Abstract
Antibiotic dosing strategies are generally based on systemic drug concentrations. However, drug concentrations at the infection site drive antimicrobial effect, and efficacy predictions and dosing strategies should be based on these concentrations. We set out to review different translational pharmacokinetic-pharmacodynamic (PK/PD) approaches from a target site perspective. The most common approach involves calculating the probability of attaining animal-derived PK/PD index targets, which link PK parameters to antimicrobial susceptibility measures. This approach is time efficient but ignores some aspects of the shape of the PK profile and inter-species differences in drug clearance and distribution, and provides no information on the PD time-course. Time–kill curves, in contrast, depict bacterial response over time. In vitro dynamic time–kill setups allow for the evaluation of bacterial response to clinical PK profiles, but are not representative of the infection site environment. The translational value of in vivo time–kill experiments, conversely, is limited from a PK perspective. Computational PK/PD models, especially when developed using both in vitro and in vivo data and coupled to target site PK models, can bridge translational gaps in both PK and PD. Ultimately, clinical PK and experimental and computational tools should be combined to tailor antibiotic treatment strategies to the site of infection.
1. Introduction
The increasing emergence and spread of antimicrobial resistance and the concurrent lack of development of novel antibiotics to counter it represents a global health risk [1,2,3,4,5]. As of September 2020, there were 43 antibiotics in clinical development pipelines, but only two of those belong to novel chemical classes and target multi-drug resistant Gram-negative pathogens categorised as critical by the World Health Organisation [5]. In light of this, it becomes increasingly important that dosing strategies for both existing and newly developed antibiotics are optimised to cure infections efficiently whilst simultaneously minimising the risk of resistance development.
Antimicrobial dosing strategies and clinical susceptibility breakpoints are predominantly set and evaluated based on drug concentrations in plasma. However, the efficacy of antimicrobials is governed by pharmacokinetic/pharmacodynamic (PK/PD) relationships at the infection site, which in most cases is not the bloodstream. Since antibiotics might be distributed unequally between the systemic circulation and different tissues, inferring antimicrobial activity solely from systemic drug concentrations is often not appropriate. Basing dosing strategies on plasma PK data may result in suboptimal or supraoptimal exposure levels at the infection site [6,7], increasing the risk of therapy failure, resistance development, or unnecessary toxicity. This approach can be especially problematic when treating patient populations for which tissue penetration of certain antibiotics has been shown to be impaired or highly variable, such as septic [8,9,10,11,12,13,14,15], other critically ill [16,17], obese [18,19,20,21,22,23,24], diabetic [14,25,26,27], or post-operative patients [28].
The importance of determining PK at the site of infection is now widely acknowledged, and also stressed by regulatory authorities [29,30]. Being the most common foci of infection, drug penetration into the lungs, urinary tract, skin and soft tissues deserve particular consideration [31]. Antimicrobial concentrations in urine, which drive bacterial killing in some types of urinary tract infections [32,33], are easily obtained. Microdialysis [34,35,36,37] and bronchoalveolar lavage [38] are the most common techniques used to sample interstitial space fluid and epithelial lining fluid (ELF), respectively.
However, target site PK should not only be measured, but subsequently needs to be used to estimate potential effects at that target site. This should not be based on the ratio between plasma and target site concentrations at a single time point, neither on the observation that concentrations at the target site exceed the minimum inhibitory concentration (MIC) of relevant pathogens at a certain time point [39], as PK profiles in tissues can have a markedly different shape compared to those in plasma. Rather, information on the entire concentration–time profile at the target site should be used to predict antimicrobial effect.
Since following the microbiological course of an infection in a quantitative way in patients is usually not feasible, we rely on the translation of preclinical data to link PK to predictions of antimicrobial effect. One approach that allows for doing so is predicting the probability of target attainment (PTA) based on PK/PD index targets. Alternatively, the time-course of bacterial response to specific PK profiles can be depicted in time–kill curves, which can be established using in vitro, in vivo, and computational modelling approaches (Figure 1). We review these approaches and discuss their strengths, limitations, and opportunities for future research, with a specific focus on their translational capacity from a target site PK/PD perspective.
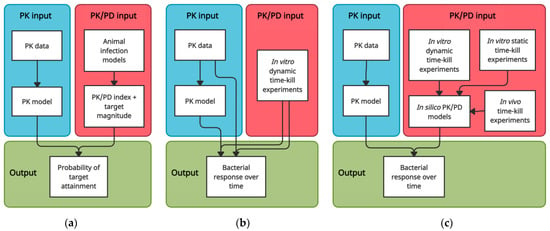
Figure 1.
Schematic overview of translational approaches to predict antimicrobial efficacy based on PK data. (a) PK/PD index—PTA approach; (b) In vitro dynamic time–kill approach; (c) Computational PK/PD modelling approaches.
2. PK/PD Index Targets and PTA Analyses
The most frequently used approach to evaluate antibiotic dosing regimens, determine susceptibility breakpoints, or predict whether observed PK profiles are likely to result in sufficient antimicrobial effect involves determining the probability of attaining a PK/PD target. The PK/PD target is usually set based on studies in animal infection models. The different animal infection models used for studying antimicrobial PK/PD and their respective advantages, limitations and challenges have been described by others in detail [40,41,42,43]. Here, we provide a brief general description of the approach (Figure 1a) and mainly focus on the translational capacity of PK/PD targets derived from animal infection models.
