quantitation of bacterial biofilm adhesion and viscoelasticity by microbead force spectroscopy. Biophys. J. 2009, 96, 2935–2948. [Google Scholar] [CrossRef]
- Wang, H.; Wilksch, J.J.; Lithgow, T.; Strugnell, R.A.; Gee, M.L. Nanomechanics measurements of live bacteria reveal a mechanism for bacterial cell protection: The polysaccharide capsule in Klebsiella is a responsive polymer hydrogel that adapts to osmotic stress. Soft Matter 2013, 9, 7560–7567. [Google Scholar] [CrossRef]
- Bailey, R.G.; Turner, R.D.; Mullin, N.; Clarke, N.; Foster, S.J.; Hobbs, J.K. The interplay between cell wall mechanical properties and the cell cycle in staphylococcus aureus. Biophys. J. 2014, 107, 2538–2545. [Google Scholar] [CrossRef] [PubMed]
- Arnal, L.; Serra, D.O.; Cattelan, N.; Castez, M.F.; Vázquez, L.; Salvarezza, R.C.; Yantorno, O.M.; Vela, M.E. Adhesin contribution to nanomechanical properties of the virulent Bordetella pertussis envelope. Langmuir 2012, 28, 7461–7469. [Google Scholar] [CrossRef]
- Roduit, C.; Sekatski, S.; Dietler, G.; Catsicas, S.; Lafont, F.; Kasas, S. Stiffness tomography by atomic force microscopy. Biophys. J. 2009, 97, 674–677. [Google Scholar] [CrossRef]
- Roduit, C.; Saha, B.; Alonso-Sarduy, L.; Volterra, A.; Dietler, G.; Kasas, S. OpenFovea: Open-source AFM data processing software. Nat. Methods 2012, 9, 774–775. [Google Scholar] [CrossRef]
- Longo, G.; Rio, L.M.; Trampuz, A.; Dietler, G.; Bizzini, A.; Kasas, S. Antibiotic-induced modifications of the stiffness of bacterial membranes. J. Microbiol. Methods 2013, 93, 1–6. [Google Scholar] [CrossRef]
- Longo, G.; Kasas, S. Effects of antibacterial agents and drugs monitored by atomic force microscopy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Stupar, P.; Dietler, G. AFM contribution to unveil pro- and eukaryotic cell mechanical properties. Semin. Cell Dev. Biol. 2018, 73, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R. Nanomechanical mapping of soft materials with the atomic force microscope: Methods, theory and applications. Chem. Soc. Rev. 2020, 49, 5850–5884. [Google Scholar] [CrossRef]
- Formosa-Dague, C.; Duval, R.E.; Dague, E. Cell biology of microbes and pharmacology of antimicrobial drugs explored by Atomic Force Microscopy. Semin. Cell Dev. Biol. 2018, 73, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.R.; Stephenson, R.J.; Welland, M.E.; Gerber, C.; Gimzewski, J.K. Photothermal spectroscopy with femtojoule sensitivity using a micromechanical device. Nature 1994, 372, 79–81. [Google Scholar] [CrossRef]
- Berger, R.; Gerber, C.; Gimzewski, J.K.; Meyer, E.; Güntherodt, H.J. Thermal analysis using a micromechanical calorimeter. Appl. Phys. Lett. 1996, 69, 40–42. [Google Scholar] [CrossRef]
- Boisen, A.; Dohn, S.; Keller, S.S.; Schmid, S.; Tenje, M. Cantilever-like micromechanical sensors. Rep. Prog. Phys. 2011, 74, 036101. [Google Scholar] [CrossRef]
- Godin, M.; Tabard-Cossa, V.; Miyahara, Y.; Monga, T.; Williams, P.J.; Beaulieu, L.Y.; Bruce Lennox, R.; Grutter, P. Cantilever-based sensing: The origin of surface stress and optimization strategies. Nanotechnology 2010, 21, 75501. [Google Scholar] [CrossRef]
- Alvarez, M.; Lechuga, L.M. Microcantilever-based platforms as biosensing tools. Analyst 2010, 135, 827. [Google Scholar] [CrossRef]
- Hansen, K.M.; Thundat, T. Microcantilever biosensors. Methods 2005, 37, 57–64. [Google Scholar] [CrossRef]
- Waggoner, P.S.; Craighead, H.G. Micro- and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip 2007, 7, 1238–1255. [Google Scholar] [CrossRef]
