Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives
Abstract
1. Introduction
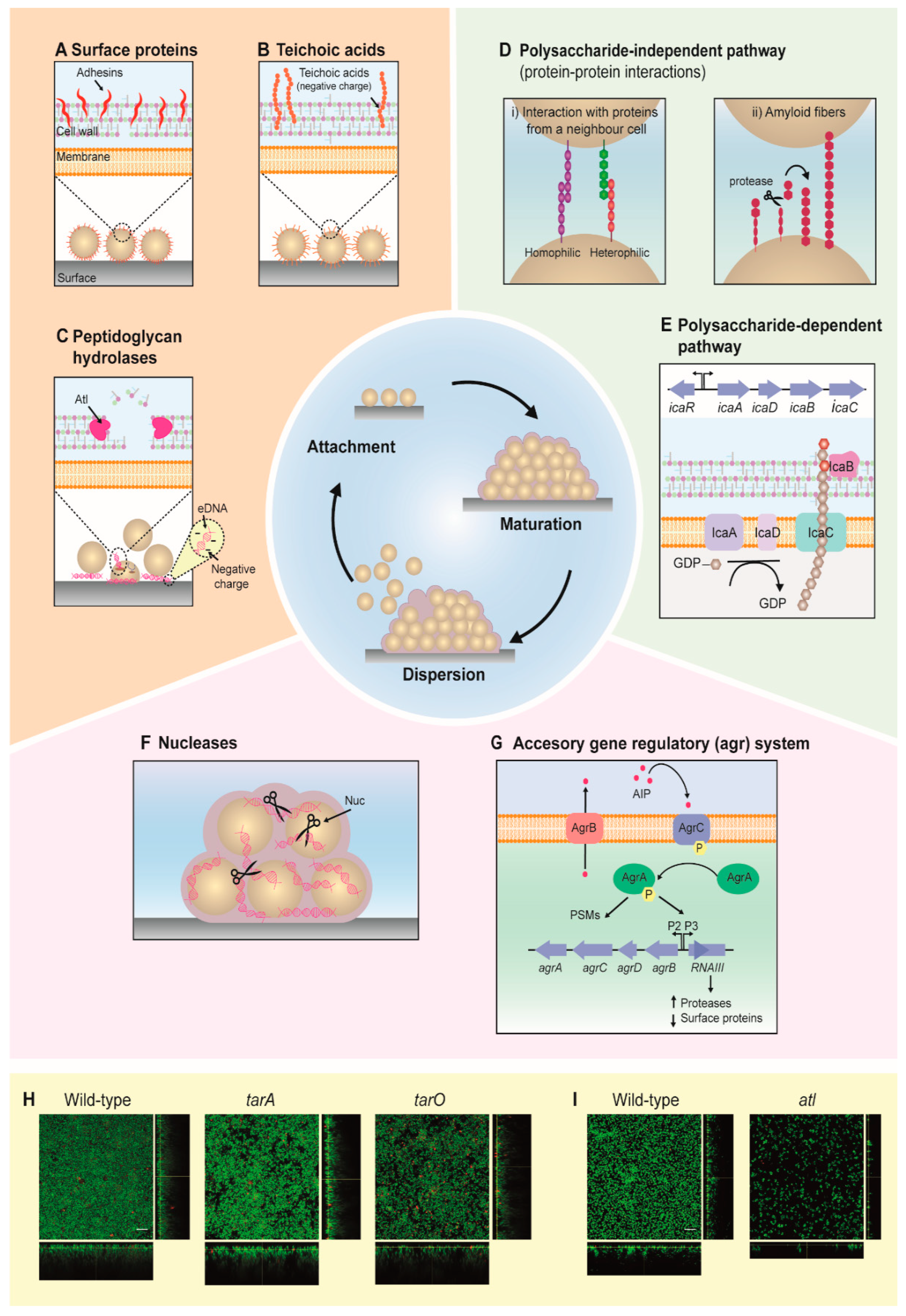
2. Development of Staphylococcal Biofilms
2.1. Attachment
2.2. Maturation
2.3. Dispersion
2.4. Biofilm Formation Is Highly Dependent on Environmental Factors
3. Hurdles in Tackling Staphylococcal Biofilms
3.1. Antibiotic Resistance among Pathogenic Staphylococci
3.2. Limited Antibiotic Penetration in Biofilms
3.3. Heterogeneous Populations in Biofilms
3.4. Biofilms as a Mediator of Horizontal Gene Transfer
4. Molecular Targets to Fight Staphylococcal Biofilms
5. Alternative Treatments of Staphylococcal Biofilms
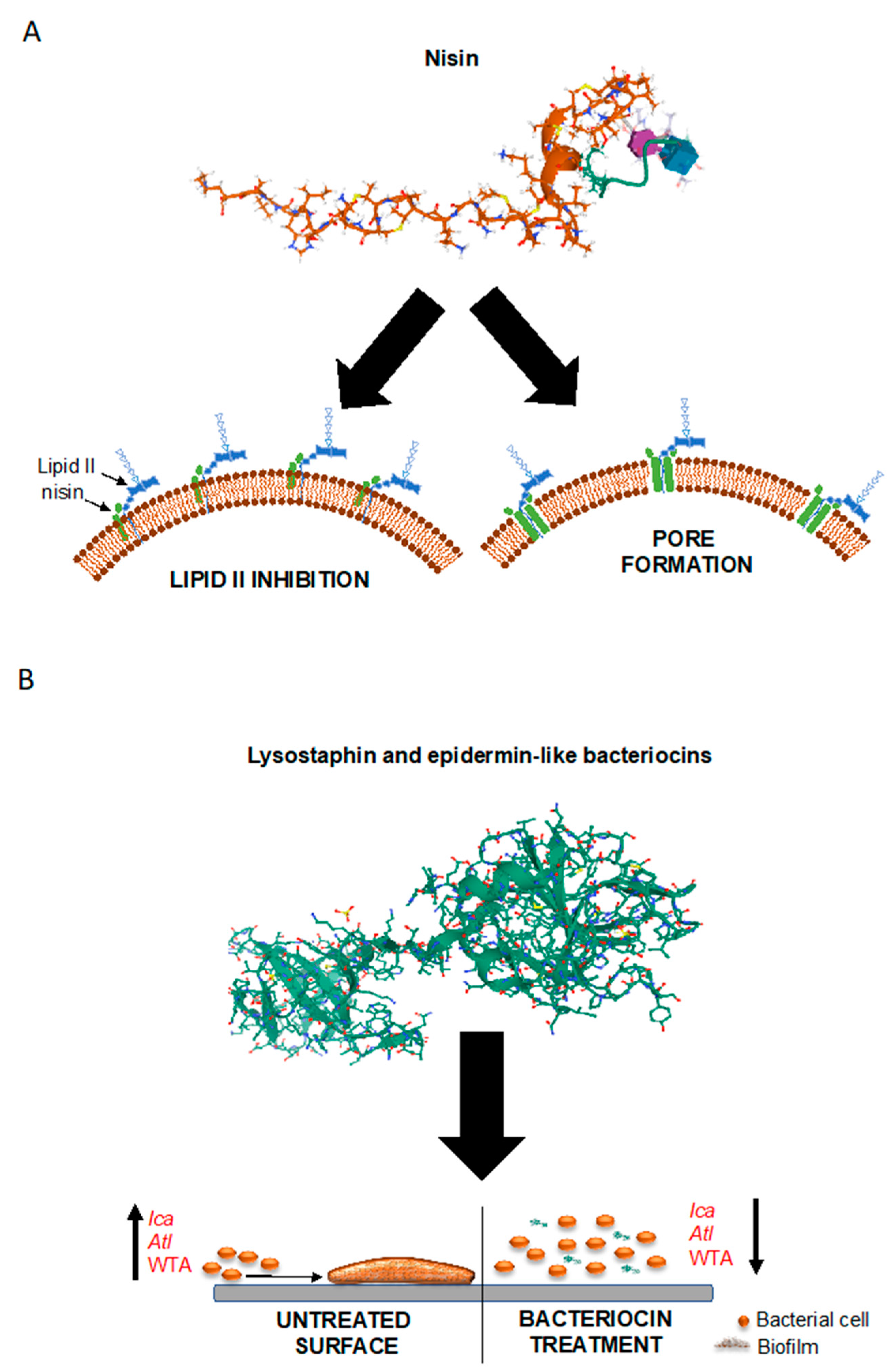
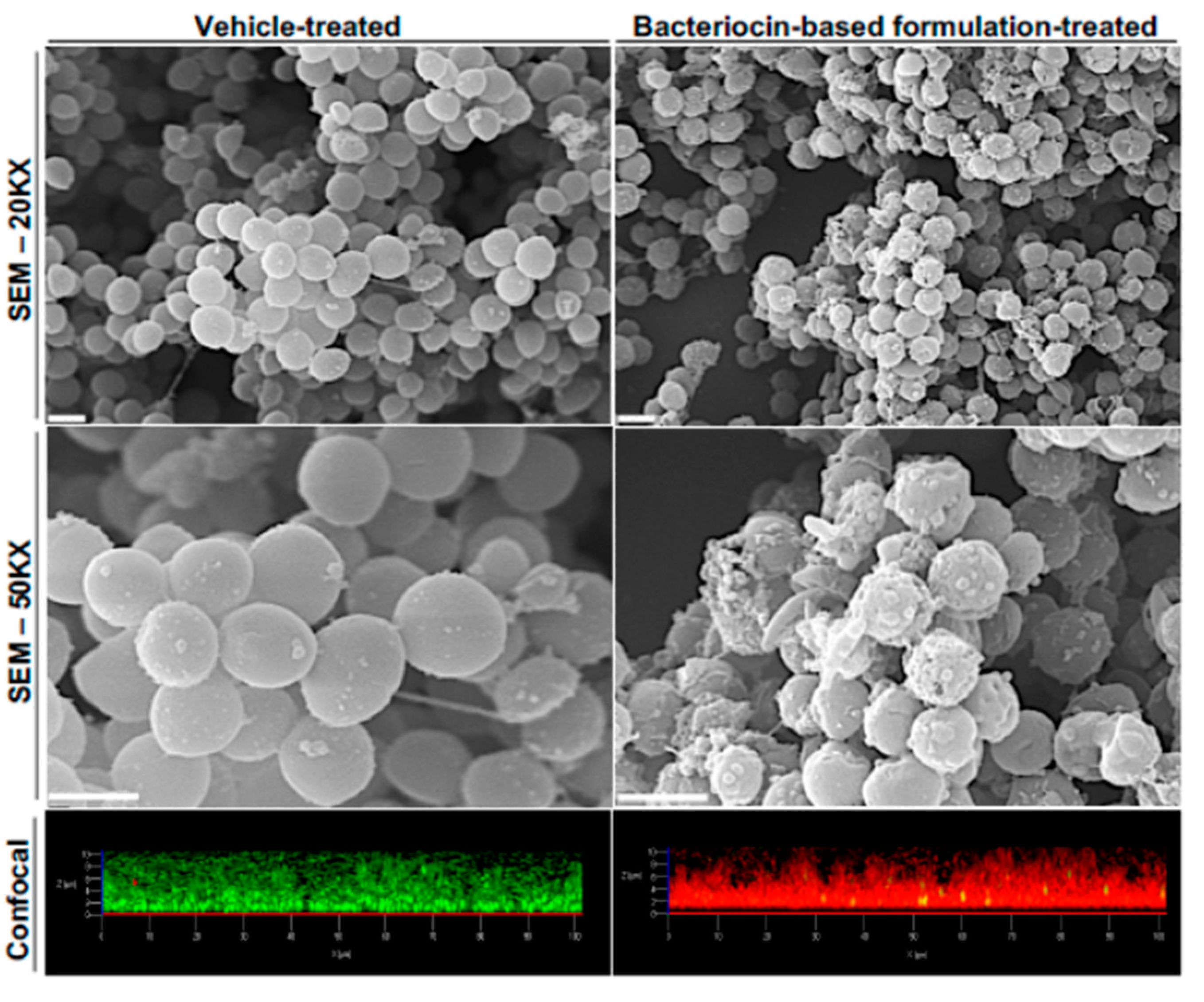
5.1. Bacteriocins
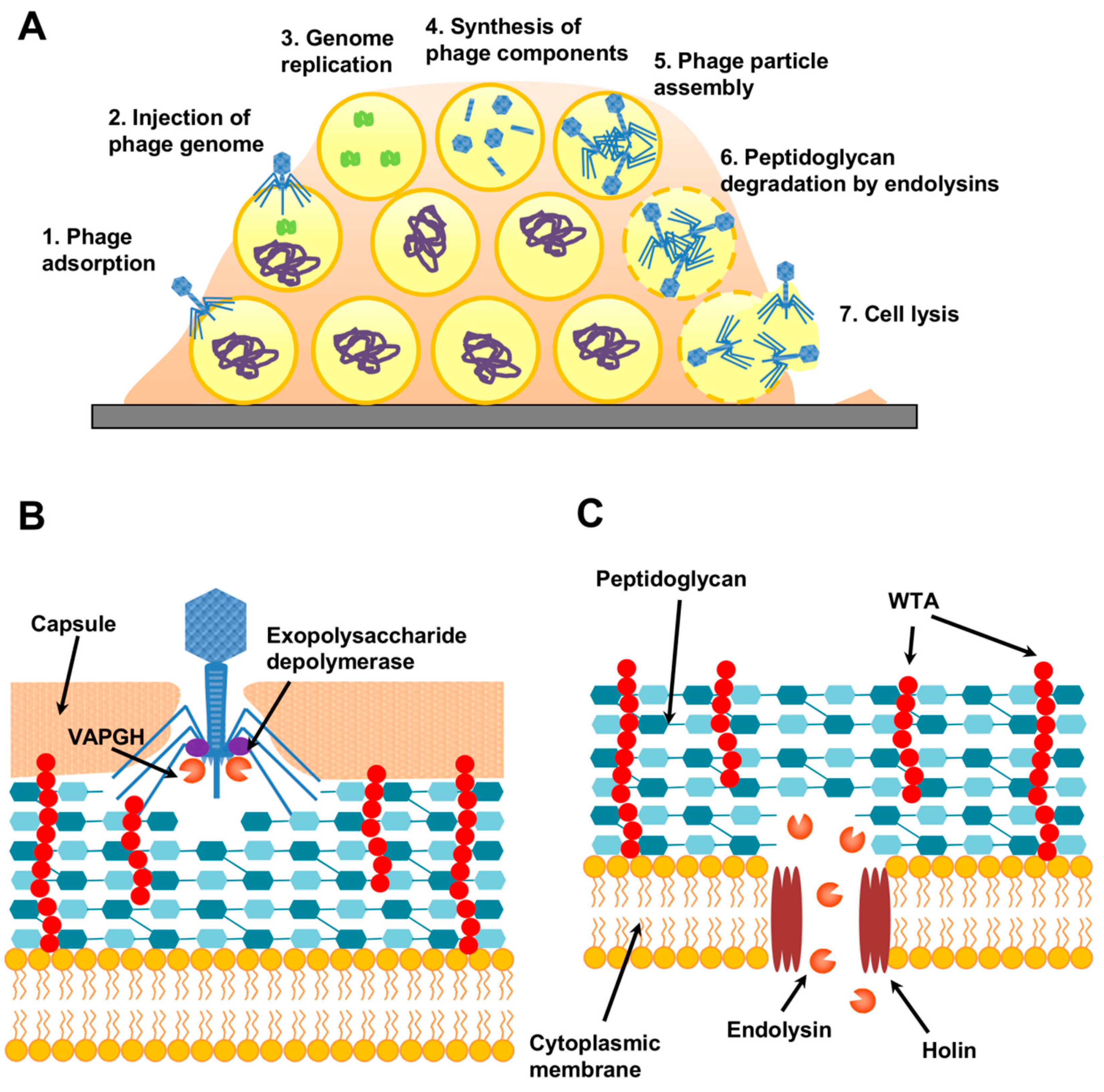
5.2. Phage-Derived Antibiofilm Strategies
5.3. Antibodies
5.4. Nanotechnology
5.5. Photoinactivation
5.6. Other Anti-Biofilm Agents
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monegro, A.F.; Muppidi, V.; Regunath, H. Hospital Acquired Infections; In StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Friedrich, A.W. Control of hospital acquired infections and antimicrobial resistance in Europe: The way to go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; E Kretzschmar, M.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Jia, K.; Fang, T.; Wang, X.; Liu, Y.; Sun, W.; Wang, Y.; Ding, T.; Wang, J.; Li, C.; Xu, D.; et al. Antibiotic Resistance Patterns of Staphylococcus aureus Isolates from Retail Foods in Mainland China: A Meta-Analysis. Foodborne Pathog. Dis. 2020, 17, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; McKimm, J.; Bin Abu Bakar, M. Health care-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef]
- Ling, M.L.; Apisarnthanarak, A.; Madriaga, G. The Burden of Healthcare-Associated Infections in Southeast Asia: A Systematic Literature Review and Meta-analysis. Clin. Infect. Dis. 2015, 60, 1690–1699. [Google Scholar] [CrossRef]
- Pinto, R.M.; Lopes-De-Campos, D.; Martins, M.C.L.; van Dijck, P.; Nunes, C.; Reis, S. Impact of nanosystems in Staphylococcus aureus biofilms treatment. FEMS Microbiol. Rev. 2019, 43, 622–641. [Google Scholar] [CrossRef]
- Dengler, V.; Foulston, L.; de Francesco, A.S.; Losick, R.M. An Electrostatic Net Model for the Role of Extracellular DNA in Biofilm Formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef]
- Fong, J.N.C.; Yildiz, F.H. Biofilm Matrix Proteins. Microbiol. Spectr. 2015, 3, 201–222. [Google Scholar] [CrossRef]
- Izano, E.A.; Amarante, M.A.; Kher, W.B.; Kaplan, J.B. Differential Roles of Poly-N-Acetylglucosamine Surface Polysaccharide and Extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis Biofilms. Appl. Environ. Microbiol. 2007, 74, 470–476. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; de Leo, F.R.; Otto, M. A Crucial Role for Exopolysaccharide Modification in Bacterial Biofilm Formation, Immune Evasion, and Virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Hurlow, J.; Couch, K.; Laforet, K.; Bolton, L.; Metcalf, D.; Bowler, P. Clinical Biofilms: A Challenging Frontier in Wound Care. Adv. Wound Care 2015, 4, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Biofilm Dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.; Humphreys, H.; O’Gara, J.P. The genetics of staphylococcal biofilm formation—Will a greater understanding of pathogenesis lead to better management of device-related infection? Clin. Microbiol. Infect. 2005, 11, 967–973. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef]
- Davidson, D.J.; Spratt, D.; Liddle, A.D. Implant materials and prosthetic joint infection: The battle with the biofilm. EFORT Open Rev. 2019, 4, 633–639. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Genet. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Chen, C.; Krishnan, V.; Macon, K.; Manne, K.; Narayana, S.V.L.; Schneewind, O. Secreted Proteases Control Autolysin-mediated Biofilm Growth of Staphylococcus aureus. J. Biol. Chem. 2013, 288, 29440–29452. [Google Scholar] [CrossRef]
- Ribeiro, M.; Monteiro, F.J.; Ferraz, M.P. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatterials 2012, 2, 176–194. [Google Scholar] [CrossRef]
- Wi, Y.M.; Patel, R. Understanding Biofilms and Novel Approaches to the Diagnosis, Prevention, and Treatment of Medical Device-Associated Infections. Infect. Dis. Clin. N. Am. 2018, 32, 915–929. [Google Scholar] [CrossRef] [PubMed]
- Balaure, P.C.; Grumezescu, A.M. Recent Advances in Surface Nanoengineering for Biofilm Prevention and Control. Part II: Active, Combined Active and Passive, and Smart Bacteria-Responsive Antibiofilm Nanocoatings. Nanomaterials 2020, 10, 1527. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Jiang, H.; Manolache, S.; Wong, A.C.L.; Denes, F.S. Plasma-Mediated Grafting of Poly(ethylene glycol) on Polyamide and Polyester Surfaces and Evaluation of Antifouling Ability of Modified Substrates. Langmuir 2007, 23, 7306–7313. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhang, J.; Wang, Z.; Chen, S. Development of robust biocompatible silicone with high resistance to protein adsorption and bacterial adhesion. Acta Biomater. 2011, 7, 2053–2059. [Google Scholar] [CrossRef]
- Chien, H.-W.; Chen, X.-Y.; Tsai, W.-P.; Lee, M. Inhibition of biofilm formation by rough shark skin-patterned surfaces. Colloids Surf. B Biointerf. 2020, 186, 110738. [Google Scholar] [CrossRef]
- Brown, A.F.; Leech, J.M.; Rogers, T.R.; McLoughlin, R.M. Staphylococcus aureus Colonization: Modulation of Host Immune Response and Impact on Human Vaccine Design. Front. Immunol. 2014, 4, 507. [Google Scholar] [CrossRef]
- Kaye, K.S.; Petty, L.A.; Shorr, A.F.; Zilberberg, M.D. Current Epidemiology, Etiology, and Burden of Acute Skin Infections in the United States. Clin. Infect. Dis. 2019, 68, S193–S199. [Google Scholar] [CrossRef]
- Suaya, J.A.; Mera, R.M.; Cassidy, A.; O’Hara, P.; Amrine-Madsen, H.; Burstin, S.; Miller, N.S. Incidence and cost of hospitalizations associated with Staphylococcus aureus skin and soft tissue infections in the United States from 2001 through 2009. BMC Infect. Dis. 2014, 14, 296. [Google Scholar] [CrossRef]
- Nouwen, J.; Schouten, J.; Schneebergen, P.; Snijders, S.; Maaskant, J.; Koolen, M.; van Belkum, A.; Verbrugh, H. Staphylococcus aureus Carriage Patterns and the Risk of Infections Associated with Continuous Peritoneal Dialysis. J. Clin. Microbiol. 2006, 44, 2233–2236. [Google Scholar] [CrossRef]
- Garrouste-Orgeas, M.; Timsit, J.-F.; Kallel, H.; Ben Ali, A.; Dumay, M.F.; Paoli, B.; Misset, B.; Carlet, J. Colonization with Methicillin-Resistant Staphylococcus aureus in ICU Patients Morbidity, Mortality, and Glycopeptide Use. Infect. Control. Hosp. Epidemiol. 2001, 22, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Genet. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Kiedrowski, M.R.; Gaston, J.R.; Kocak, B.R.; Coburn, S.L.; Lee, S.; Pilewski, J.M.; Myerburg, M.M.; Bomberger, J.M. Staphylococcus aureus Biofilm Growth on Cystic Fibrosis Airway Epithelial Cells Is Enhanced during Respiratory Syncytial Virus Coinfection. mSphere 2018, 3, e00341-18. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 2018, 76, fty003. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Weigel, L.M.; Donlan, R.M.; Shin, D.H.; Jensen, B.; Clark, N.C.; McDougal, L.K.; Zhu, W.; Musser, K.A.; Thompson, J.; Kohlerschmidt, D.; et al. High-Level Vancomycin-Resistant Staphylococcus aureus Isolates Associated with a Polymicrobial Biofilm. Antimicrob. Agents Chemother. 2006, 51, 231–238. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus Form Polymicrobial Biofilms: Effects on Antimicrobial Resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Magalhães, A.P.; Lopes, S.P.; Pereira, M.O. Insights into Cystic Fibrosis Polymicrobial Consortia: The Role of Species Interactions in Biofilm Development, Phenotype, and Response to In-Use Antibiotics. Front. Microbiol. 2017, 7, 2146. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, G.; Jensen, E.R.; Lenoy, E.; Schneewind, O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 2000, 97, 5510–5515. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penadés, J.R. Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Schroeder, K.; Jularic, M.; Horsburgh, S.M.; Hirschhausen, N.; Neumann, C.; Bertling, A.; Schulte, A.; Foster, S.; Kehrel, B.E.; Peters, G.; et al. Molecular Characterization of a Novel Staphylococcus aureus Surface Protein (SasC) Involved in Cell Aggregation and Biofilm Accumulation. PLoS ONE 2009, 4, e7567. [Google Scholar] [CrossRef]
