Abstract
The incidence of Extended-spectrum β-lactamase (ESBL)-encoding genes (blaCTX-M and blaTEM) among Gram-negative multidrug-resistant pathogens collected from three different countries was investigated. Two hundred and ninety-two clinical isolates were collected from Egypt (n = 90), Saudi Arabia (n = 162), and Sudan (n = 40). Based on the antimicrobial sensitivity against 20 antimicrobial agents from 11 antibiotic classes, the most resistant strains were selected and identified using the Vitek2 system and 16S rRNA gene sequence analysis. A total of 85.6% of the isolates were found to be resistant to more than three antibiotic classes. The ratios of the multidrug-resistant strains for Egypt, Saudi Arabia, and Sudan were 74.4%, 90.1%, and 97.5%, respectively. Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa showed inconstant resistance levels to the different classes of antibiotics. Escherichia coli and Klebsiella pneumoniae had the highest levels of resistance against macrolides followed by penicillins and cephalosporin, while Pseudomonas aeruginosa was most resistant to penicillins followed by classes that varied among different countries. The isolates were positive for the presence of the blaCTX-M and blaTEM genes. The blaCTX-M gene was the predominant gene in all isolates (100%), while blaTEM was detected in 66.7% of the selected isolates. This work highlights the detection of multidrug-resistant bacteria and resistant genes among different countries. We suggest that the medical authorities urgently implement antimicrobial surveillance plans and infection control policies for early detection and effective prevention of the rapid spread of these pathogens.
1. Introduction
Widespread use of antibiotics is often accompanied by increased bacterial resistance [1]. This can lead to the emergence of new antibiotic-resistant mechanisms that adversely affecting our ability to treat diseases. Increased antimicrobial resistance to antibiotics leads to longer treatment periods and higher health care costs. Several reports have indicated that multi-drug resistance exists in different countries such as the United States [2,3] and European countries [4]. However, resistance to microbial agents is growing very rapidly in developing countries in Africa [5] and Asia [6]. Multidrug-resistant (MDR) bacteria are non-sensitive to one or more antimicrobial agents, while extensively-drug-resistant (XDR) bacteria are non-sensitive to at least one or two agents, and pan-drug-resistant (PDR) bacteria are non-sensitive to all agents in all antimicrobial categories [7].
Extended-spectrum beta-lactamases (ESBLs) are enzymes that can hydrolyze β-lactam antibiotics. Their production, mainly depends on expression of bla genes, is one of the most prevalent resistance mechanisms in Gram-negative bacteria [1]. Several ESBL variants have been detected and grouped into different families including TEM, CTX-M, SHV, and OXA [8]. Among those, CTX-M and TEM are the most common ESBLs. The bla genes may be placed on transferable elements (plasmids or transposons) that can facilitate a horizontal spread of antibiotic resistance among different strains [3]. Besides, MDR clinical isolates are particularly worrisome to patients and clinicians and they are rapidly spreading across countries with the least resources to tackle them. Additionally, the possession of ESBLs may complicate infection control in hospital [5,6,7]. Due to the noticeable geographical differentiation of bla genes among ESBL-producers, examination of the prevalence of these genes among ESBL-producing strains is of great importance for clinical care and for developing optimal infection control measures in hospital [2,5].
Escherichia coli is the most common bacterial species in clinical laboratories, and has been implicated as playing a role in human infectious diseases [8]. E. coli antibiotic-resistance is very worrying because it is the most pathogenic Gram-negative bacterium. It causes urinary tract infections and both society- and hospital-acquired bacteremia and diarrhea [9]. The emergence and spread of E. coli resistance to several antibiotics has been reported in several regions [10].
Klebsiella pneumoniae is also a Gram-negative pathogen that has a large genome of plasmids and chromosomal genes. Some Klebsiella strains are opportunistic pathogens, infecting immunocompromised patients and causing health-care-associated infections involving pneumoniae, bloodstream infections, and urinary tract infections [11]. Other strains are hyper-virulent, infecting healthy people and causing acute infections involving endophthalmitis, pyogenic liver abscess, and meningitis [11]. Control of antimicrobial resistance in multi-drug-resistant-Klebsiella pneumoniae (MDR-KP) is a great challenge for clinicians. The optimal medication option for MDR-KP infections has still not been well determined. Combination therapies, including high-dose meropenem, fosfomycin, colistin, tigecycline, and aminoglycosides, are widely used, with suboptimal results [12]. The expansion of extended-spectrum β-lactamase (ESBL) production by Klebsiella pneumoniae (50% in some countries) is associated with high rates of mortality [13].
Pseudomonas aeruginosa is an opportunistic pathogen that is a major cause of morbidity and mortality in immunocompromised individuals. Elimination of P. aeruginosa has become increasingly difficult due to its remarkable ability to resist antibiotics [14]. Pseudomonas aeruginosa strains use their high levels of intrinsic and gained resistance mechanisms against most antibiotics. Also, the adaptive antibiotic resistance of P. aeruginosa is a recently characterized mechanism, whereby P. aeruginosa has developed biofilm-mediated resistance as well as forming multidrug-tolerant cells that are accountable for the recalcitrance and retrogression of infections [14].
Therefore, it is assumed that there should be continuous follow-up to reveal the extent of resistance of these pathogens to antibiotics, and results should be compared with those of preceding studies. In our previous study, we isolated different bacterial pathogens from Egypt and surrounding countries and studied their prevalence [15]. The present study was conducted to investigate the prevalence of multidrug-resistant pathogens and the antimicrobial resistance of different resistant genes in the highly resistant strains. The differences in antibiotic-resistance among strains spreading in different regions were compared.
2. Results
2.1. Study Cohort and Atrain Identification
A total of 292 different clinical isolates were obtained from various discharged private laboratories in Egypt, Saudi Arabia, and Sudan from April 2015 to July 2016. Our study included 110 (37.7%) males and 182 (62.3%) females [15]. In a previous study, the most frequently isolated organism was Escherichia coli (n = 103, 35.2%), followed by Pseudomonas aeruginosa (n = 83, 28.3%), Klebsiella pneumoniae (n = 47, 16%), and Staphylococcus aureus (n = 45, 15.4%). The least frequently isolated species was Proteus meribalis (n = 14, 4.8%) [15] (Figure 1).
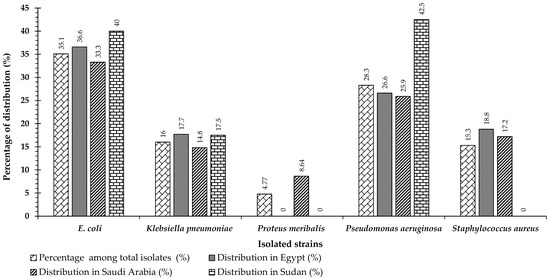
Figure 1.
Percentages of pathogenic strains isolated from various clinical samples (collected from three different countries: Egypt, Saudi Arabia, and Sudan) among all isolates identified and the distribution of strains in each country (modified from Azab et al., [15]).
The distribution of isolates varied greatly among the three countries according to the sample size of each country. For Escherichia coli, the distribution percentage was 36.7% (n = 33), 33.3% (n = 54), and 40% (n = 16) for Egypt, Saudi Arabia, and Sudan, respectively. The percentages of Klebsiella pneumoniae were 17.8% (n = 16), 14.8% (n = 24), and 17.5% (n = 7), while the percentages of Pseudomonas aeruginosa were 26.7% (n = 24), 25.9% (n = 42), and 42.5% (n = 17), respectively. The distribution percentage of Staphylococcus aureus, (Gram-positive) was 18.9% (n = 17) and 17.3% (n = 28, 17.3%) for Egypt and Saudi Arabia, respectively. Finally, the percentage of Proteus meribalis was 8.6% (n = 14) for Saudi Arabia, but this species was not detected in the samples from Egypt and Sudan (Figure 1). Thus, Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae were the most repeated Gram-negative isolates in the three countries in the current investigation.