- Braun, T.; Ghatkesar, M.K.; Backmann, N.; Grange, W.; Boulanger, P.; Letellier, L.; Lang, H.-P.; Bietsch, A.; Gerber, C.; Hegner, M. Quantitative time-resolved measurement of membrane protein–ligand interactions using microcantilever array sensors. Nat. Nanotechnol. 2009, 4, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Ilic, B.; Czaplewski, D.; Craighead, H.G.; Neuzil, P.; Campagnolo, C.; Batt, C. Mechanical resonant immunospecific biological detector. Appl. Phys. Lett. 2000, 77, 450–452. [Google Scholar] [CrossRef]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Güntherodt, H.J.; Gerber, C.; Gimzewski, J.K. Translating biomolecular recognition into nanomechanics. Science 2000, 288, 316–318. [Google Scholar] [CrossRef]
- Fritz, J. Cantilever biosensors. Analyst 2008, 133, 855–863. [Google Scholar] [CrossRef]
- Willaert, R.; Kasas, S.; Devreese, B.; Dietler, G. Yeast Nanobiotechnology. Fermentation 2016, 2, 18. [Google Scholar] [CrossRef]
- Lang, H.P.; Baller, M.K.; Berger, R.; Gerber, C.; Gimzewski, J.K.; Battiston, F.M.; Fornaro, P.; Ramseyer, J.P.; Meyer, E.; Güntherodt, H.J. An artificial nose based on a micromechanical cantilever array. Anal. Chim. Acta 1999, 393, 59–65. [Google Scholar] [CrossRef]
- Braun, T.; Barwich, V.; Ghatkesar, M.K.; Bredekamp, A.H.; Gerber, C.; Hegner, M.; Lang, H.P. Micromechanical mass sensors for biomolecular detection in a physiological environment. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2005, 72, 031907. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, S.; Chiyoma, T.; Ikeuchi, A.; Okano, H.; Sone, H.; Izumi, T. Possibility of a femtogram mass biosensor using a self-sensing cantilever. Curr. Appl. Phys. 2006, 6, 384–388. [Google Scholar] [CrossRef]
- Godin, M.; Delgado, F.F.; Son, S.; Grover, W.H.; Bryan, A.K.; Tzur, A.; Jorgensen, P.; Payer, K.; Grossman, A.D.; Kirschner, M.W.; et al. Using buoyant mass to measure the growth of single cells. Nat. Methods 2010, 7, 387–390. [Google Scholar] [CrossRef]
- Ndieyira, J.W.; Watari, M.; Barrera, A.D.; Zhou, D.; Vögtli, M.; Batchelor, M.; Cooper, M.A.; Strunz, T.; Horton, M.A.; Abell, C.; et al. Nanomechanical detection of antibiotic-mucopeptide binding in a model for superbug drug resistance. Nat. Nanotechnol. 2008, 3, 691–696. [Google Scholar] [CrossRef]
- Liu, Y.; Schweizer, L.M.; Wang, W.; Reuben, R.L.; Schweizer, M.; Shu, W. Label-free and real-time monitoring of yeast cell growth by the bending of polymer microcantilever biosensors. Sens. Actuators B Chem. 2013, 178, 621–626. [Google Scholar] [CrossRef]
- Cermak, N.; Olcum, S.; Delgado, F.F.; Wasserman, S.C.; Payer, K.R.; Murakami, M.A.; Knudsen, S.M.; Kimmerling, R.J.; Stevens, M.M.; Kikuchi, Y.; et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat. Biotechnol. 2016, 34, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Burg, T.P.; Godin, M.; Knudsen, S.M.; Shen, W.; Carlson, G.; Foster, J.S.; Babcock, K.; Manalis, S.R. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 2007, 446, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.K.; Goranov, A.; Amon, A.; Manalis, S.R. Measurement of mass, density, and volume during the cell cycle of yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Jang, J.; Irimia, D.; Sturgis, J.; Lee, J.; Robinson, J.P.; Toner, M.; Bashir, R. “Living cantilever arrays” for characterization of mass of single live cells in fluids. Lab Chip 2008, 8, 1034–1041. [Google Scholar] [CrossRef]
- Bryan, A.K.; Hecht, V.C.; Shen, W.; Payer, K.; Grover, W.H.; Manalis, S.R. Measuring single cell mass, volume, and density with dual suspended microchannel resonators. Lab Chip 2014, 14, 569–576. [Google Scholar] [CrossRef]
- Nugaeva, N.; Gfeller, K.Y.; Backmann, N.; Lang, H.P.; Düggelin, M.; Hegner, M. Micromechanical cantilever array sensors for selective fungal immobilization and fast growth detection. Biosens. Bioelectron. 2005, 21, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Khan, M.F.; Kaur, K.; Thundat, T. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Sarduy, L.; De Los Rios, P.; Benedetti, F.; Vobornik, D.; Dietler, G.; Kasas, S.; Longo, G. Real-Time Monitoring of Protein Conformational Changes Using a Nano-Mechanical Sensor. PLoS ONE 2014, 9, e103674. [Google Scholar] [CrossRef]