- Greene, C.; McDevitt, D.; Francois, P.; Vaudaux, P.; Lew, D.; Poster, T. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 1995, 17, 1143–1152. [Google Scholar] [CrossRef]
- Conlon, B.P.; Geoghegan, J.A.; Waters, E.M.; McCarthy, H.; Rowe, S.E.; Davies, J.R.; Schaeffer, C.R.; Foster, T.J.; Fey, P.D.; O’Gara, J.P. Role for the A Domain of Unprocessed Accumulation-Associated Protein (Aap) in the Attachment Phase of the Staphylococcus epidermidis Biofilm Phenotype. J. Bacteriol. 2014, 196, 4268–4275. [Google Scholar] [CrossRef]
- Veenstra, G.J.; Cremers, F.F.; van Dijk, H.; Fleer, A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J. Bacteriol. 1996, 178, 537–541. [Google Scholar] [CrossRef]
- Timmerman, C.P.; Fleer, A.; Besnier, J.M.; de Graaf, L.; Cremers, F.; Verhoef, J. Characterization of a proteinaceous adhesin of Staphylococcus epidermidis which mediates attachment to polystyrene. Infect. Immun. 1991, 59, 4187–4192. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Genet. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key Role of Teichoic Acid Net Charge in Staphylococcus aureus Colonization of Artificial Surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef]
- Holland, L.M.; Conlon, B.P.; O’Gara, J.P. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiol. 2011, 157, 408–418. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D.; Singh, A.K.; Drolia, R.; Bai, X.; Tenguria, S.; Bhunia, A.K. Tunicamycin Mediated Inhibition of Wall Teichoic Acid Affects Staphylococcus aureus and Listeria monocytogenes Cell Morphology, Biofilm Formation and Virulence. Front. Microbiol. 2018, 9, 1352. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bose, J.L.; Horswill, A.R.; Bayles, K.W. Temporal and Stochastic Control of Staphylococcus aureus Biofilm Development. mBio 2014, 5, e01341-14. [Google Scholar] [CrossRef]
- Biswas, R.; Voggu, L.; Simon, U.K.; Hentschel, P.; Thumm, G.; Götz, F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 2006, 259, 260–268. [Google Scholar] [CrossRef]
- Heilmann, C.; Hussain, M.; Peters, G.; Götz, F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 1997, 24, 1013–1024. [Google Scholar] [CrossRef]
- Bose, J.L.; Lehman, M.K.; Fey, P.D.; Bayles, K.W. Contribution of the Staphylococcus aureus Atl AM and GL Murein Hydrolase Activities in Cell Division, Autolysis, and Biofilm Formation. PLoS ONE 2012, 7, e42244. [Google Scholar] [CrossRef]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential Role for the Major Autolysin in the Fibronectin-Binding Protein-Mediated Staphylococcus aureus Biofilm Phenotype. Infect. Immun. 2010, 79, 1153–1165. [Google Scholar] [CrossRef]
- Hirschhausen, N.; Schlesier, T.; Peters, G.; Heilmann, C. Characterization of the Modular Design of the Autolysin/Adhesin Aaa from Staphylococcus aureus. PLoS ONE 2012, 7, e40353. [Google Scholar] [CrossRef]
- Oshida, T.; Sugai, M.; Komatsuzawa, H.; Hong, Y.M.; Suginaka, H.; Tomasz, A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: Cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 1995, 92, 285–289. [Google Scholar] [CrossRef]
- Kajimura, J.; Fujiwara, T.; Yamada, S.; Suzawa, Y.; Nishida, T.; Oyamada, Y.; Hayashi, I.; Yamagishi, J.-I.; Komatsuzawa, H.; Sugai, M. Identification and molecular characterization of anN-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation ofStaphylococcus aureus. Mol. Microbiol. 2005, 58, 1087–1101. [Google Scholar] [CrossRef]
- Thalsø-Madsen, I.; Torrubia, F.R.; Xu, L.; Petersen, A.; Jensen, C.; Frees, D. The Sle1 Cell Wall Amidase Is Essential for β-Lactam Resistance in Community-Acquired Methicillin-Resistant Staphylococcus aureus USA300. Antimicrob. Agents Chemother. 2019, 64, e01931-19. [Google Scholar] [CrossRef]
- Hirschhausen, N.; Schlesier, T.; Schmidt, M.A.; Götz, F.; Peters, G.; Heilmann, C. A novel staphylococcal internalization mechanism involves the major autolysin Atl and heat shock cognate protein Hsc70 as host cell receptor. Cell. Microbiol. 2010, 12, 1746–1764. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef]
- Mashruwala, A.A.; Gries, C.M.; Scherr, T.D.; Kielian, T.; Boyd, J.M. SaeRS Is Responsive to Cellular Respiratory Status and Regulates Fermentative Biofilm Formation in Staphylococcus aureus. Infect. Immun. 2017, 85, e00157-17. [Google Scholar] [CrossRef]
- Mack, D.; Haeder, M.; Siemssen, N.; Laufs, R. Association of Biofilm Production of Coagulase-Negative Staphylococci with Expression of a Specific Polysaccharide Intercellular Adhesin. J. Infect. Dis. 1996, 174, 881–883. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The Intercellular Adhesion (ica) Locus Is Present in Staphylococcus aureus and Is Required for Biofilm Formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef]
- Gerke, C.; Kraft, A.; Süßmuth, R.; Schweitzer, O.; Götz, F. Characterization of the N-Acetylglucosaminyltransferase Activity Involved in the Biosynthesis of the Staphylococcus epidermidis Polysaccharide Intercellular Adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef]
- Pokrovskaya, V.; Poloczek, J.; Little, D.J.; Griffiths, H.; Howell, P.L.; Nitz, M. Functional Characterization of Staphylococcus epidermidis IcaB, a De-N-acetylase Important for Biofilm Formation. Biochemistry 2013, 52, 5463–5471. [Google Scholar] [CrossRef]
- Cerca, N.; Jefferson, K.K.; Maira-Litrán, T.; Pier, D.B.; Kelly-Quintos, C.; Goldmann, D.A.; Azeredo, J.; Pier, G.B. Molecular Basis for Preferential Protective Efficacy of Antibodies Directed to the Poorly Acetylated Form of Staphylococcal Poly-N-Acetyl-β-(1–6)-Glucosamine. Infect. Immun. 2007, 75, 3406–3413. [Google Scholar] [CrossRef]
- Atkin, K.E.; Macdonald, S.J.; Brentnall, A.S.; Potts, J.R.; Thomas, G.H. A different path: Revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014, 588, 1869–1872. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A Novel Staphylococcus aureus Biofilm Phenotype Mediated by the Fibronectin-Binding Proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- Paharik, A.E.; Kotasinska, M.; Both, A.; Hoang, T.-M.N.; Büttner, H.; Roy, P.; Fey, P.D.; Horswill, A.R.; Rohde, H. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol. Microbiol. 2017, 103, 860–874. [Google Scholar] [CrossRef]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef]
- Olson, M.E.; Nygaard, T.K.; Ackermann, L.; Watkins, R.L.; Zurek, O.W.; Pallister, K.B.; Griffith, S.; Kiedrowski, M.R.; Flack, C.E.; Kavanaugh, J.S.; et al. Staphylococcus aureus Nuclease Is an SaeRS-Dependent Virulence Factor. Infect. Immun. 2013, 81, 1316–1324. [Google Scholar] [CrossRef]
- Delmain, E.A.; Moormeier, D.E.; Endres, J.L.; Hodges, R.E.; Sadykov, M.R.; Horswill, A.R.; Bayles, K.W. Stochastic Expression of Sae-Dependent Virulence Genes during Staphylococcus aureus Biofilm Development Is Dependent on SaeS. mBio 2020, 11. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Periasamy, S.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. Phenol-soluble modulins in staphylococci. Commun. Integr. Biol. 2012, 5, 275–277. [Google Scholar] [CrossRef]