2.2. Antimicrobial Susceptibility Testing
The relation between antibiotic resistance and clinical specimen types was firstly investigated. Table 1 summarizes the resistance patterns of antibiotics regards the clinical specimen types. For the strains isolated from abscesses, the resistance was most commonly exhibited toward tetracycline (85%), followed by cephalosporins 2nd (82%), cephalosporins 3rd and penicillin (81%), cephalosporins 1st and macrolides (70%), quinolones (24%), and carbapenem (6%). For blood samples, the highest resistance was against macrolides (90%), followed by penicillin (82%), sulfonamide (69%), and quinolones (37%). Regarding samples isolated from the middle ear, the highest resistance was against penicillin (86%), followed by nitrofurans (77%), cephalosporins 3rd (73%), quinolones (18%), and carbapenem (9%). The Tracheal tube samples showed the highest resistance against penicillin and nitrofurans (100%), followed by cephalosporins 1st and cephalosporins 2nd (63), and tetracycline (33%). For the nasal swabs, the highest resistant were against penicillin, cephalosporins 1st and cephalosporins 2nd (100%), followed by cephalosporins 3rd, quinolones and carbapenem (67%), and finally macrolides, sulfonamides, and nitrofurans (50%). The sputum samples had the ability to resist both penicillin and the cephalosporins 2nd (88%), followed by cephalosporins 1st (86%), cephalosporins 3rd, and quinolones (75%), sulfonamide and aminoglycoside (63%), and carbapenem (50%). For throat swabs, the highest resistance was against penicillin, cephalosporins 1st and cephalosporins 2nd, macrolides, sulfonamide, and nitrofurans (100%), and aminoglycosides (33%). The results demonstrated that the samples were isolated from urethral swabs didn’t showed any resistance. For urine, the highest resistance was against penicillin (88%), followed by cephalosporins 1st (81%), tetracycline (71%), nitrofurans (29%), and carbapenem (4%). Finally, the strains isolated from vaginal swabs showed the highest resistance against macrolides (84%), followed by cephalosporins 1st (79%), penicillin (79%), and nitrofurans (31%).

Table 1.
Summarizes the percentage antibiotic resistance patterns for strains isolated from various clinical specimen types.
The patterns of the susceptibility of antibiotic classes to pathogens isolated from our clinical samples are shown in Figure 2. Most strains showed multidrug-resistance phenotypes to at least three different antimicrobial classes. In the present study, 3.4% (n = 10) of all strains were sensitive to all antibiotic classes, while 4.5% (n = 13) were resistant to one antibiotic class, and 6.5% (n = 19) were resistant to two antibiotic classes. Surprisingly, 85.6% (n = 250) of the isolates were multidrug-resistant (MDR), out of which 2.8% (n = 7) were extensively-drug-resistant (XDR) (Figure 2).
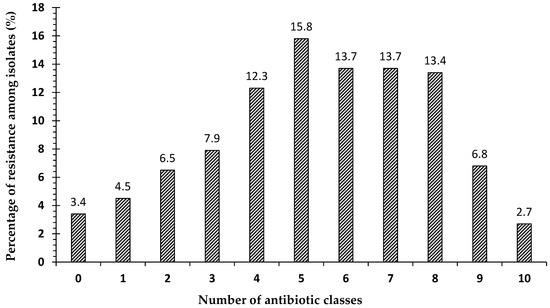
Figure 2.
The percentage of strains resistant to antimicrobial agents from 11 antibiotic classes of the 292 bacterial strains obtained from clinical specimens: 0 means not resistant to any antibiotic group (sensitive to all groups); 1–10 indicate the numbers of classes to which the bacterial strains showed resistance; 1 indicates the percentage of strains that were resistant to only one class of antibiotic; 2 indicates the percentage of strains that were resistant to two classes of antibiotics, etc.
The distribution of antibiotic resistance varied among strains obtained from the three countries. Among the 90 strains obtained from Egypt, 4.4% (n = 4) were sensitive to all antibiotics, while 10% (n = 9) were resistant to one antibiotic class, and 11.1% (n = 10) were resistant to two antibiotic classes. A total of 74.5% (n = 67) strains were classified as MDR, of which 1.5% (n = 1) were XDR. On the other hand, of the 162 strains obtained from Saudi Arabia, only 2.5% (n = 4) were sensitive to all antibiotic classes, while 1.9% (n = 3) were resistant to one antibiotic class, and 5.6% (n = 9) were resistant to two classes. A total of 90% (n = 146) were classified as MDR, of which 2.7% (n = 4) were XDR. Among the 40 strains obtained from Sudan, no isolates were sensitive to all antibiotics, while 3.0% (n = 1) of the strains were resistant to only one class. A total of 97.5% (n = 39) of all strains were classified as MDR, of which 7.7% (n = 3) were XDR (Figure 3).
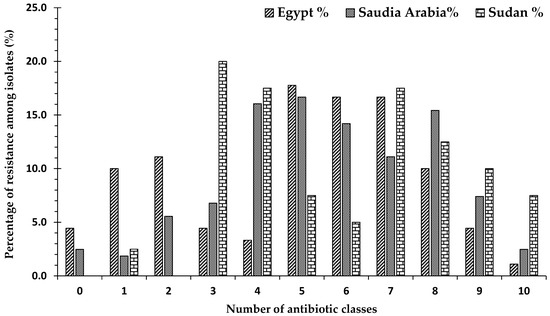
Figure 3.
The percentages of strains resistant to antimicrobial agents from 11 antibiotic classes from all bacterial strains isolated from Egypt, Sudan, and Saudi Arabia: 0 means the percentage of strains obtained from a country that were not resistant to any antibiotic group (sensitive to all groups); numbers 1–10 indicate the numbers of classes that the bacterial strains showed resistance to; 1 indicates the percentage of strains obtained from this country that showed resistance to only one class of antibiotic; 2 indicates the percentage of strains obtained from a country that showed resistance to two classes of antibiotics amongst the 11 well known antibiotic classes, etc.
The diversity of drug-resistance also varied according to strain. Of the 103 E. coli isolates, 1.9% (n = 2) were sensitive to all antibiotic classes, while 2.9% (n = 3) and 5.8% (n = 6) of the E. coli strains were resistant to one or two classes, respectively. The percentage of E. coli strains that were MDR was 89.3% (n = 92), of which 1.9% (n = 1) were XDR. Of a total of 47 K. pneumoniae strains, 6.4% (n = 3) and 4.3% (n = 2) showed resistance to one or two classes, while no isolates were sensitive to all classes. A total of 89.3% (n = 42) of the K. pneumoniae strains were MDR, of which 4.7% (n = 2) were XDR. On the other hand, no P. meribalis strains were resistant to less than three classes, but all P. meribalis strains (n = 14) were MDR, of which 7.1% (n = 1) were XDR. Among the 83 P. aeruginosa strains, no strains were found to be sensitive to all classes, while 3.6% (n = 3) were resistant to either one or two classes of antibiotics. A total of 92.8% (n = 77) of P. aeruginosa strains were MDR, of which 3.9% (n = 3) were XDR. For the 45 obtained S. aureus strains, 13.3% (n = 6) were found to be sensitive to all classes, while 8.9% (n = 4) and 17.8% (n = 8) of the strains exhibited resistance to one or two antibiotic classes, respectively. A total of 60% (n = 27) were MDR, but no XDR strains were obtained (Figure 4). Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae were chosen for further molecular characterization and study, as they were the most frequently identified strains in the three countries under study.
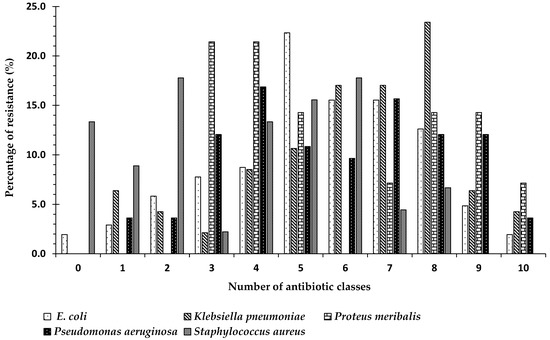
Figure 4.