- Stupar, P.; Opota, O.; Longo, G.; Prod’hom, G.; Dietler, G.; Greub, G.; Kasas, S. Nanomechanical sensor applied to blood culture pellets: A fast approach to determine the antibiotic susceptibility against agents of bloodstream infections. Clin. Microbiol. Infect. 2017, 23, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Lissandrello, C.; Inci, F.; Francom, M.; Paul, M.R.; Demirci, U.; Ekinci, K.L. Nanomechanical motion of Escherichia coli adhered to a surface. Appl. Phys. Lett. 2014, 105, 113701. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Cuervo, A.; Carrascosa, J.L. Nanomechanical detection of: Escherichia coli infection by bacteriophage T7 using cantilever sensors. Nanoscale 2019, 11, 17689–17698. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. USA 2015, 112, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Mustazzolu, A.; Venturelli, L.; Dinarelli, S.; Brown, K.; Floto, R.A.; Dietler, G.; Fattorini, L.; Kasas, S.; Girasole, M.; Longo, G. A rapid unraveling of the activity and antibiotic susceptibility of mycobacteria. Antimicrob. Agents Chemother. 2019, 63, e02194-18. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.-C.; Venturelli, L.; Kannan, A.; Sanglard, D.; Dietler, G.; Willaert, R.; Kasas, S. Yeast Nanometric Scale Oscillations Highlights Fibronectin Induced Changes in C. albicans. Fermentation 2020, 6, 28. [Google Scholar] [CrossRef]
- Stupar, P.; Chomicki, W.; Maillard, C.; Mikeladze, D.; Kalauzi, A.; Radotić, K.; Dietler, G.; Kasas, S. Mitochondrial activity detected by cantilever based sensor. Mech. Sci. 2017, 8, 23–28. [Google Scholar] [CrossRef]
- Wu, S.; Liu, X.; Zhou, X.; Liang, X.M.; Gao, D.; Liu, H.; Zhao, G.; Zhang, Q.; Wu, X. Quantification of cell viability and rapid screening anti-cancer drug utilizing nanomechanical fluctuation. Biosens. Bioelectron. 2016, 77, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Stupar, P.; Longo, G.; Dietler, G. Détecter la vie grâce à la microscopie à force atomique. Médecine/Sciences 2015, 31, 369–371. [Google Scholar] [CrossRef][Green Version]
- Venturelli, L.; Kohler, A.C.; Stupar, P.; Villalba, M.I.; Kalauzi, A.; Radotic, K.; Bertacchi, M.; Dinarelli, S.; Girasole, M.; Pešić, M.; et al. A perspective view on the nanomotion detection of living organisms and its features. J. Mol. Recognit. 2020, 33, e2849. [Google Scholar] [CrossRef]
- Bennett, I.; Pyne, A.L.B.; McKendry, R.A. Cantilever Sensors for Rapid Optical Antimicrobial Sensitivity Testing. ACS Sens. 2020, 5, 3133–3139. [Google Scholar] [CrossRef] [PubMed]
| Method | Characteristics | Reference |
|---|---|---|
| Culture-based AST methods | ||
| Broth dilution assay | Macro- or microdilution of medium–antibiotic solution and growth evaluation based on turbidity or colorimetric differences. | [3,4,7,16,17,18,19,20,21,22,23,24,25,26,27] |
| Disk diffusion | Optical analysis of the resulting colony is based on the growth. MIC determination. | [3,9,10,11] |
| Gradient diffusion | Similar to the disk diffusion method using a plastic strip. | [38] |
| Time-kill test | Reveals a time- or concentration-dependent antimicrobial effect drugs synergism or antagonism. | [28,29,30,31,32] |
| Optical-based AST methods | ||
| Optical tracking of cell division | Single-cell division tracking associated with large volume imaging. | [39] |
| Multiplexed automated digital microscopy | Optical imaging of cells with quantification of growth rates in the presence of antibiotics. | [33,34,35] |
| oCelloscope | Estimate the growth of bacterial cells with an optical microscope. | [36] |
| Single-cell morphological analysis (SCMA) | Imaging changes of the morphology of single cells upon antibiotic treatment. | [37] |