- Wang, H.; Claveau, D.; Vaillancourt, J.P.; Roemer, T.; Meredith, T.C. High-frequency transposition for determining antibacterial mode of action. Nat. Chem. Biol. 2011, 7, 720–729. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. agr-Mediated Dispersal of Staphylococcus aureus Biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef]
- Mootz, J.M.; Malone, C.L.; Shaw, L.N.; Horswill, A.R. Staphopains Modulate Staphylococcus aureus Biofilm Integrity. Infect. Immun. 2013, 81, 3227–3238. [Google Scholar] [CrossRef]
- Abraham, N.M.; Jefferson, K.K. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 2012, 158, 1504–1512. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Kavanaugh, J.S.; Malone, C.L.; Mootz, J.M.; Voyich, J.M.; Smeltzer, M.S.; Bayles, K.W.; Horswill, A.R. Nuclease Modulates Biofilm Formation in Community-Associated Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2011, 6, e26714. [Google Scholar] [CrossRef]
- Beenken, K.E.; Spencer, H.; Griffin, L.M.; Smeltzer, M.S. Impact of Extracellular Nuclease Production on the Biofilm Phenotype of Staphylococcus aureus under in Vitro and in Vivo Conditions. Infect. Immun. 2012, 80, 1634–1638. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Crosby, H.A.; Hernandez, F.J.; Malone, C.L.; Ii, J.O.M.; Horswill, A.R. Staphylococcus aureus Nuc2 Is a Functional, Surface-Attached Extracellular Nuclease. PLoS ONE 2014, 9, e95574. [Google Scholar] [CrossRef]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial Interference Caused by Autoinducing Peptide Variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef]
- Lina, G.; Jarraud, S.; Ji, G.; Greenland, T.; Pedraza, A.; Etienne, J.; Novick, R.P.; Vandenesch, F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 1998, 28, 655–662. [Google Scholar] [CrossRef]
- Cisar, E.A.G.; Geisinger, E.; Muir, T.W.; Novick, R.P. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol. Microbiol. 2009, 74, 44–57. [Google Scholar] [CrossRef]
- Koenig, R.L.; Ray, J.L.; Maleki, S.J.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus AgrA Binding to the RNAIII-agr Regulatory Region. J. Bacteriol. 2004, 186, 7549–7555. [Google Scholar] [CrossRef]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4, 4. [Google Scholar] [CrossRef]
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus Pathogenicity and Polymicrobial Interactions: Lessons beyond Koch’s Postulates. J. Fungi 2019, 5, 81. [Google Scholar] [CrossRef]
- Nadell, C.D.; Xavier, J.B.; Foster, K.R. The sociobiology of biofilms. FEMS Microbiol. Rev. 2008, 33, 206–224. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Federle, M.J.; Bassler, B.L. Interspecies communication in bacteria. J. Clin. Investig. 2003, 112, 1291–1299. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Dunny, G.M.; Leonard, B.A.B. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 1997, 51, 527–564. [Google Scholar] [CrossRef]
- Mashruwala, A.A.; van de Guchte, A.; Boyd, J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife 2017, 6, e23845. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Methicillin Resistance in Staphylococcus aureus: Mechanisms and Modulation. Sci. Prog. 2002, 85, 57–72. [Google Scholar] [CrossRef]
- Chambers, H.F.; de Leo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Genet. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Ito, T.; Hiramatsu, K. MRSA (methicillin-resistant Staphylococcus aureus). Nihon Rinsho 2003, 61 (Suppl. 3), 164–170. [Google Scholar]
- Matsuhashi, M.; Song, M.D.; Ishino, F.; Wachi, M.; Doi, M.; Inoue, M.; Ubukata, K.; Yamashita, N.; Konno, M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 1986, 167, 975–980. [Google Scholar] [CrossRef]
- Song, M.D.; Wachi, M.; Doi, M.; Ishino, F.; Matsuhashi, M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987, 221, 167–171. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oguri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef]
- Chang, S.; Sievert, D.M.; Hageman, J.C.; Boulton, M.L.; Tenover, F.C.; Downes, F.P.; Shah, S.; Rudrik, J.T.; Pupp, G.R.; Brown, W.J.; et al. Infection with Vancomycin-Resistant Staphylococcus aureus Containing the vanA Resistance Gene. New Engl. J. Med. 2003, 348, 1342–1347. [Google Scholar] [CrossRef]
- Tenover, F.C.; Weigel, L.M.; Appelbaum, P.C.; McDougal, L.K.; Chaitram, J.; McAllister, S.; Clark, N.; Killgore, G.; O’Hara, C.M.; Jevitt, L.; et al. Vancomycin-Resistant Staphylococcus aureus Isolate from a Patient in Pennsylvania. Antimicrob. Agents Chemother. 2004, 48, 275–280. [Google Scholar] [CrossRef]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Futur. Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef]
- Sieradzki, K.; Pinho, M.G.; Tomasz, A. Inactivated pbp4 in Highly Glycopeptide-resistant Laboratory Mutants of Staphylococcus aureus. J. Biol. Chem. 1999, 274, 18942–18946. [Google Scholar] [CrossRef]
- Weigel, L.M.; Clewell, D.B.; Gill, S.R.; Clark, N.C.; McDougal, L.K.; Flannagan, S.E.; Kolonay, J.F.; Shetty, J.; Killgore, G.E.; Tenover, F.C. Genetic Analysis of a High-Level Vancomycin-Resistant Isolate of Staphylococcus aureus. Science 2003, 302, 1569–1571. [Google Scholar] [CrossRef]
- Adam, B.; Baillie, G.S.; Douglas, L.J. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 2002, 51, 344–349. [Google Scholar] [CrossRef]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Andes, D.; van Dijck, P.; Jabra-Rizk, M.A. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. mBio 2016, 7, e01365-16. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; van Dijck, P.; Jabra-Rizk, M.A. Modulation of Staphylococcus aureus Response to Antimicrobials by the Candida albicans Quorum Sensing Molecule Farnesol. Antimicrob. Agents Chemother. 2017, 61, e01573-17. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8, e00873-17. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; Ruoff, K.L.; O’Toole, G.A. Pseudomonas aeruginosa Increases the Sensitivity of Biofilm-Grown Staphylococcus aureus to Membrane-Targeting Antiseptics and Antibiotics. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Trizna, E.Y.; Yarullina, M.N.; Baidamshina, D.R.; Mironova, A.V.; Akhatova, F.S.; Rozhina, E.V.; Fakhrullin, R.F.; Khabibrakhmanova, A.M.; Kurbangalieva, A.R.; Bogachev, M.I.; et al. Bidirectional alterations in antibiotics susceptibility in Staphylococcus aureus—Pseudomonas aeruginosa dual-species biofilm. Sci. Rep. 2020, 10, 14849. [Google Scholar] [CrossRef] [PubMed]
- Cag, Y.; Caskurlu, H.; Fan, Y.; Cao, B.; Vahaboglu, H. Resistance mechanisms. Ann. Transl. Med. 2016, 4, 326. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.-G.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef]
- Stefani, S.; Campanile, F.; Santagati, M.; Mezzatesta, M.L.; Cafiso, V.; Pacini, G. Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: A review of the available evidence. Int. J. Antimicrob. Agents 2015, 46, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats; National Library of Medicine: Bethesda, MD, USA, 2015; Volume 40, pp. 277–283. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Heal. 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Miliya, T.; Chansamouth, V.; Aung, M.T.; Karkey, A.; Teparrukkul, P.; Batra, R.; Lan, N.P.H.; Stelling, J.; Turner, P.; et al. Automating the generation of antimicrobial resistance surveillance reports: A proof-of-concept study in seven hospitals in seven countries (Preprint). J. Med. Internet Res. 2020, 22, e19762. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Yu, X.; Zhou, H.; Li, B.; Chen, G.; Ye, Z.; Wang, Y.; Cui, X.; Zheng, Y.; et al. Impact of antimicrobial stewardship managed by clinical pharmacists on antibiotic use and drug resistance in a Chinese hospital, 2010–2016: A retrospective observational study. BMJ Open 2019, 9, e026072. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, H.; Wang, Z. Distribution of acquired antibiotic resistance genes among Enterococcus spp. isolated from a hospital in Baotou, China. BMC Res. Notes 2019, 12, 27. [Google Scholar] [CrossRef]