The percentage specific bacterial species that were resistant to antimicrobial agents from 11 antibiotic classes among isolated strains of the same species (obtained from 3 different countries: Egypt, Sudan, and Saudi Arabia): 0 means the percentage of strains that were not resistant to any antibiotic group (sensitive to all groups); numbers 1–10 indicate the numbers of classes to which the bacterial strain showed resistance; 1 indicates the percentage of strains that showed resistance to only one class of antibiotic; 2 indicates the percentage of strains that showed resistance to two classes of antibiotics amongst the 11 well known antibiotic classes, etc.
2.3. Antibiotic Susceptibility Testing of Escherichia coli Strains
Table 2 summarizes the resistance patterns of E. coli strains isolated from clinical specimens of patients from the three countries. For the 33 E. coli strains obtained from Egypt, resistance was most commonly exhibited toward macrolides (90.9%), followed by penicillins (87.9%), cephalosporins 3rd (75.8%), cephalosporins 1st (75.0%), cephalosporins 2nd (72.7%), tetracyclines (66.7%), quinolones (57.6%), sulfonamide (48.5%), aminoglycosides (41.9%), and nitrofuran (4.5%). For the 54 E. coli strains isolated from Saudi Arabia, the highest level of resistance was toward macrolides (98.1%), followed by cephalosporins 1st (92.5.0%), penicillins (90.7%), tetracyclines (82.4%), sulfonamide (61.5%), cephalosporins 2nd (51.9%), aminoglycosides (48.1%), cephalosporins 3rd (44.4%), quinolones (35.2%), nitrofuran (11.1 %), and carbapenem (2.3%). Also, the level of highest resistance for the 16 E. coli strains obtained from Sudan was exhibited toward macrolides (100%), followed by penicillins (87.5%), cephalosporins 1st (80%), tetracyclines (80%), cephalosporins 2nd (62.5%), cephalosporins 3rd (56.3%), quinolones (50%), aminoglycosides (21.4%), and nitrofuran (6.3%).

Table 2.
Summary of the patterns of resistance to antibiotic classes for Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae strains obtained from three different countries.
2.4. Antibiotic Susceptibility of Klebsiella pneumoniae Strains
The resistance patterns of K. pneumoniae strains obtained from clinical specimens of patients from three countries are shown in Table 2. All 16 K. pneumoniae strains obtained from Egypt exhibited resistance to macrolides (100%), with 85.7% being resistance to nitrofuran, 81.3% to penicillins, 81.3% to cephalosporins 2nd, 81.3% to quinolones, 80% to cephalosporins 1st, 75% to cephalosporins 3rd, 62.5% to aminoglycosides, 56.3% to sulfonamide, 56.3% to carbapenem, and 33.3% to tetracyclines.
For the 24 K. pneumoniae strains obtained from Saudi Arabia, the highest number of strains were resistant to macrolides (100%), followed by penicillins (83.3%), tetracyclines (73.9%), cephalosporins 1st (70.8%), sulfonamide (54.2%), aminoglycosides (50%), cephalosporins 3rd (50%), nitrofuran (42.1%), cephalosporins 2nd (41.7%), and quinolones (33.3%). For the 7 K. pneumoniae strains obtained from Sudan, the highest number of strains were resistant to macrolides (100%), penicillins (100%), cephalosporins 1st (100%), cephalosporins 2nd (100%), cephalosporins 3rd (100%), quinolones (100%), followed by nitrofuran (85.7%), aminoglycosides (71.4%), sulfonamide (28.6%), and tetracyclines (28.6%)
2.5. Antibiotic Susceptibility of Pseudomonas aeruginosa Strains
The resistance patterns of P. aeruginosa isolated from clinical specimens of patients in the three countries are shown in Table 2. For the 24 K. pneumoniae strains obtained from Egypt, the highest number of strains were resistant to macrolides (100%), nitrofuran (100%), and penicillins (100%), followed by tetracyclines (83.3%), cephalosporins 2nd (79.2%), sulfonamide (78.3%), cephalosporins 3rd (66.7%), cephalosporins 1st (61.5%), quinolones (47.8%), aminoglycosides (41.7%), and carbapenem (25%). For the 42 P. aeruginosa strains obtained from Saudi Arabia, the highest number of strains were resistant to cephalosporins 1st (95.1%), followed by tetracyclines (92.7%), penicillins (90.5%), macrolides (90.5%), cephalosporins 2nd (66.8%), sulfonamide (66%), nitrofuran (54.8%), aminoglycosides (50%), cephalosporins 3rd (42.9%), quinolones (19.7%), and carbapenem (4.9%). For the 17 strains of P. aeruginosa isolated from Sudan, the highest number of strains were resistant to cephalosporins 2nd (100%), followed by penicillins (94.1%), cephalosporins 3rd (70.6%), cephalosporins 1st (58.9%), sulfonamide (58.8%), macrolides (35.3%), tetracyclines (29.4%), nitrofuran (18.8%), aminoglycosides (12.5%), and quinolones (11.8%).
2.6. Molecular Characterization of Isolates
Molecular characterization of the two most significant strains obtained from each country was done with 16S rRNA gene fragments. These were chosen for further molecular identification and investigation of gene resistance based on their frequency among the three countries and their high resistance to the majority of antibiotic classes. A 16S rRNA gene fragment showed that one isolate belonged to the genus Escherichia and was identified as the Escherichia coli strain and the second isolate belonged to the genus Klebsiella and was identified as the Klebsiella pneumoniae strain for all countries under study. A phylogenetic analysis based on 16S rRNA gene sequencing and alignment revealed that the two isolates were clustered with Escherichia coli and Klebsiella pneumoniae strains. The first isolates were confirmed to be E. coli strains (reference no. NR_114042.1) with similarity levels of 99.3%, 99.6%, and 98.6% for the Egypt, Saudi Arabia, and Sudan isolates, respectively. The second isolates were confirmed to be K. pneumoniae strains (reference strain nos. NR_114715.1, NR_117686.1, and NR_119276.1) with similarity levels of 98.8%, 96.7%, and 96.9% for Egypt, Saudi Arabia, and Sudan isolates, respectively (Figure 5).
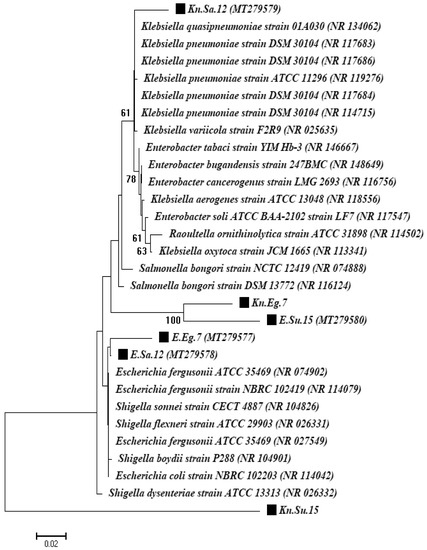
Figure 5.
A phylogenetic tree based on 16S rRNA gene sequencing for the Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae strains.
2.7. Frequency of Antibiotic Resistance
Our results showed that all selected strains were resistant to macrolides, nitrofuran, penicillins, tetracyclines, cephalosporins 1st, cephalosporins 2nd, cephalosporins 3rd, quinolones, aminoglycosides, sulfonamide, and carbapenem. These strains were positive for the presence of blaCTX-M and blaTEM genes. The predominant gene in this study was blaCTX-M (size 500 bp), which was found in all selected isolates (n = 6, 100%). The blaTEM gene (size 600 bp) was found in most of the selected isolates (n = 4, 66.7%). blaCTX-M was detected in E. coli and K. pneumoniae obtained from the three countries included in this study, while the blaTEM gene was not detected in the E. coli and K. pneumoniae strains obtained from Saudi Arabia and Egypt, respectively (Figure 6).
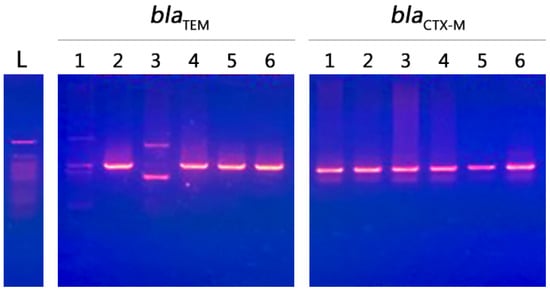
Figure 6.