| Surface plasmon resonance (SPR) | A SPR biosensor was used to determine the susceptibility of Staphylococcus aureus clinical isolates. | [40] |
| Electrical-based AST methods | ||
| Electric resistance | Growth of cells in a microchannel is directly proportional to the measured resistance change. | [41] |
| Impedance-based Fast Antimicrobial Susceptibility Test (IFAST) | Changes in biophysical properties of bacteria measured by impedance cytometry. | [42] |
| Electrochemical | Measurement of the change in current due to electrochemical reactions. | [43,44,45] |
| Electrical AST (e-AST) | Growth of cells is monitored by detecting capacitance change of bacteria bound to 60 aptamer-functionalized capacitance sensors | [46] |
| Mechanical-based AST methods | ||
| Asynchronous magnetic bead rotation | Detects bacterial growth, based on the rotation of a cluster of magnetic microparticles. | [47] |
| Method | Characteristics | Reference |
|---|---|---|
| 16S rRNA identification | Influence of antibiotic on growth by measurement of 16S rRNA. | [76] |
| ATP bioluminescence | ATP quantification as an estimate of the microbial population metabolic activity. | [69,70,71,72,73,74] |
| DNA microarrays | DNA microarray using 70mer oligonucleotide. probes to detect resistance genes. | [49] |
| Real-Time PCR | Detection of resistance genes. | [50,51,52,53] |
| MALDI-TOF MS and broth dilution | Combination of microbial identification with an established AST method. | [65,66] |
| MALDI-TOF MS and SILAC | Identification of metabolic inactive microorganisms upon antibiotic treatment. | [68] |
| Microorganisms | Remark | Antimicrobial | Reference |
|---|---|---|---|
| Escherichia coli | Motile bacterium, rapidly growing bacterium | Ampicillin, ceftriaxone, ciprofloxacin | [77,141,142,143] |
| Bacteriophage T7 | [143] | ||
| Bordetella pertussis | Non-motile bacterium, slowly growing bacterium | Clarithromycin, ampicillin | [94] |
| Staphylococcus aureus | Non-motile bacterium, rapidly growing bacterium | Ciprofloxacin | [144] |
| Mycobacterium abscessus | Non-motile bacterium, rapidly growing bacterium | Rifampicin, isoniazid, amikacin | [145] |
| Bacillus Calmette-Guérin | Non-motile bacterium, slowly growing bacterium | Rifampicin, isoniazid, amikacin | [145] |
| Candida albicans | Yeast (candidiasis) | Caspofungin | [144,146] |
| Cell/Protein | Remark | Killing/ Neutralizing Agent | Reference |
|---|---|---|---|
| Topoisomerase II | Protein conformational changes are detected | AMPPNP 1, aclarubicin | [140] |
| Mitochondria | Intracellular organelle oscillation detection | Rotenon | [147] |
| Osteoblasts | Mammalian cell | Glutaraldehyde | [144] |
| Neurons | Mammalian cell | Osmotic shock | [144] |
| Breast cancer cells | Mammalian cell | Paclitaxe, doxorubicin | [148,149] |
| Arabidopsis thaliana | Plant cell | Absence of light | [144] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasas, S.; Malovichko, A.; Villalba, M.I.; Vela, M.E.; Yantorno, O.; Willaert, R.G. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. https://doi.org/10.3390/antibiotics10030287
Kasas S, Malovichko A, Villalba MI, Vela ME, Yantorno O, Willaert RG. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics. 2021; 10(3):287. https://doi.org/10.3390/antibiotics10030287
Chicago/Turabian StyleKasas, Sandor, Anton Malovichko, Maria Ines Villalba, María Elena Vela, Osvaldo Yantorno, and Ronnie G. Willaert. 2021. "Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing" Antibiotics 10, no. 3: 287. https://doi.org/10.3390/antibiotics10030287
APA StyleKasas, S., Malovichko, A., Villalba, M. I., Vela, M. E., Yantorno, O., & Willaert, R. G. (2021). Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics, 10(3), 287. https://doi.org/10.3390/antibiotics10030287