- Deguchi, H.; Kitazawa, K.; Kayukawa, K.; Kondoh, E.; Fukumoto, A.; Yamasaki, T.; Kinoshita, S.; Sotozono, C. The trend of resistance to antibiotics for ocular infection of Staphylococcus aureus, coagulase-negative staphylococci, and Corynebacterium compared with 10-years previous: A retrospective observational study. PLoS ONE 2018, 13, e0203705. [Google Scholar] [CrossRef]
- Ramsamy, Y.; Essack, S.Y.; Sartorius, B.; Patel, M.; Mlisana, K. Antibiotic resistance trends of ESKAPE pathogens in Kwazulu-Natal, South Africa: A five-year retrospective analysis. Afr. J. Lab. Med. 2018, 7, 8. [Google Scholar] [CrossRef]
- Refaat, M.; Zakka, P.; Khoury, M.; Chami, H.; Mansour, S.; Harbieh, B.; Abi-Saleh, B.; Bizri, A.R. Cardiac implantable electronic device infections. Medicine 2019, 98, e14906. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K.; National Healthcare Safety Network Team and Participating National Healthcare Safety Network Facilities. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Annual Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control. Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Cassini, A.; Plachouras, D.; Eckmanns, T.; Abu-Sin, M.; Blank, H.-P.; Ducomble, T.; Haller, S.; Harder, T.; Klingeberg, A.; Sixtensson, M.; et al. Burden of Six Healthcare-Associated Infections on European Population Health: Estimating Incidence-Based Disability-Adjusted Life Years through a Population Prevalence-Based Modelling Study. PLoS Med. 2016, 13, e1002150. [Google Scholar] [CrossRef]
- Gatermann, S.G.; Koschinski, T.; Friedrich, S. Distribution and expression of macrolide resistance genes in coagulase-negative staphylococci. Clin. Microbiol. Infect. 2007, 13, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Butin, M.; Martins-Simões, P.; Pichon, B.; Leyssene, D.; Bordes-Couecou, S.; Meugnier, H.; Rouard, C.; Lemaitre, N.; Schramm, F.; Kearns, A.; et al. Emergence and dissemination of a linezolid-resistant Staphylococcus capitisclone in Europe. J. Antimicrob. Chemother. 2016, 72, 1014–1020. [Google Scholar] [CrossRef]
- Dimitriou, J.; Levivier, M.; Gugliotta, M. Comparison of Complications in Patients Receiving Different Types of Intracranial Pressure Monitoring: A Retrospective Study in a Single Center in Switzerland. World Neurosurg. 2016, 89, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Chaves, F.; García-Álvarez, M.; Sanz, F.; Alba, C.; Otero, J.R. Nosocomial Spread of a Staphylococcus hominis subsp. novobiosepticus Strain Causing Sepsis in a Neonatal Intensive Care Unit. J. Clin. Microbiol. 2005, 43, 4877–4879. [Google Scholar] [CrossRef]
- Laurent, F.; Butin, M. Staphylococcus capitis and NRCS-A clone: The story of an unrecognized pathogen in neonatal intensive care units. Clin. Microbiol. Infect. 2019, 25, 1081–1085. [Google Scholar] [CrossRef]
- Decousser, J.W.; Desroches, M.; Bourgeois-Nicolaos, N.; Potier, J.; Jehl, F.; Lina, G.; Cattoir, V.; Vandenesch, F.; Doucet-Populaire, F. Susceptibility trends including emergence of linezolid resistance among coagulase-negative staphylococci and meticillin-resistant Staphylococcus aureus from invasive infections. Int. J. Antimicrob. Agents 2015, 46, 622–630. [Google Scholar] [CrossRef]
- May, L.; Klein, E.Y.; Rothman, R.E.; Laxminarayan, R. Trends in Antibiotic Resistance in Coagulase-Negative Staphylococci in the United States, 1999 to 2012. Antimicrob. Agents Chemother. 2013, 58, 1404–1409. [Google Scholar] [CrossRef]
- Becker, K.; Both, A.; Weißelberg, S.; Heilmann, C.; Rohde, H. Emergence of coagulase-negative staphylococci. Expert Rev. Anti-Infect. Ther. 2020, 18, 349–366. [Google Scholar] [CrossRef]
- Soumya, K.R.; Philip, S.; Sugathan, S.; Mathew, J.; Krishnankutty, R.E. Virulence factors associated with Coagulase Negative Staphylococci isolated from human infections. 3 Biotech. 2017, 7, 140. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-Negative Staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- Seng, R.; Kitti, T.; Thummeepak, R.; Kongthai, P.; Leungtongkam, U.; Wannalerdsakun, S.; Sitthisak, S. Biofilm formation of methicillin-resistant coagulase negative staphylococci (MR-CoNS) isolated from community and hospital environments. PLoS ONE 2017, 12, e0184172. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutiérrez, J.M.; Alonso, C.A.; Pérez-Moreno, M.O.; Aspiroz, C.; Bellés, A.; Schwarz, S.; et al. Mechanisms of Linezolid Resistance among Clinical Staphylococcus spp. in Spain: Spread of Methicillin- and Linezolid-Resistant S. epidermidis ST2. Microb. Drug Resist. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.H.; Monk, I.R.; da Silva, A.G.; Seemann, T.; Chua, K.Y.L.; Kearns, A.; Hill, R.; Woodford, N.; Bartels, M.D.; Strommenger, B.; et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 2018, 3, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Enhanced production of exopolysaccharide matrix and biofilm by a menadione-auxotrophic Staphylococcus aureus small-colony variant. J. Med. Microbiol. 2010, 59, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Siala, W.; Mingeot-Leclercq, M.-P.; Tulkens, P.M.; Hallin, M.; Denis, O.; van Bambeke, F. Comparison of the Antibiotic Activities of Daptomycin, Vancomycin, and the Investigational Fluoroquinolone Delafloxacin against Biofilms from Staphylococcus aureus Clinical Isolates. Antimicrob. Agents Chemother. 2014, 58, 6385–6397. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.E.; Christensen, H.; Aarestrup, F.M. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 2006, 57, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Martineau, F.; Picard, F.J.; Lansac, N.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000, 44, 231–238. [Google Scholar] [CrossRef]
- Fluit, A.C.; Visser, M.R.; Schmitz, F.-J. Molecular Detection of Antimicrobial Resistance. Clin. Microbiol. Rev. 2001, 14, 836–871. [Google Scholar] [CrossRef]
- Vega, N.M.; Gore, J. Collective antibiotic resistance: Mechanisms and implications. Curr. Opin. Microbiol. 2014, 21, 28–34. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Genet. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Pabst, B.; Pitts, B.; Lauchnor, E.; Stewart, P.S. Gel-Entrapped Staphylococcus aureus Bacteria as Models of Biofilm Infection Exhibit Growth in Dense Aggregates, Oxygen Limitation, Antibiotic Tolerance, and Heterogeneous Gene Expression. Antimicrob. Agents Chemother. 2016, 60, 6294–6301. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.A.; Pitts, B.; Beyenal, H.; Veluchamy, R.A.; Lewandowski, Z.; Davison, W.M.; Buckingham-Meyer, K.; Stewart, P.S. Spatial Patterns of DNA Replication, Protein Synthesis, and Oxygen Concentration within Bacterial Biofilms Reveal Diverse Physiological States. J. Bacteriol. 2007, 189, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Genet. 2016, 14, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1, 1–7. [Google Scholar] [CrossRef]
- Waters, E.M.; Rowe, S.E.; O’Gara, J.P.; Conlon, B.P. Convergence of Staphylococcus aureus Persister and Biofilm Research: Can Biofilms Be Defined as Communities of Adherent Persister Cells? PLoS Pathog. 2016, 12, e1006012. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Lewis, K. Persister Cells in Biofilm Associated Infections. Adv. Exp. Med. Biol. 2015, 831, 1–9. [Google Scholar] [CrossRef]