The presence of blaCTX-M and blaTEM genes. blaCTX-M genes in multidrug-resistant isolates obtained from three different countries are shown. The DNA ladder is 50 bp and the blaTEM gene (size 600 bp) was found in most of the selected isolates (n = 4); it was not detected in E. coli and K. pneumoniae strains obtained from Saudi Arabia and Egypt, respectively. The blaCTX-M gene (size 500 bp) was found in all selected isolates (n = 6). The blaCTX-M gene was detected in E. coli and K. pneumoniae obtained from the three countries included in this study; 1, 2, and 5 are codes for the selected E coli strains from Saudi Arabia, Egypt, and Sudan, respectively; 3, 4, and 6 are codes for the selected K. pneumoniae strains from Egypt, Saudi Arabia, and Sudan, respectively.
3. Discussion
Investigating the prevalence of antimicrobial resistance rates is important for creating empirical treatment strategies and evaluating the existing guidelines. The frequency and number of types of infection caused by Gram-negative bacteria have increased dramatically in the past few decades, with disparities between reports from different institutions and countries [16]. Infection with multidrug-resistant Gram-negative bacteria is a global problem that is correlated with increased morbidity and mortality rates [17]. Bacteria can obtain antibiotic resistance through mutational changes or by gaining resistance genes through horizontal gene transfer [18]. The prevalence of antimicrobial resistance is increasing worldwide, particularly in developing countries, because of greater access to antibiotic drugs. This increase in microbial drug resistance is mainly caused by the use of antimicrobials in humans and other animals [19].
In our previous study, a total of 292 different clinical isolates obtained from various discharged private laboratories from countries including Egypt, Saudi Arabia, and Sudan were evaluated to determine the pathogen distribution. In that study, the most frequently isolated organism was Escherichia coli, followed by Pseudomonas aeruginosa, Klebsiella pneumoniae, and Staphylococcus aureus. The least frequently isolated organism was Proteus mirabilis [15]. Our reported incidences were used to compare the prevalence of pathogens in different countries in the same region and to study multidrug resistance.
In the current study, we evaluated antimicrobial susceptibility patterns, and the results showed that a small percentage (3.4%) of the total isolated organisms were sensitive to all classes of antibiotics and a small percentage (2.7%) were resistant to ten classes of a total of eleven classes (extensively drug-resistant). The most common level of resistance was resistance to five classes, followed by resistance to six, seven, and eight classes, respectively. We found that 85.6% of all isolates were resistant to three or more (≥3) antibiotic classes, so the majority of isolates were multidrug-resistant (MDR). The highest frequency of multidrug-resistant isolates was detected in Sudan with (97.5%) followed by Saudi Arabia (90.1%) and Egypt (74.4%), while 50.3% of all strains were resistant to six antibiotic classes or more (≥6): Percentages of 48.9%, 50.6% and 49.9% for Egypt, Saudi Arabia, and Sudan, respectively. This high frequency is comparable with those found in other reports. The highest percentage of organisms that are sensitive to all antibiotic classes was found in Egypt, and the lowest rate of resistance was found in Saudi Arabia, while no organisms were detected to be sensitive to all groups in Sudan. In contrast, Sudan had the highest percentage of strains that were resistant to nine and ten groups, followed by Saudi Arabia and Egypt.
Our findings are in agreement with previous studies conducted in Sudan, which showed that E. coli, K. pneumoniae, P. aeruginosa, S. aureus, and Proteus spp. are the most commonly encountered organisms, and most of these strains have been found to be highly resistant to several antibiotic classes [20]. Inconsistent with another study, we showed that most isolates were resistant to many antibiotics, and Gram-negative isolates were resistant to 67% of the examined antibiotics [21]. This is in contrast to earlier publications that demonstrated that the most common pathogen isolated from different clinical specimens was K. pneumoniae followed by P. aeruginosa and then E. coli, and about 22.3% were resistant to three or more classes of antibiotics [22].
Our findings agree with previous studies conducted in Egypt, which showed that 93.6% of the isolated Gram-negative bacteria were multidrug-resistant [23]. Other studies have reported that 71.1% of isolates studied exhibited multidrug-resistance [24], and 91% of all E. coli strains were resistant to three or more antibiotics [25]. In another study conducted on bacterial isolates in drinking water in Cairo, the majority of the tested strains (62.4 to 98%) exhibited multiple antibiotic resistance [26]. Previous data from two major hospitals in Makkah, KSA showed that 24.6% of E. coli strains, 34.4% of K. pneumoniae strains, and 52.7% of P. aeruginosa strains were resistant to ceftazidime (cephalosporin 3rd generation) [27]. They documented the increasing prevalence of extended-spectrum beta-lactamase (ESBL) producing isolates from Saudi Arabia; in some institutes, 29% of E. coli strains and 65% of K. pneumoniae strains were ESBL [28].
In the present study, the species with the highest number strains that exhibited resistance to ≥3 antibiotic classes (multidrug-resistant) was P. aeruginosa (92.6%), followed by K. pneumoniae (89.3%), and E. coli (89.2%). The E. coli strains obtained in our study showed inconsistent resistance levels to all classes of antibiotics except carbapenem between countries. The most common pathogen for E. coli to be resistant to was macrolides (96.3% of all strains) followed by penicillins (88.7%), cephalosporin 1st (82.5%), tetracyclines (76.4%), cephalosporin 2nd (62.4%) and cephalosporin 3rd (58.8%), while low levels of resistance were found for nitrofuran (7.3%) and carbapenem (2.3%).
Our findings are inconsistent with the results of previous studies conducted in Sudan, which showed that multidrug-resistant E. coli strains were frequently resistant to macrolides [29], penicillins [21,30,31], cephalosporins [21,22,31,32], and tetracyclines [21,31], while they were less frequently resistant to quinolones, aminoglycosides [22,31], and carbapenems [22]. Other studies recorded complete sensitivity toward carbapenems [30,32], and others recorded frequent resistance against carbapenems [33]. In contrast, another study in Saudi Arabia showed that a high percentage of E. coli strains were resistant to penicillin [34,35], followed by cephalosporins [34], macrolides, and aminoglycosides [35], whereas few strains were resistant to aminoglycosides and nitrofuran [34], and all were sensitive to carbapenems [34]. Other studies in Egypt reported that a high percentage of E. coli strains were highly resistant to beta-lactams [36], followed by penicillins [37,38], cephalosporines [38], macrolides, tetracyclines, and quinolones [37,38], and aminoglycosides [37]. The lowest level of resistance was exhibited against nitrofuran and carbapenems, and aminoglycosides [38]. In contrast, another study showed that all isolated E. coli strains were sensitive to carbapenem [36].
In the current study, K. pneumoniae strains showed inconstant resistance levels to all classes of antibiotics except carbapenem. K. pneumoniae strains were most commonly resistant to macrolides followed by penicillins and then cephalosporin 1st, but the results for other groups were variable. In Saudi Arabia and Sudan, the lowest number of strains were resistant to carbapenem, but in Egypt, resistance was least commonly observed for tetracycline. However, this is in contrast to earlier publications from Sudan that demonstrated that a high number of K. pneumoniae strains were resistant to penicillins [21], followed by cephalosporines [21,22,30], nitrofuran, carbapenem, and quinolones [21]. Resistance was least commonly found for carbapenem, quinolones [22], aminoglycosides [21,22], and tetracyclines [21]. Other reported studies showed complete sensitivity toward carbapenems [30,39]. Studies in Saudi Arabia showed a high degree of resistance of K. pneumoniae to penicillins and tetracyclines [39,40], cephalosporins, carbapenems, aminoglycosides, and nitrofuran [40]. Resistance was least commonly found toward aminoglycosides [39,40], carbapenems [40], and quinolones [39]. Previous studies in Egypt showed that K. pneumoniae were highly resistant to cephalosporins [41,42,43], followed by penicillins, carbapenem, quinolones, and aminoglycosides [41,42].