- Wang, Z.; de la Fuente-Núñez, C.; Shen, Y.; Haapasalo, M.; Hancock, R.E.W. Treatment of Oral Multispecies Biofilms by an Anti-Biofilm Peptide. PLoS ONE 2015, 10, e0132512. [Google Scholar] [CrossRef]
- Sahukhal, G.S.; Pandey, S.; Elasri, M.O. msaABCR operon is involved in persister cell formation in Staphylococcus aureus. BMC Microbiol. 2017, 17, 218. [Google Scholar] [CrossRef]
- Kelly, B.; Vespermann, A.; Bolton, D. Horizontal gene transfer of virulence determinants in selected bacterial foodborne pathogens. Food Chem. Toxicol. 2009, 47, 969–977. [Google Scholar] [CrossRef]
- Lindsay, J.A. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 2014, 304, 103–109. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Loeffler, A.; Witney, A.A.; Gould, K.A.; Lloyd, D.H.; Lindsay, J.A. Extensive Horizontal Gene Transfer during Staphylococcus aureus Co-colonization in Vivo. Genome Biol. Evol. 2014, 6, 2697–2708. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Clark, N.C.; McDougal, L.K.; Hageman, J.; McDonald, L.C.; Patel, J.B. Vancomycin-Resistant Staphylococcus aureus Isolates Associated with Inc18-Like vanA Plasmids in Michigan. Antimicrob. Agents Chemother. 2007, 52, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Appropriate Targets for Antibacterial Drugs. Cold Spring Harb. Perspect. Med. 2016, 6, a030239. [Google Scholar] [CrossRef]
- Monserrat-Martinez, A.; Gambin, Y.; Sierecki, E. Thinking Outside the Bug: Molecular Targets and Strategies to Overcome Antibiotic Resistance. Int. J. Mol. Sci. 2019, 20, 1255. [Google Scholar] [CrossRef]
- Jiang, Y.; Geng, M.; Bai, L. Targeting Biofilms Therapy: Current Research Strategies and Development Hurdles. Microorganisms 2020, 8, 1222. [Google Scholar] [CrossRef]
- Oniga, S.D.; Araniciu, C.; Palage, M.D.; Popa, M.; Chifiriuc, M.C.; Marc, G.; Pîrnău, A.; Stoica, C.I.; Lagoudis, I.; Dragoumis, T.; et al. New 2-Phenylthiazoles as Potential Sortase A Inhibitors: Synthesis, Biological Evaluation and Molecular Docking. Molecules 2017, 22, 1827. [Google Scholar] [CrossRef]
- Nițulescu, G.; Zanfirescu, A.; Octavian-Tudorel, O.; Nicorescu, I.M.; Nițulescu, G.M.; Margină, D. Structural Analysis of Sortase A Inhibitors. Molecules 2016, 21, 1591. [Google Scholar] [CrossRef]
- Cascioferro, S.; Cusimano, M.G.; Schillaci, D. Antiadhesion agents against Gram-positive pathogens. Futur. Microbiol. 2014, 9, 1209–1220. [Google Scholar] [CrossRef]
- Cascioferro, S.; Totsika, M.; Schillaci, D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014, 77, 105–112. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, Y.; Yestrepsky, B.D.; Sorenson, R.J.; Chen, M.; Larsen, S.D.; Sun, H. Novel Inhibitors of Staphylococcus aureus Virulence Gene Expression and Biofilm Formation. PLoS ONE 2012, 7, e47255. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, Y.; Okuda, K.-I.; Yamada, S.; Nagakura, M.; Sugimoto, S.; Nagano, T.; Okabe, T.; Kojima, H.; Iwamoto, T.; Kuwano, K.; et al. Norgestimate inhibits staphylococcal biofilm formation and resensitizes methicillin-resistant Staphylococcus aureus to β-lactam antibiotics. NPJ Biofilms Microbiomes 2017, 3, 1–9. [Google Scholar] [CrossRef]
- Zoll, S.; Pätzold, B.; Schlag, M.; Götz, F.; Kalbacher, H.; Stehle, T. Structural Basis of Cell Wall Cleavage by a Staphylococcal Autolysin. PLoS Pathog. 2010, 6, e1000807. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Wermser, C.; Lopez, D. Identification of Staphylococcus aureus genes involved in the formation of structured macrocolonies. Microbiol. 2018, 164, 801–815. [Google Scholar] [CrossRef]
- Sugimoto, S.; Sato, F.; Miyakawa, R.; Chiba, A.; Onodera, S.; Hori, S.; Mizunoe, Y. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci. Rep. 2018, 8, 2254. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Nguyen, T.H.; Otto, M. The staphylococcal exopolysaccharide PIA—Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 2020, 18, 3324–3334. [Google Scholar] [CrossRef]
- Rumbaugh, K.P.; Sauer, K. Biofilm dispersion. Nat. Rev. Genet. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Genet. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, Ecology, and Application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef]
- Dykes, G.A. Bacteriocins: Ecological and evolutionary significance. Trends Ecol. Evol. 1995, 10, 186–189. [Google Scholar] [CrossRef]
- Tagg, J.R.; Dajani, A.S.; Wannamaker, L.W. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 1976, 40, 722–756. [Google Scholar] [CrossRef] [PubMed]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.; Sood, S.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Takala, T.M.; Reunanen, J.; Saris, O.; Saris, P.E.J. Attachment of Escherichia coli to Listeria monocytogenes for Pediocin-Mediated Killing. Curr. Microbiol. 2014, 70, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Kjos, M.; Nes, I.; Diep, D.; Lotfipour, F. Natural antimicrobial peptides from bacteria: Characteristics and potential applications to fight against antibiotic resistance. J. Appl. Microbiol. 2012, 113, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Appleyard, A.N.; Choi, S.; Read, D.M.; Lightfoot, A.; Boakes, S.; Hoffmann, A.; Chopra, I.; Bierbaum, G.; Rudd, B.A.; Dawson, M.J.; et al. Dissecting Structural and Functional Diversity of the Lantibiotic Mersacidin. Chem. Biol. 2009, 16, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Connor, P.M.O.; Zabetakis, I.; Hill, C.; Ross, R.P. The generation of nisin variants with enhanced activity against specific Gram-positive pathogens. Mol. Microbiol. 2008, 69, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Begley, M.; O’Connor, P.M.; Daly, K.M.; Hugenholtz, F.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineered Nisin A Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS ONE 2012, 7, e46884. [Google Scholar] [CrossRef]
- Rink, R.; Wierenga, J.; Kuipers, A.; Kluskens, L.; Driessen, A.J.M.; Kuipers, O.P.; Moll, G.N. Dissection and Modulation of the Four Distinct Activities of Nisin by Mutagenesis of Rings A and B and by C-Terminal Truncation. Appl. Environ. Microbiol. 2007, 73, 5809–5816. [Google Scholar] [CrossRef]
- Rouse, S.; Field, D.; Daly, K.M.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineered nisin derivatives with enhanced activity in complex matrices. Microb. Biotechnol. 2012, 5, 501–508. [Google Scholar] [CrossRef]
- Rollema, H.S.; Kuipers, O.P.; Both, P.; de Vos, W.M.; Siezen, R.J. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 1995, 61, 2873–2878. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhang, Z.-Z.; Chen, X.-Z.; Yang, W.; Huan, L.-D. Site-directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl. Microbiol. Biotechnol. 2004, 64, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineering Lantibiotics for Therapeutic Success. Front. Microbiol. 2015, 6, 1363. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Cotter, P.D.; Ross, R.P.; Hill, C. Bioengineering of the model lantibiotic nisin. Bioengineered 2015, 6, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, K.V.; Kranjec, C.; Thorstensen, T.; Carlsen, H.; Diep, D.B. Successful Development of Bacteriocins into Therapeutic Formulation for Treatment of MRSA Skin Infection in a Murine Model. Antimicrob. Agents Chemother. 2020, 64, 64. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Ovchinnikov, K.V.; Grønseth, T.; Ebineshan, K.; Srikantam, A.; Diep, D.B. A bacteriocin-based antimicrobial formulation to effectively disrupt the cell viability of methicillin-resistant Staphylococcus aureus (MRSA) biofilms. NPJ Biofilms Microbiomes 2020, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Bierbaum, G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- Chatterjee, C.; Paul, M.; Xie, L.; van der Donk, W.A. Biosynthesis and Mode of Action of Lantibiotics. Chem. Rev. 2005, 105, 633–684. [Google Scholar] [CrossRef]
- McAuliffe, O.; Ross, R.P.; Hill, C. Lantibiotics: Structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 2001, 25, 285–308. [Google Scholar] [CrossRef]
- Breukink, E.; de Kruijff, B. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta Biomembr. 1999, 1462, 223–234. [Google Scholar] [CrossRef]
- Okuda, K.-I.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of Bacteriocins on Methicillin-Resistant Staphylococcus aureus Biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef] [PubMed]
- Corbin, A.; Pitts, B.; Parker, A.; Stewart, P.S. Antimicrobial Penetration and Efficacy in an in Vitro Oral Biofilm Model. Antimicrob. Agents Chemother. 2011, 55, 3338–3344. [Google Scholar] [CrossRef] [PubMed]
- Davison, W.M.; Pitts, B.; Stewart, P.S. Spatial and Temporal Patterns of Biocide Action against Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother. 2010, 54, 2920–2927. [Google Scholar] [CrossRef] [PubMed]
- Bastos, M.; Ceotto, H.; Coelho, M.L.V.; Nascimento, J. Staphylococcal Antimicrobial Peptides: Relevant Properties and Potential Biotechnological Applications. Curr. Pharm. Biotechnol. 2009, 10, 38–61. [Google Scholar] [CrossRef]
- Newstead, L.L.; Varjonen, K.; Nuttall, T.; Paterson, G.K. Staphylococcal-Produced Bacteriocins and Antimicrobial Peptides: Their Potential as Alternative Treatments for Staphylococcus aureus Infections. Antibiotics 2020, 9, 40. [Google Scholar] [CrossRef]
- Fontana, M.B.C.; Dd Bastos, M.D.C.F.; Brandelli, A. Bacteriocins Pep5 and Epidermin Inhibit Staphylococcus epidermidis Adhesion to Catheters. Curr. Microbiol. 2006, 52, 350–353. [Google Scholar] [CrossRef]
- Schindler, C.A.; Schuhardt, V.T. Lysostaphin: A New Bacteriolytic Agent for the Staphylococcus. Proc. Natl. Acad. Sci. USA 1964, 51, 414–421. [Google Scholar] [CrossRef]
- Kokai-Kun, J.F.; Chanturiya, T.; Mond, J.J. Lysostaphin eradicates established Staphylococcus aureus biofilms in jugular vein catheterized mice. J. Antimicrob. Chemother. 2009, 64, 94–100. [Google Scholar] [CrossRef]
- Wu, J.A.; Kusuma, C.; Mond, J.J.; Kokai-Kun, J.F. Lysostaphin Disrupts Staphylococcus aureus and Staphylococcus epidermidis Biofilms on Artificial Surfaces. Antimicrob. Agents Chemother. 2003, 47, 3407–3414. [Google Scholar] [CrossRef]
- Johnson, C.T.; Wroe, J.A.; Agarwal, R.; Martin, K.E.; Guldberg, R.E.; Donlan, R.M.; Westblade, L.F.; García, A.J. Hydrogel delivery of lysostaphin eliminates orthopedic implant infection by Staphylococcus aureus and supports fracture healing. Proc. Natl. Acad. Sci. USA 2018, 115, E4960–E4969. [Google Scholar] [CrossRef] [PubMed]
- Windolf, C.D.; Lögters, T.; Scholz, M.; Windolf, J.; Flohé, S. Lysostaphin-Coated Titan-Implants Preventing Localized Osteitis by Staphylococcus aureus in a Mouse Model. PLoS ONE 2014, 9, e115940. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.; Jung, G.; Horner, T.; Zahner, H.; Schnell, N.; Entian, K.-D.; Gotz, F. Gallidermin: A new lanthionine-containing polypeptide antibiotic. J. Biol. Inorg. Chem. 1988, 177, 53–59. [Google Scholar] [CrossRef]
- Bengtsson, T.; Lönn, J.; Khalaf, H.; Palm, E. The lantibiotic gallidermin acts bactericidal against Staphylococcus epidermidis and Staphylococcus aureus and antagonizes the bacteria-induced proinflammatory responses in dermal fibroblasts. Microbiology 2018, 7, e00606. [Google Scholar] [CrossRef] [PubMed]
- Saising, J.; Dube, L.; Ziebandt, A.-K.; Voravuthikunchai, S.P.; Nega, M.; Götz, F. Activity of Gallidermin on Staphylococcus aureus and Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother. 2012, 56, 5804–5810. [Google Scholar] [CrossRef]
- Field, D.; Connor, R.O.; Cotter, P.D.; Ross, R.P.; Hill, C. In Vitro Activities of Nisin and Nisin Derivatives Alone and In Combination with Antibiotics against Staphylococcus Biofilms. Front. Microbiol. 2016, 7, 508. [Google Scholar] [CrossRef]
- Tong, Z.; Ni, L.; Ling, J. Antibacterial peptide nisin: A potential role in the inhibition of oral pathogenic bacteria. Peptides 2014, 60, 32–40. [Google Scholar] [CrossRef]
- Dosler, S.; Mataraci, E. In vitro pharmacokinetics of antimicrobial cationic peptides alone and in combination with antibiotics against methicillin resistant Staphylococcus aureus biofilms. Peptides 2013, 49, 53–58. [Google Scholar] [CrossRef]
- Mataraci, E.; Dosler, S. In VitroActivities of Antibiotics and Antimicrobial Cationic Peptides Alone and in Combination against Methicillin-Resistant Staphylococcus aureus Biofilms. Antimicrob. Agents Chemother. 2012, 56, 6366–6371. [Google Scholar] [CrossRef]
- Ceotto-Vigoder, H.; Marques, S.L.S.; Santos, I.N.S.; Alves, M.D.B.; Barrias, E.S.; Potter, A.; Alviano, D.S.; Bastos, M.C.F. Nisin and lysostaphin activity against preformed biofilm of Staphylococcus aureus involved in bovine mastitis. J. Appl. Microbiol. 2016, 121, 101–114. [Google Scholar] [CrossRef]
- Pimentel-Filho, N.D.J.; Martins, M.C.D.F.; Nogueira, G.B.; Mantovani, H.C.; Vanetti, M.C.D. Bovicin HC5 and nisin reduce Staphylococcus aureus adhesion to polystyrene and change the hydrophobicity profile and Gibbs free energy of adhesion. Int. J. Food Microbiol. 2014, 190, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Powell, A.M.; Becker, S.C.; Camp, M.J.; Donovan, D.M. Chimeric Phage Lysins Act Synergistically with Lysostaphin To Kill Mastitis-Causing Staphylococcus aureus in Murine Mammary Glands. Appl. Environ. Microbiol. 2012, 78, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, L.; Donovan, D.M.; García, P. Enhanced Staphylolytic Activity of the Staphylococcus aureus Bacteriophage vB_SauS-phiIPLA88 HydH5 Virion-Associated Peptidoglycan Hydrolase: Fusions, Deletions, and Synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Foster-Frey, J.; Donovan, D.M. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008, 287, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, K.V.; Chi, H.; Mehmeti, I.; Holo, H.; Nes, I.F.; Diep, D.B. Novel Group of Leaderless Multipeptide Bacteriocins from Gram-Positive Bacteria. Appl. Environ. Microbiol. 2016, 82, 5216–5224. [Google Scholar] [CrossRef]
- Ciufolini, M.A.; Lefranc, D. Micrococcin P1: Structure, biology and synthesis. Nat. Prod. Rep. 2010, 27, 330–342. [Google Scholar] [CrossRef]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; García, P.; Rodriguez, A. The Perfect Bacteriophage for Therapeutic Applications—A Quick Guide. Antibiotics 2019, 8, 126. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Gutiérrez, D.; Campelo, A.B.; Rodríguez, A.; García, P. Analysis of Different Parameters Affecting Diffusion, Propagation and Survival of Staphylophages in Bacterial Biofilms. Front. Microbiol. 2018, 9, 2348. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Campelo, A.B.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. The Behavior of Staphylococcus aureus Dual-Species Biofilms Treated with Bacteriophage phiIPLA-RODI Depends on the Accompanying Microorganism. Appl. Environ. Microbiol. 2016, 83, 83. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two Phages, phiIPLA-RODI and phiIPLA-C1C, Lyse Mono- and Dual-Species Staphylococcal Biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.; Son, B.; Ryu, S. Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol. 2019, 84, 103245. [Google Scholar] [CrossRef] [PubMed]