In the present study, P. aeruginosa strains showed inconstancy in their resistance level to all classes of antibiotics, except carbapenem. The pathogen that strains were most commonly resistant to varied from macrolides to cephalosporin among countries, and the pathogens associated with the highest level of sensitivity were carbapenem and tetracycline. Studies in Sudan have shown that P. aeruginosa strains are most frequently resistant toward penicillins [21], followed by cephalosporines [22,44], quinolones, nitrofuran, and tetracyclines [21], and aminoglycosides [44]. Resistance was least commonly found for carbapenem, aminoglycosides, and quinolones [22]. Other studies in Saud Arabia have shown frequent resistance of P. aeruginosa to penicillins and carbapenems [45,46] and a low frequency of resistance toward aminoglycosides and cephalosporins 4th generation [45,46]. In contrast with our findings, other studies in Egypt showed that P. aeruginosa most frequently exhibited resistance against carbapenems [47,48,49], followed by extended-spectrum beta-lactamases [49], penicillins, cephalosporins, macrolides, and tetracyclines [47]. Resistance was least frequently exhibited toward carbapenems [50]. Other studies showed widespread occurrence of Gram-negative bacteria with resistance to beta-lactam and carbapenem in Middle Eastern hospitals [51].
The frequencies and types of infections caused by extended-spectrum β-lactamase (ESBL) producing pathogens have increased dramatically in the past few decades with disparities between different regions and countries. Since the beginning of the new millennium, Gram-negative bacteria have become the most commonly isolated ESBL-producing bacteria worldwide with blaCTX-M ESBLs being the most frequently isolated type. Thus, we evaluated the prevalence of blaCTX-M and blaTEM resistant gene traits from clinical specimens within different countries. Resistant genes were characterized at the molecular level. In the current study, the selected isolates were found to be positive for the presence of blaCTX-M and blaTEM genes. The predominant gene identified in this study was blaCTX-M, which was found in all selected isolates (100%). The blaTEM gene was found in most isolates (66.7%), the blaCTX-M was detected in E. coli and K.pneumoniae for all countries included in our study, while the blaTEM gene was not detected in E. coli and K. pneumoniae strains isolated from Saudi Arabia and Egypt, respectively. Our findings are in agreement with previous studies that showed that blaCTX-M is more prevalent among tested strains of E. coli, K. pneumoniae, and P. mirabilis as compared with blaTEM [52]. A study in Egypt revealed 46 E. coli isolates and showed that all extended-spectrum beta-lactamase (ESBL) producers were positive for blaCTX-M and blaTEM genes [53]. Another study carried out on K. pneumoniae showed that blaCTX-M and blaTEM genes were detected in 52.8% and 53.3% of ESBL isolates, respectively [54,55]. In another study, blaCTX-M and blaTEM genes were detected in K. pneumoniae more frequently than E. coli, and blaTEM genes were detected more frequently than blaCTX-M [38]. In agreement with our findings, studies conducted in different regions of Sudan have shown that E. coli is the most predominant isolate with the blaCTX-M genotype, followed by K. pneumoniae [56,57]. In another study, the blaTEM gene was detected in 55.1% of E. coli strains and 58% of K. pneumoniae strains, but the blaCTX-M gene was detected in 71.4% of E. coli strains and 68.4% of K. pneumoniae strains [54]. Other studies conducted in Saudi Arabia detected both genes in a large number of K. pneumoniae strains isolated from two different hospitals. The results showed that the isolates contained the blaCTX-M and blaTEM genes [53,58].
Our results present alarming evidence of a severe spread of ESBL genes in the studied countries, especially the epidemiological blaCTX-M and blaTEM genes. There is potential for the dissemination of MDR strains in different regions. ESBLs are plasmid-mediated β-lactamases that have been recognized for their ability to hydrolyze different generations of cephalosporins (oxyimino-cephalosporin) and other antibiotic classes. These enzymes are repressed by β-lactamase inhibitors such as clavulanic acid and tazobactam. Public health efforts should focus on the correct use of antibiotics to limit their dissemination. In addition, further investigation of the molecular epidemiology of ESBLs in various clinical specimens is needed to obtain a better database of ESBL-producing pathogens present in different countries.
4. Methods
4.1. Study Design and Specimen Collection
A total of 292 clinical specimens were collected over the period from April 2015 to July 2016. Bacterial isolates of Gram-negative pathogens were obtained from various discharged private laboratories; the clinical specimens were collected from three countries (Egypt, Saudi Arabia, and Sudan). Of the 292 samples, 90 (30.8%) were from Egypt, 162 (55.5%) were from Saudi Arabia, and 40 (13.7%) were from Sudan. Bacterial strains were collected from different clinical specimens (urine (n = 168), vaginal swab (n = 43), ear swab (n = 22), blood (n = 19), abscess (n = 17), endotracheal tube (n = 8), sputum (n = 8), throat swab (n = 3), nasal swab (n = 3), and urethral swab (n = 1)). The study protocol was approved by the ethics committee, and the research followed the principles of the Declaration of Helsinki. All specimens were collected aseptically and transported to the microbiology laboratory, where they were immediately processed according to the standard microbiological procedures [59].
4.2. Isolation and Identification of Isolates
The samples were cultured by the streaking on blood agar [60], cysteine-lactose-electrolyte-deficient agar [61], chocolate agar [62], MacConkey agar [63], and Thayer martin agar [64], and then incubated at a 37 °C for 24 hours. The bacterial colonies were purified and identified according to the colonies’ morphology. The pathogenic isolates were identified basely by physiological and different biochemical tests [65].
Vitek2 system was used for the identification of isolates (bioMerieux, Vitek2 Compact System Version: 07.01, GN card), HCWW- Reference Lab. Microbiology Lab. GN card is based on 64 biochemical methods and newly developed substrates that measuring carbon source utilization, enzymatic activities, and resistance [66].
4.3. Antimicrobial Susceptibility Testing
The antimicrobial sensitivity of pathogenic isolates to different antimicrobial agents was determined using the Kirby–Bauer disc diffusion method; each isolate was seeded on Mueller–Hinton agar following the Clinical and Laboratory Standards Institute [67] guidelines. Several antimicrobial discs representing different classes of antimicrobial agents were included in the presented study. These discs including the following: Penicillin combination, which was Amoxicillin-clavulanic acid (30 μg) and Ampicillin sulbactam (20 μg), Cephalosporin 1st generation which represented Cephalexin (30 μg), Cephalosporin 2nd generation which involved Cefuroxime (30 μg) and Cefaclor (30 μg), Cephalosporin 3rd generation containing Cefixime (5 μg), Cefotaxime (30 μg), Ceftriaxone (30 μg) and Ceftazidime (30 μg), Macrolides including Erythromycin (15 μg), Clarithromycin (15 μg) and Azithromycin (15 μg), Gentamicin (10 μg) as Aminoglycosides, Tetracycline /Doxycycline (30 μg), Quinolones/Ciprofloxacin (5 μg), Levofloxacin (5 μg), and Ofloxacin (5 μg), Sulfonamides/Trimethoprim/sulfamethoxazole (Co-trimoxazole) (25 μg), Nitrofurantoin (300 μg), and imipenem (10 μg). All antibiotic discs were produced by Oxoid (Oxoid, UK). A colony from each isolate was grown in Mueller Hinton broth (Oxoid, UK) at 37 °C. The discs of antibiotics were applied using fine-end forceps, and plates were incubated at 37 °C for 24 hrs. Inhibition zones were measured and interpreted as sensitive (S), intermediate (I), or resistant (R) according to Clinical and Laboratory Standards criteria [68]. Isolates that show resistance to at least three different classes of antimicrobial agents were considered as MDR.