- Son, J.S.; Kim, E.B.; Lee, S.J.; Jun, S.Y.; Yoon, S.J.; Kang, S.H.; Choi, Y.J. Characterization of Staphylococcus aureus derived from bovine mastitis and isolation of two lytic bacteriophages. J. Gen. Appl. Microbiol. 2010, 56, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.R.; Gaudion, A.; Bean, J.E.; Esteban, P.P.; Arnot, T.C.; Harper, D.R.; Kot, W.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. Combined Use of Bacteriophage K and a Novel Bacteriophage To Reduce Staphylococcus aureus Biofilm Formation. Appl. Environ. Microbiol. 2014, 80, 6694–6703. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; França, A.; Brandão, A.; Sillankorva, S.; Cerca, N.; Azeredo, J. Assessment of Sep1 virus interaction with stationary cultures by transcriptional and flow cytometry studies. FEMS Microbiol. Ecol. 2018, 94, fiy143. [Google Scholar] [CrossRef]
- Kelly, D.; McAuliffe, O.; Ross, R.; Coffey, A. Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage K and modified derivatives. Lett. Appl. Microbiol. 2012, 54, 286–291. [Google Scholar] [CrossRef]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci. Rep. 2017, 7, 40965. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G.M. Formation of biofilms under phage predation: Considerations concerning a biofilm increase. Biofouling 2013, 29, 457–468. [Google Scholar] [CrossRef]
- Olsen, N.M.C.; Thiran, E.; Hasler, T.; Vanzieleghem, T.; Belibasakis, G.N.; Mahillon, J.; Loessner, M.J.; Schmelcher, M. Synergistic Removal of Static and Dynamic Staphylococcus aureus Biofilms by Combined Treatment with a Bacteriophage Endolysin and a Polysaccharide Depolymerase. Viruses 2018, 10, 438. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective Removal of Staphylococcal Biofilms by the Endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Oliveira, H.A.M.; Melo, L.D.R.; Santos, S.R.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L. Molecular Aspects and Comparative Genomics of Bacteriophage Endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Fernández, L.; Rodriguez, A.; García, P. Are Phage Lytic Proteins the Secret Weapon to Kill Staphylococcus aureus? mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Y.; Huang, Y.; Li, Y.; Wei, H. Degradation of methicillin-resistant Staphylococcus aureus biofilms using a chimeric lysin. Biofouling 2014, 30, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Sass, P.; Bierbaum, G. Lytic Activity of Recombinant Bacteriophage φ11 and φ12 Endolysins on Whole Cells and Biofilms of Staphylococcus aureus. Appl. Environ. Microbiol. 2006, 73, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Khan, B.K.; Raz, A.; Rotolo, J.A.; Wittekind, M. Bacteriophage Lysin CF-301, a Potent Antistaphylococcal Biofilm Agent. Antimicrob. Agents Chemother. 2017, 61, e02666-16. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Oh, M.-D.; Choi, Y.-J.; Lee, W.J.; Kong, J.-C.; Seol, J.G.; Kang, S.H. Antibacterial properties of a pre-formulated recombinant phage endolysin, SAL-1. Int. J. Antimicrob. Agents 2013, 41, 156–161. [Google Scholar] [CrossRef]
- Fenton, M.; Keary, R.; McAuliffe, O.; Ross, R.P.; O’Mahony, J.; Coffey, A. Bacteriophage-Derived Peptidase CHAP(K) Eliminates and Prevents Staphylococcal Biofilms. Int. J. Microbiol. 2013, 2013, 1–8. [Google Scholar] [CrossRef]
- Linden, S.B.; Zhang, H.; Heselpoth, R.D.; Shen, Y.; Schmelcher, M.; Eichenseher, F.; Nelson, D. Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2014, 99, 741–752. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, H.; Wang, J.; Yu, J.; Wei, H. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 40182. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Garrido, V.; Fernández, L.; Portilla, S.; Rodríguez, A.; Grilló, M.J.; García, P. Phage Lytic Protein LysRODI Prevents Staphylococcal Mastitis in Mice. Front. Microbiol. 2020, 11, 7. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Dickey, J.; Perrot, V. Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS ONE 2019, 14, e0209390. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.-B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, D.; Taha, M.; Yi, Q.; Ramirez-Arcos, S.; Diallo, J.-S.; Carli, A.; Abdelbary, H. Does Treatment Order Matter? Investigating the Ability of Bacteriophage to Augment Antibiotic Activity against Staphylococcus aureus Biofilms. Front. Microbiol. 2018, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Agún, S.; Fernández, L.; González-Menéndez, E.; Martínez, B.; Rodriguez, A.; García, P. Study of the Interactions between Bacteriophage phiIPLA-RODI and Four Chemical Disinfectants for the Elimination of Staphylococcus aureus Contamination. Viruses 2018, 10, 103. [Google Scholar] [CrossRef]
- Nair, S.; Poonacha, N.; Desai, S.; Hiremath, D.; Tuppad, D.; Mohan, T.; Chikkamadaiah, R.; Durgaiah, M.; Kumar, S.; Channabasappa, S.; et al. Restoration of sensitivity of a diverse set of drug-resistant Staphylococcus clinical strains by bactericidal protein P128. J. Med. Microbiol. 2018, 67, 296–307. [Google Scholar] [CrossRef]
- Colque-Navarro, P.; Jacobsson, G.; Andersson, R.; Flock, J.-I.; Möllby, R. Levels of Antibody against 11 Staphylococcus aureus Antigens in a Healthy Population. Clin. Vaccine Immunol. 2010, 17, 1117–1123. [Google Scholar] [CrossRef]
- Dryla, A.; Prustomersky, S.; Gelbmann, D.; Hanner, M.; Bettinger, E.; Kocsis, B.; Kustos, T.; Henics, T.; Meinke, A.; Nagy, E. Comparison of Antibody Repertoires against Staphylococcus aureus in Healthy Individuals and in Acutely Infected Patients. Clin. Diagn. Lab. Immunol. 2005, 12, 387–398. [Google Scholar] [CrossRef]
- Thomer, L.; Emolo, C.; Thammavongsa, V.; Kim, H.K.; McAdow, M.E.; Yu, W.; Kieffer, M.; Schneewind, O.; Missiakas, D. Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J. Exp. Med. 2016, 213, 293–301. [Google Scholar] [CrossRef]
- Kumar, A.; Ray, P.; Kanwar, M.; Sharma, M.; Varma, S. A comparative analysis of antibody repertoire against Staphylococcus aureus antigens in Patients with Deep-Seated versus Superficial staphylococcal Infections. Int. J. Med. Sci. 2005, 2, 129–136. [Google Scholar] [CrossRef]
- Giersing, B.K.; Dastgheyb, S.S.; Modjarrad, K.; Moorthy, V. Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine 2016, 34, 2962–2966. [Google Scholar] [CrossRef] [PubMed]
- Belyi, Y.; Rybolovlev, I.; Polyakov, N.; Chernikova, A.; Tabakova, I.; Gintsburg, A. Staphylococcus Aureus Surface Protein G is An Immunodominant Protein and a Possible Target in An Anti-Biofilm Drug Development. Open Microbiol. J. 2018, 12, 94–106. [Google Scholar] [CrossRef]
- Domanski, P.J.; Patel, P.R.; Bayer, A.S.; Zhang, L.; Hall, A.E.; Syribeys, P.J.; Gorovits, E.L.; Bryant, D.; Vernachio, J.H.; Hutchins, J.T.; et al. Characterization of a Humanized Monoclonal Antibody Recognizing Clumping Factor A Expressed by Staphylococcus aureus. Infect. Immun. 2005, 73, 5229–5232. [Google Scholar] [CrossRef]
- Tkaczyk, C.; Kasturirangan, S.; Minola, A.; Jones-Nelson, O.; Gunter, V.; Shi, Y.Y.; Rosenthal, K.; Aleti, V.; Semenova, E.; Warrener, P.; et al. Multimechanistic Monoclonal Antibodies (MAbs) Targeting Staphylococcus aureus Alpha-Toxin and Clumping Factor A: Activity and Efficacy Comparisons of a MAb Combination and an Engineered Bispecific Antibody Approach. Antimicrob. Agents Chemother. 2017, 61, e00629-17. [Google Scholar] [CrossRef] [PubMed]
- Varshney, A.K.; Kuzmicheva, G.A.; Bowling, R.A.; Sunley, K.M.; Bowling, R.A.; Kwan, T.-Y.; Mays, H.R.; Rambhadran, A.; Zhang, Y.; Martin, R.L.; et al. A natural human monoclonal antibody targeting Staphylococcus Protein A protects against Staphylococcus aureus bacteremia. PLoS ONE 2018, 13, e0190537. [Google Scholar] [CrossRef] [PubMed]
- França, A.; Vilanova, M.; Cerca, N.; Pier, G.B. Monoclonal Antibody Raised against PNAG Has Variable Effects on Static S. epidermidis Biofilm Accumulation in Vitro. Int. J. Biol. Sci. 2013, 9, 518–520. [Google Scholar] [CrossRef] [PubMed]