4.4. Molecular Identification of Bacterial Isolates
Molecular identification was performed by the extraction of genomic DNA using the modified method as previously described [69,70]. The 16S rRNA gene fragments were analyzed using forward and reverse universal primers as 27f (5 GAGTTTGATCACTGGCTCAG-3) and 1492r (5-TACGGCTACCTTGTTACGACTT-3). Genomic DNA was used as a template for polymerase chain reaction (PCR). The PCR mixture contained the folloing: PCR buffer (1×), dNTP (0.25 mM), MgCl2 (0.5 mM), Taq DNA polymerase [QIAGEN] (2.5 U), primers (0.5 μM of each), and genomic DNA (1 μg of). The PCR was conducted in DNA Engine Thermal Cycler (PTC-200, Bio-Rad, Hercules, CA, USA) at the following conditions: Hot starting performed at 94 °C (3 min), then 30 cycles of 94 °C (0.5 min), 55 °C (0.5 min), and 72 °C (1 min). The extension was performed at 72 °C for 10 min. The PCR products were commercially sequenced at Sigma Company (Cairo, Egypt) using an ABI 3730xl DNA sequencer. Through BLASTN, The sequences were compared with sequences available at GenBank database. Multiple sequence alignments were then performed by ClustalX 1.8 software package and the phylogenetic tree was established using a neighbor-joining method of Kimura 2-parameter model to calculate genetic distance as the transitional and transversional substitution rates using MEGA (version 6.1) software. The level of confidence for each branch at 1000 repeats was tested by bootstrap analysis. Detection of chimeras (>3% diverged from the closest sequences) was conducted using Uchime2_NCBI tool. 16S rRNA gene sequences of all strains have been deposited in NCBI Gene Bank using Bankit program and assigned with accession numbers MT279577- MT279580.
4.5. Molecular Screening of Resistant Genes
Bacterial template DNA was prepared using the boiled lysate method for molecular screening of two different resistance genes (blaTEM and blaCTX-M genes). For blaTEM gene, the forward primer was blaTEM-F (5′-GCTCACCCAGAAACGCTGGT-3′), and the reverse primer was blaTEM-R (5′-CCATCTGGCCCCAGTGCTGC-3′). For blaCTX-M gene, the forward primer was blaCTXM- F (5′-SCSATGTGCAGYACCA GTAA-3′), and reverse primer was blaCTX-M-R (5′-ACCAGAAYVAGCGGBGC-3′). The oligonucleotide primers blaTEM was designed based on the nucleotide sequence of blaTEM gene reported in the National Center for Biotechnology Information (NCBI) Gene Bank database, while blaCTX-M primers were synthesized according to a previously published protocol [71]. The PCR mixture (25 μ) contained 1 μL of each primer (10 pmol/μL), 12.5 μL of 2× Taq PCR RED Master Mix (Ampliqon, Denmark), 3 μL of template DNA, and 7.5 μL of ultra pure water. The PCR conditions for amplification of the blaTEM and blaCTX-M genes were selected based on the properties of the primers and were as follows, initial denaturation for 5 min at 94 °C, 35 cycles of denaturation for 45 s at 94 °C, annealing for 30 s at 54 °C, and extension for 60 s at 72 °C with a final extension for 5 min at 72 °C; and initial denaturation for 3 min at 94 °C, 35 cycles of denaturation for 60 s at 94 °C, annealing for 30 s at 58 °C, and extension for 60 s at 72 °C with a final extension for 10 min at 72 °C, respectively. The PCR products were subjected to agarose gel (1.2%) electrophoresis in in borate buffer (TBE, Tris-Borate-EDTA) at 80 V for 2 h and was visualized by a UV illumination after ethidium bromide (0.5 μg/mL) staining. The molecular weight was estimated by comparing with 50 bp DNA Ladder (InVitrogen) marker.
4.6. Statistical Analysis
Collected data was analyzed using Statistical Package for Social Sciences (SPSS; Version 23) software to calculate frequencies and percentages for all categories of organisms and resistance of isolated organisms against different antibiotics among the three countries.
5. Conclusions
This work highlights the detection of multidrug-resistant bacteria and resistant genes in different countries. These bacteria and genes compromise the effectiveness of different antibiotic classes. Thus, implementation of antimicrobial surveillance plans and infection control policies by medical authorities to ensure early detection and effectively halt the rapid spread of these pathogens is required.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. K.S.M.A., Investigation, Methodology, Writing—original draft; M.A.A.-R., Conceptualization, Formal analysis, Investigation, Methodology, review and editing; H.H.E.-S., Formal analysis, Investigation, Writing—review and editing; E.A., A.A.G., Formal analysis, Writing—review and editing; A.A.G., Formal analysis, Investigation, Writing—review and editing; M.M.S.F., Conceptualization, Formal analysis, Investigation, Methodology, review and editing; E.A., A.A.G., Funding. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Informed Consent Statement
The study protocol was approved by the ethics committee, and the research was conducted following the principles of the Declaration of Helsinki. All specimens were collected aseptically and transported to the microbiology laboratory, where they were immediately processed according to the standard microbiological procedures. We would like to confirm that this material is the authors’ own original work, which has not been previously published elsewhere. The paper is not currently being considered for publication elsewhere. The paper reflects the authors’ own research and analysis in a truthful and complete manner. The paper properly credits the meaningful contributions of co-authors and co-researchers. The results are appropriately placed in the context of prior and existing research. All authors have been personally and actively involved in substantive work leading to the manuscript and will hold themselves jointly and individually responsible for its content.
Data Availability Statement
The datasets used and analyzed in the current study are available from the corre-sponding author upon reasonable request.
Acknowledgments
We thank Taif University Researchers Supporting Project number (TURSP-2020/13), Taif University, Taif, Saudi Arabia. We thank Albaraa S. El-Saied, Associate professor at Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt, for his support and assistance with statistical analysis and great contribution through this work.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
bla: β-lactamase; bp: Base pair; DNA: Deoxyribonucleic acid; ESBL: Extended-spectrum β-lactamase; F: Forward; GN: Gram-negative; HCWW: Holding Company for water and waste Water; KSA: Kingdom of Saudi Arabia; MDR: Multidrug-resistant; MDR-KB: multi-drug-resistant-Klebsiella pneumoniae; n: Number; NCBI: National Center for Biotechnology Information; PCR: Polymerase chain reaction; PDR: Pandrug resistant; R: Reverse; rRNA: Ribosomal ribonucleic acid; SPSS: Statistical Package for Social Sciences; XDR: Extensively drug-resistant.
References
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Sahm, D.F.; Thornsberry, C.; Mayfield, D.C.; Jones, M.E.; Karlowsky, J.A. Multidrug-resistant urinary tract isolates of Escherichia coli: Prevalence and patient demographics in the United States in 2000. Antimicrob. Agents Chemother. 2001, 45, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Jones, M.E.; Draghi, D.C.; Thornsberry, C.; Sahm, D.F.; Volturo, G.A. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Van De Sande-Bruinsma, N.; Grundmann, H.; Verloo, D.; Tiemersma, E.; Monen, J.; Goossens, H.; Ferech, M.; European Antimicrobial Resistance Surveillance System Group; European Surveillance of Antimicrobial Consumption Project Group. Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 2008, 14, 1722–1730. [Google Scholar] [CrossRef]
- Okeke, I.N.; Aboderin, O.A.; Byarugaba, D.K.; Ojo, K.K.; Opintan, J.A. Growing problem of multidrug-resistant enteric pathogens in Africa. Emerg. Infect. Dis. 2007, 13, 1640. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Hsueh, P.-R.; Badal, R.E.; Hawser, S.P.; Hoban, D.J.; Bouchillon, S.K.; Ni, Y.; Paterson, D.L. Antimicrobial susceptibility profiles of aerobic and facultative Gram-negative bacilli isolated from patients with intra-abdominal infections in the Asia-Pacific region according to currently established susceptibility interpretive criteria. J. Infect. 2011, 62, 280–291. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Koneman, E.W.; Allen, S.D.; Janda, W.; Schreckenberger, P.; Winn, W. Diagnostic microbiology. In The Nonfermentative Gram-Negative Bacilli; Lippincott-Raven Publishers: Philedelphia, PA, USA, 1997; pp. 253–320. [Google Scholar]
- Coleman, B.L.; Louie, M.; Salvadori, M.I.; McEwen, S.A.; Neumann, N.; Sibley, K.; Irwin, R.J.; Jamieson, F.B.; Daignault, D.; Majury, A.; et al. Contamination of Canadian private drinking water sources with antimicrobial resistant Escherichia coli. Water Res. 2013, 47, 3026–3036. [Google Scholar] [CrossRef]
- Nys, S.; Okeke, I.N.; Kariuki, S.; Dinant, G.J.; Driessen, C.; Stobberingh, E.E. Antibiotic resistance of faecal Escherichia coli from healthy volunteers from eight developing countries. J. Antimicrob. Chemother. 2004, 54, 952–955. [Google Scholar] [CrossRef]
- Martin, R.M.; Bachman, M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E.; Carnelutti, A.; Graziano, E.; Russo, A. Multidrug-resistant Klebsiella pneumoniae: Challenges for treatment, prevention and infection control. Expert Rev. Anti-Infect. Ther. 2018, 16, 749–761. [Google Scholar] [CrossRef]
- Paterson, D.L.; Ko, W.-C.; Von Gottberg, A.; Mohapatra, S.; Casellas, J.M.; Goossens, H.; Mulazimoglu, L.; Trenholme, G.; Klugman, K.P.; Bonomo, R.A.; et al. Antibiotic Therapy for Klebsiella pneumoniae Bacteremia: Implications of production of extended-spectrum β-Lactamases. Clin. Infect. Dis. 2004, 39, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Azab, K.S.M.; Abdel-Rahman, M.A.; Farag, M.M.S.; El-Sheikh, H.H. Identification and distribution of pathogenic bacteria in clinical specimens within Egypt, Saudi Arabia, and Sudan. Al-Azhar Bull. Sci. 2020, 10, 25–34. [Google Scholar]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef]
- Higgins, P.; Wisplinghoff, H.; Krut, O.; Seifert, H. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin. Microbiol. Infect. 2007, 13, 1199–1201. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Hamid, M.I.; Bendary, M.M.; Merwad, A.M.A.; Elsohaby, I.; Ghaith, D.M.; Alshareef, W.A. What is behind phylogenetic analysis of hospital-, community- and livestock-associated methicillin-resistant Staphylococcus aureus? Transbound. Emerg. Dis. 2019, 66, 1506–1517. [Google Scholar] [CrossRef] [PubMed]
- Abass, A.M.; Ahmed, M.E.; Ibrahim, I.G.; Yahia, S.A. Bacterial resistance to antibiotics: Current situation in Sudan. J. Adv. Microbiol. 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Saeed, A.; Hamid, S.A.; Bayoumi, M.; Shanan, S.; Alouffi, S.; Alharbi, S.A.; Alshammari, F.D.; Abd, H. Elevated antibiotic resistance of Sudanese urinary tract infection bacteria. EXCLI J. 2017, 16, 1073–1080. [Google Scholar]
- Elbadawi, H.S.; Elhag, K.M.; Mahgoub, E.; Altayb, H.N.; Hamid, M.M.A. Antimicrobial resistance surveillance among Gram-negative bacterial isolates from patients in hospitals in Khartoum State, Sudan. F1000Research 2019, 8, 156. [Google Scholar] [CrossRef]
- Khalifa, H.O.; Soliman, A.M.; Ahmed, A.M.; Shimamoto, T.; Nariya, H.; Matsumoto, T.; Shimamoto, T. High prevalence of antimicrobial resistance in Gram-negative bacteria isolated from clinical settings in Egypt: Recalling for judicious use of conventional antimicrobials in developing nations. Microb. Drug Resist. 2019, 25, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Shehab El-Din, E.M.R.; El-Sokkary, M.M.A.; Bassiouny, M.R.; Hassan, R. Epidemiology of neonatal sepsis and implicated pathogens: A study from Egypt. BioMed Res. Int. 2015, 2015, 509484. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.M.; Ahmed, S.F.; Klena, J.D.; Mohamed, Z.K.; Moussa, T.; Ghenghesh, K.S. Enteroaggregative Escherichia coli in diarrheic children in Egypt: Molecular characterization and antimicrobial susceptibility. J. Infect. Dev. Ctries. 2014, 8, 589–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El-Zanfaly, H.T.; Kassim, E.-S.A.-A.; Badr-Eldin, S.M. Incidence of antibiotic resistant bacteria in drinking water in Cairo. Water Air Soil Pollut. 1987, 32, 123–128. [Google Scholar] [CrossRef]
- Zowawi, H.M. Antimicrobial resistance in Saudi Arabia. An urgent call for an immediate action. Saudi Med. J. 2016, 37, 935–940. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. β-Lactamase production in key Gram-negative pathogen isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef]
- Elmofti, H.A.; Almofti, Y.; Abuelhassan, N.N.; Omer, N.N. Identification and antibiotic resistance patterns of Escherichia coli isolated from broilers farms in Bahri locality/Sudan. Acta Sci. Nutr. Health 2019, 3, 197–203. [Google Scholar] [CrossRef]
- Mekki, A.; Hassan, A.; Eldin, D.; Elsayed, M. Extended spectrum beta lactamases among multi drug resistant Escherichia coli and Klebsiella species causing urinary tract infections in Khartoum. J. Bacteriol. Res. 2010, 2, 18–21. [Google Scholar]
- Ibrahim, M.; Bilal, N.; Hamid, M. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr. Health Sci. 2013, 12, 368–375. [Google Scholar] [CrossRef]
- Ahmed, O.B.; Omar, A.O.; Asghar, A.H.; Elhassan, M.M. Increasing prevalence of ESBL-producing Enterobacteriaceae in Sudan community patients with UTIs. Egypt. Acad. J. Biol. Sci. G Microbiol. 2013, 5, 17–24. [Google Scholar] [CrossRef]
- Satir, S.; Elkhalifa, A.; Ali, M.; Rahim, A.; Elhussein, A.; Elkhidir, I.; Enan, K. Detection of carbepenem resistance genes among selected Gram negative bacteria isolated from patients in-Khartoum state, Sudan. Clin. Microbiol. 2016, 5, 6. [Google Scholar] [CrossRef]
- Yasir, M.; Ajlan, A.M.; Shakil, S.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Farman, M.; Baazeem, Z.M.; Baabdullah, R.; Alawi, M.; Al-Abdullah, N.; et al. Molecular characterization, antimicrobial resistance and clinico-bioinformatics approaches to address the problem of extended-spectrum β-lactamase-producing Escherichia coli in western Saudi Arabia. Sci. Rep. 2018, 8, 14847. [Google Scholar] [CrossRef] [PubMed]
- Hemeg, H.A. Molecular characterization of antibiotic resistant Escherichia coli isolates recovered from food samples and outpatient Clinics, KSA. Saudi J. Biol. Sci. 2018, 25, 928–931. [Google Scholar] [CrossRef]
- Aly, M.E.; Essam, T.M.; Amin, M.A. Antibiotic resistance profile of E. coli strains isolated from clinical specimens and food samples in Egypt. Int. J. Microbiol. Res. 2012, 3, 176–182. [Google Scholar]