- Pastrana, F.R.; Neef, J.; Koedijk, D.G.A.M.; de Graaf, D.; Duipmans, J.; Jonkman, M.F.; Engelmann, S.; Van Dijl, J.M.; Buist, G. Human antibody responses against non-covalently cell wall-bound Staphylococcus aureus proteins. Sci. Rep. 2018, 8, 3234. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Simon, J.K. Applications of nanotechnology for immunology. Nat. Rev. Immunol. 2013, 13, 592–605. [Google Scholar] [CrossRef]
- Natan, M.; Banin, E. From Nano to Micro: Using nanotechnology to combat microorganisms and their multidrug resistance. FEMS Microbiol. Rev. 2017, 41, 302–322. [Google Scholar] [CrossRef]
- Banerjee, S.; Ghosh, D.; Vishakha, K.; Das, S.; Mondal, S.; Ganguli, A. Photodynamic antimicrobial chemotherapy (PACT) using riboflavin inhibits the mono and dual species biofilm produced by antibiotic resistant Staphylococcus aureus and Escherichia coli. Photodiagnosis Photodyn. Ther. 2020, 32, 102002. [Google Scholar] [CrossRef]
- Ramasamy, M.; Lee, J. Recent Nanotechnology Approaches for Prevention and Treatment of Biofilm-Associated Infections on Medical Devices. BioMed Res. Int. 2016, 2016, 1851242. [Google Scholar] [CrossRef] [PubMed]
- Malaekeh-Nikouei, B.; Bazzaz, B.S.F.; Mirhadi, E.; Tajani, A.S.; Khameneh, B. The role of nanotechnology in combating biofilm-based antibiotic resistance. J. Drug Deliv. Sci. Technol. 2020, 60, 101880. [Google Scholar] [CrossRef]
- Tran, H.M.; Tran, H.; Booth, M.A.; Fox, K.; Nguyen, T.-H.; Tran, N.; Tran, P.A. Nanomaterials for Treating Bacterial Biofilms on Implantable Medical Devices. Nanomaterials 2020, 10, 2253. [Google Scholar] [CrossRef] [PubMed]
- Reynoso, E.; Ferreyra, D.D.; Durantini, E.N.; Spesia, M.B. Photodynamic inactivation to prevent and disrupt Staphylococcus aureus biofilm under different media conditions. Photodermatol. Photoimmunol. Photomed. 2019, 35, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.M.; Hamblin, M.R.; Yow, C.M.N. A comparative in vitro photoinactivation study of clinical isolates of multidrug-resistant pathogens. J. Infect. Chemother. 2007, 13, 87–91. [Google Scholar] [CrossRef]
- Teixeira, C.G.D.S.; Sanitá, P.V.; Ribeiro, A.P.D.; Dias, L.M.; Jorge, J.H.; Pavarina, A.C. Antimicrobial photodynamic therapy effectiveness against susceptible and methicillin-resistant Staphylococcus aureus biofilms. Photodiagnosis Photodyn. Ther. 2020, 30, 101760. [Google Scholar] [CrossRef]
- Ferrer-Espada, R.; Liu, X.; Goh, X.S.; Dai, T. Antimicrobial Blue Light Inactivation of Polymicrobial Biofilms. Front. Microbiol. 2019, 10, 721. [Google Scholar] [CrossRef]
- Halstead, F.D.; Thwaite, J.E.; Burt, R.; Laws, T.R.; Raguse, M.; Moeller, R.; Webber, M.A.; Oppenheim, B.A. Antibacterial Activity of Blue Light against Nosocomial Wound Pathogens Growing Planktonically and as Mature Biofilms. Appl. Environ. Microbiol. 2016, 82, 4006–4016. [Google Scholar] [CrossRef]
- Yang, H.; Xu, S.; Huang, K.; Xu, X.; Hu, F.; He, C.; Shu, W.; Wang, Z.; Gong, F.; Zhang, C.; et al. Anti-staphylococcus Antibiotics Interfere With the Transcription of Leucocidin ED Gene in Staphylococcus aureus Strain Newman. Front. Microbiol. 2020, 11, 265. [Google Scholar] [CrossRef]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Genet. Genom. 1989, 219, 480–485. [Google Scholar] [CrossRef]
- Cirioni, O.; Mocchegiani, F.; Cacciatore, I.; Vecchiet, J.; Silvestri, C.; Baldassarre, L.; Ucciferri, C.; Orsetti, E.; Castelli, P.; Provinciali, M.; et al. Quorum sensing inhibitor FS3-coated vascular graft enhances daptomycin efficacy in a rat model of staphylococcal infection. Peptides 2013, 40, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; Malachowa, N.; Whitney, A.R.; Braughton, K.R.; Gardner, D.J.; Long, D.; Wardenburg, J.B.; Schneewind, O.; Otto, M.; de Leo, F.R. Comparative Analysis of USA300 Virulence Determinants in a Rabbit Model of Skin and Soft Tissue Infection. J. Infect. Dis. 2011, 204, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Cirioni, O.; Giacometti, A.; Ghiselli, R.; Braunstein, J.B.; Silvestri, C.; Mocchegiani, F.; Saba, V.; Scalise, G. Treatment of Staphylococcus aureus Biofilm Infection by the Quorum-Sensing Inhibitor RIP. Antimicrob. Agents Chemother. 2007, 51, 2226–2229. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Giacometti, A.; Cirioni, O.; Gov, Y.; Ghiselli, R.; Mocchegiani, F.; Viticchi, C.; del Prete, M.S.; Saba, V.; Scalise, G.; et al. Use of the Quorum-Sensing Inhibitor RNAIII-Inhibiting Peptide to Prevent Biofilm Formation In Vivo by Drug-Resistant Staphylococcus epidermidis. J. Infect. Dis. 2003, 187, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Eberhardt, K.J.; Chung, E.; Yeaman, M.R.; Sullam, P.M.; Ramos, M.; Bayer, A.S. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 1994, 94, 1815–1822. [Google Scholar] [CrossRef]
- Abdelnour, A.; Arvidson, S.; Bremell, T.; Rydén, C.; Tarkowski, A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 1993, 61, 3879–3885. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Malek, I.; Schmitz, R.A. Metagenomic quorum quenching enzymes affect biofilm formation of Candida albicans and Staphylococcus epidermidis. PLoS ONE 2019, 14, e0211366. [Google Scholar] [CrossRef]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef]
- Piecuch, A.; Obłąk, E.; Guz-Regner, K. Antibacterial Activity of Alanine-Derived Gemini Quaternary Ammonium Compounds. J. Surfact. Deterg. 2015, 19, 275–282. [Google Scholar] [CrossRef]
- Pursey, E.; Sünderhauf, D.; Gaze, W.H.; Westra, E.R.; van Houte, S. CRISPR-Cas antimicrobials: Challenges and future prospects. PLoS Pathog. 2018, 14, e1006990. [Google Scholar] [CrossRef]
- Cobb, L.H.; Park, J.; Swanson, E.A.; Beard, M.C.; McCabe, E.M.; Rourke, A.S.; Seo, K.S.; Olivier, A.K.; Priddy, L.B. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS ONE 2019, 14, e0220421. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. mBio 2013, 4, e00537-12. [Google Scholar] [CrossRef] [PubMed]
- Santiago, M.; Matano, L.M.; Moussa, S.H.; Gilmore, M.; Walker, S.; Meredith, T.C. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Stamsås, G.A.; Myrbråten, I.S.; Straume, D.; Salehian, Z.; Veening, J.-W.; Håvarstein, L.S.; Kjos, M. CozEa and CozEb play overlapping and essential roles in controlling cell division in Staphylococcus aureus. Mol. Microbiol. 2018, 109, 615–632. [Google Scholar] [CrossRef] [PubMed]
- DeFrancesco, A.S.; Masloboeva, N.; Syed, A.K.; Deloughery, A.; Bradshaw, N.; Li, G.-W.; Gilmore, M.S.; Walker, S.; Losick, R.M. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, E5969–E5978. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, E.; Koch, G.; Wagner, R.M.; Fekete, A.; Stengel, S.T.; Schneider, J.; Mielich-Süss, B.; Geibel, S.; Markert, S.M.; Stigloher, C.; et al. Membrane Microdomain Disassembly Inhibits MRSA Antibiotic Resistance. Cell 2017, 171, 1354–1367. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics 2021, 10, 131. https://doi.org/10.3390/antibiotics10020131
Kranjec C, Morales Angeles D, Torrissen Mårli M, Fernández L, García P, Kjos M, Diep DB. Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics. 2021; 10(2):131. https://doi.org/10.3390/antibiotics10020131
Chicago/Turabian StyleKranjec, Christian, Danae Morales Angeles, Marita Torrissen Mårli, Lucía Fernández, Pilar García, Morten Kjos, and Dzung B. Diep. 2021. "Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives" Antibiotics 10, no. 2: 131. https://doi.org/10.3390/antibiotics10020131
APA StyleKranjec, C., Morales Angeles, D., Torrissen Mårli, M., Fernández, L., García, P., Kjos, M., & Diep, D. B. (2021). Staphylococcal Biofilms: Challenges and Novel Therapeutic Perspectives. Antibiotics, 10(2), 131. https://doi.org/10.3390/antibiotics10020131