- Amer, M.M.; Mekky, H.M.; Amer, A.M.; Fedawy, H.S. Antimicrobial resistance genes in pathogenic Escherichia coli isolated from diseased broiler chickens in Egypt and their relationship with the phenotypic resistance characteristics. Vet. World 2018, 11, 1082–1088. [Google Scholar] [CrossRef]
- Mohammed, H.; Elsadek Fakhr, A.; Al Johery, S.a.E.; Hassanein, W.A.G. Spread of TEM, VIM, SHV, and CTX-M β-lactamases in imipenem-resistant Gram-negative bacilli isolated from Egyptian hospitals. Int. J. Microbiol. 2016, 2016, 8382605. [Google Scholar] [CrossRef]
- Ahmad, S.; Al-Juaid, N.F.; Alenzi, F.Q.; Mattar, E.H.; Bakheet, O.E.-S. Prevalence, antibiotic susceptibility pattern and production of extended-spectrum beta-lactamases amongst clinical isolates of Klebsiella pneumoniae at Armed Forces Hospital in Saudi Arabia. J. Coll. Physicians Surg. Pak. 2009, 19, 264–265. [Google Scholar] [PubMed]
- Azim, N.S.A.; Nofal, M.Y.; Alharbi, M.A.; Al-Zaban, M.I.; Somily, A.M. Molecular-diversity, prevalence and antibiotic susceptibility of pathogenic Klebsiella pneumoniae under Saudi Condition. Pak. J. Biol. Sci. 2019, 22, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Shawky, S.M.; Abdallah, A.; El Kholy, M. Antimicrobial activity of Colistin and Tigecycline against carbapenem resistant Klebsiella pneumoniae clinical isolates in Alexandria, Egypt. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 731–742. [Google Scholar]
- Wasfi, R.; Elkhatib, W.F.; Ashour, H.M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci. Rep. 2016, 6, 38929. [Google Scholar] [CrossRef] [PubMed]
- El-Badawy, M.F.; Tawakol, W.M.; El-Far, S.W.; Maghrabi, I.A.; Al-Ghamdi, S.A.; Mansy, M.S.; Ashour, M.S.; Shohayeb, M.M. Molecular identification of Aminoglycoside-modifying enzymes and plasmid-mediated Quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int. J. Microbiol. 2017, 2017, 8050432. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.D.; Taee, S.; Dezfuli, A.A.; Meghdadi, H.; Shafie, F. Investigation of the prevalence of genes conferring resistance to carbapenems in Pseudomonas aeruginosa isolates from burn patients. Infect. Drug Resist. 2019, 12, 1153–1159. [Google Scholar] [CrossRef]
- Alkeshan, Y. Antimicrobial Resistance Pattern of Pseudomonas aeruginosa in Regional Tertiary Care Hospitals of Saudi Arabia. IOSR J. Dent. Med. Sci. 2014, 5, 54–62. [Google Scholar]
- Khan, M.A.; Faiz, A. Antimicrobial resistance patterns of Pseudomonas aeruginosa in tertiary care hospitals of Makkah and Jeddah. Ann. Saudi Med. 2016, 36, 23–28. [Google Scholar] [CrossRef]
- Gad, G.F.; El-Domany, R.A.; Zaki, S.; Ashour, H.M. Characterization of Pseudomonas aeruginosa isolated from clinical and environmental samples in Minia, Egypt: Prevalence, antibiogram and resistance mechanisms. J. Antimicrob. Chemother. 2007, 60, 1010–1017. [Google Scholar] [CrossRef]
- Elshafiee, E.A.; Nader, S.M.; Dorgham, S.M.; Hamza, D.A. Carbapenem-resistant Pseudomonas aeruginosa originating from farm animals and people in Egypt. J. Vet. Res. 2019, 63, 333–337. [Google Scholar] [CrossRef]
- Farhan, S.M.; Ibrahim, R.A.; Mahran, K.M.; Hetta, H.F.; El-Baky, R.M.A. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect. Drug Resist. 2019, 12, 2125–2133. [Google Scholar] [CrossRef]
- Helmy, O.M.; Kashef, M.T. Different phenotypic and molecular mechanisms associated with multidrug resistance in Gram-negative clinical isolates from Egypt. Infect. Drug Resist. 2017, 10, 479–498. [Google Scholar] [CrossRef]
- Dandachi, I.; Chaddad, A.; Hanna, J.; Matta, J.; Daoud, Z. Understanding the epidemiology of multi-drug resistant Gram-negative bacilli in the Middle East using a one health approach. Front. Microbiol. 2019, 10, 1941. [Google Scholar] [CrossRef]
- Ojdana, D.; Sacha, P.; Wieczorek, P.; Czaban, S.; Michalska-Falkowska, A.; Jaworowska, J.; Jurczak, A.; Poniatowski, B.; Tryniszewska, E. The occurrence of blaCTX-M, blaSHV, and blaTEM genes in extended-spectrum β-Lactamase-positive strains of Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis in Poland. J. Antibiot. 2014, 2014, 7. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.M.; Ashour, M.S.E.-D.; Wiegand, I. First description of CTX-M β-lactamase-producing clinical Escherichia coli isolates from Egypt. Int. J. Antimicrob. Agents 2006, 27, 545–548. [Google Scholar]
- Ahmed, O.; Omar, A.; Asghar, A.; El hassan, M. Prevalence of TEM, SHV and CTX-M genes in Escherichia coli and Klebsiella spp urinary isolates from Sudan with confirmed ESBL phenotype. Life Sci. J. 2013, 10, 191–195. [Google Scholar]
- Elkhateeb, H.; Abdel-Mongy, M.; Othman, A. In vitro Detection of Antibacterial Activity of Glycyrrhizic Acid Nanoparticle against ESBL Producing Klebsiella pneumoniae Strains. Egypt. J. Microbiol. 2018, 53, 193–205. [Google Scholar] [CrossRef]
- Osman, A.E.M.A.E.; Hashim, S.O.; Musa, M.A.; Tahir, O.M. Detection of CTX-M, TEM and SHV Genes in Gram negative bacteria isolated from nosocomial patients at Port Sudan Teaching Hospital. Eur. J. Clin. Biomed. Sci. 2017, 3, 101. [Google Scholar] [CrossRef][Green Version]
- Altayb, H.N.; Siddig, M.; El Amin, N.M.; Maowia, A.I.H.; Mukhtar, M. Molecular Characterization of CTX-M ESBLs among Pathogenic Enterobacteriaceae isolated from different regions in Sudan. Glob. Adv. Res. J. Microbiol. 2018, 7, 40–47. [Google Scholar]
- El Hassan, M.; Ozbazk, H.A.; Hemeg, H.A.; Ahmed, A. Dissemination of CTX-M extended-spectrum β-lactamases (ESBLs) among Escherichia coli and Klebsiella pneumoniae in Al-Madenah Al-Monawwarah region, Saudi Arabia. Int. J. Clin. Exp. Med. 2016, 9, 11051–11057. [Google Scholar]
- Winn, W.C.; Allen, S.D.; Janda, W.M.; Koneman, E.W.; Procop, G.; Schreckenberger, P.; Woods, G.L. Color Atlas and Textbook of Diagnostic Microbiology, 6th ed.; Jones and Bartlett Learning: Burlington, MA, USA, 2005. [Google Scholar]
- Spector, W.S. Handbook of Toxicology; National Academy Of Sciences—National Research Council: Washington, DC, USA, 1955; Volume 1. [Google Scholar]
- Sandys, G.H. A new method of preventing swarming of Proteus sp. with a description of a new medium suitable for use in routine laboratory practice. J. Med. Lab. Technol. 1960, 17, 224–233. [Google Scholar]
- McLeod, J.W.; Wheatley, B.; Phelon, H.V. On Some of the Unexplained Difficulties met with in cultivating the Gonococcus: The part played by the amino-acids. Br. J. Exp. Pathol. 1927, 8, 25–37. [Google Scholar]
- MacConkey, A.T. Note on some Cases of Food-poisoning. In Epidemiology and Infection; Cambridge University Press: Cambridge, UK, 2009; Volume 6, pp. 570–573. [Google Scholar]
- Thayer, J.D.; Martin, J.E., Jr. A selective medium for the cultivation of N. gonorrhoeae and N. meningitidis. Public Health Rep. 1964, 79, 49–57. [Google Scholar] [CrossRef]
- Sutherland, C. District Laboratory Practice in Tropical Countries 2nd edition, Part 1. Monica Cheesbrough 454 pp Price £50 ISBN 0521676304 Cambridge: CUP, 2005. Trop. Dr. 2008, 38, 132. [Google Scholar] [CrossRef]
- Pincus, D.H.; Zbinden, R.; Bosshard, P.P.; Bottger, E.C. New Vitek 2 Colorimetric GN Card for Identification of Gram-Negative Nonfermentative Bacilli. J. Clin. Microbiol. 2007, 45, 4094–4095. [Google Scholar] [CrossRef][Green Version]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 25th Informational Supplemen; CLSI Document M100–S25; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Ling, C.L.; Ciesielczuk, H.L.; Lockwood, J.; Hopkins, S.; McHugh, T.D.; Gillespie, S.H.; Kibbler, C.C. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: Comparison of two different approaches in clinical practice. J. Med. Microbiol. 2012, 61, 483–488. [Google Scholar] [CrossRef]
- Sundsfjord, A.; Simonsen, G.S.; Haldorsen, B.C.; Haaheim, H.; Hjelmevoll, S.-O.; Littauer, P.; Dahl, K.H. Genetic methods for detection of antimicrobial resistance. Acta Pathol. Microbiol. Immunol. Scand. 2004, 112, 815–837. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).