2.1. Selecting PK/PD Targets and Performing PTA Analyses
The most common infection models to establish PK/PD targets are murine thigh or lung infection models. The mice are often rendered neutropenic to be able to study the drug–pathogen interactions without antimicrobial contributions of the immune system, and are therefore said to be reflective of antimicrobial effect in a worst-case scenario (i.e., in severely immunocompromised patients). After introducing an infection into the tissue, dose fractionation studies are performed, in which a range of total daily doses, given either as a single dose or divided over different numbers of doses, is administered to different groups of animals. Blood samples to determine drug concentrations are taken at regular time intervals. The infected tissue is removed and homogenised at the end of the study period, usually 24 h, and bacterial density (expressed in colony-forming units [CFU]/mL) in these tissues is determined. The antimicrobial effect, defined as the difference in bacterial density in the infected tissue relative to the start of the treatment, is then correlated to three PK/PD indices: the fraction of time the unbound drug concentration exceeds the MIC (fT > MIC); the unbound drug concentration area under the curve to MIC ratio (fAUC/MIC); and the peak unbound drug concentration to MIC ratio (fCmax/MIC). The PK/PD target is then determined based on the PK/PD index that correlates best with antimicrobial effect and the magnitude of that index required to achieve a certain PD target, e.g., stasis or a 1- or 2-log10 reduction in bacterial count in the infected tissue after 24 h of drug exposure [44,45,46].
PK/PD targets have also been derived clinically. However, doing so is complicated due to the fact that usually a single dosing regimen is administered; a dose fractionation design is not possible in the clinic. Moreover, the endpoints in these studies are generally clinical cure and microbiological eradication. These endpoints, despite ultimately being the most relevant for antibiotic therapy, are difficult to measure, not necessarily attributable to the antibiotic effect or the presence of infection, and in the case of clinical cure partially subjective. It may also be difficult to include a sufficient number of patients with the relevant infection types and pathogens, and the range of MICs of these pathogens may be limited. Nonetheless, clinically derived PK/PD targets have generally been in good agreement with those derived from animal models [47]. This has generated confidence in the translational value of PK/PD targets derived from animal infection models.
In a subsequent translational step, PK/PD targets can be combined with observed or predicted human PK profiles to predict whether drug exposure in humans is likely to be effective. The PK input in such analyses usually comes from a population PK model that describes the structural trends and variability in PK data. Using the model, stochastic (Monte Carlo) simulations, which take into account the estimated variability, are performed to generate a large number of PK profiles. Based on these simulations, the probability that a given dosing regimen results in the PK/PD target being attained across a clinically relevant distribution of MICs of the pathogen of interest can be determined [48].
Since PK/PD targets play a central role in the development of antimicrobials and are pivotal in gaining regulatory approval [29,30,49], they are available for the majority of antibiotics. This, together with the fact that PK/PD targets capture the requirements for a PK profile to be deemed effective in a single summary parameter for both PK (e.g., the AUC) and PD (i.e., the MIC), means that PTAs can be calculated in a relatively straightforward, efficient, and economical way once new PK data, such as in special patient populations or certain tissues, become available.
2.2. Influence of the Shape of the PK Profile and Study Design
PTAs can be calculated based on summary PK parameters, but it is not necessarily true that antimicrobial effect is solely related to the magnitude of a PK/PD index, regardless of the shape of the PK profile. This was neatly demonstrated by Nielsen et al., who simulated dose fractionation studies using population PK models and PK/PD models based on in vitro time–kill data for six antibiotics of different classes. In their simulations, the selection of the PK/PD index and its target magnitude were sensitive to alterations in PK, e.g., when reduced drug clearance or PK in a different patient population or peripheral compartment were simulated [50]. Dhaese et al. performed a systematic review and meta-regression of data from preclinical infection models, which showed that the mode of infusion of β-lactam antibiotics influences PK/PD target selection [51]. These studies show that a given PK/PD index magnitude may not result in the same antimicrobial effect at all infection sites, due to the different shapes of the PK profile at these sites. Using a single PK/PD target based on plasma PK data may therefore be inappropriate.
The shape of the PK profile potentially affecting PK/PD target selection also has implications for the translatability of PK/PD targets obtained in animal infection models. This is because the shape of the PK profile in animals such as mice and rats may be considerably different from that in humans, as drug clearance in these smaller animals is often much quicker [46,52,53]. Some approaches to correct for this have been described, for example through chemically impairing renal function [54,55,56], or administering small extra drug doses in regular intervals [52,57] or through infusion pumps [58,59,60]. Indeed, Craig et al. showed that in mice with impaired renal function AUC/MIC correlated best with the effect of amikacin against most pathogens, whereas in mice with normal renal function fT > MIC was the dominant PK/PD index [54]. However, it should be noted that approaches to prolong drug half lives in animals are not routinely applied in the dose fractionation studies used to determine PK/PD targets.
Study design also impacts PK/PD target selection. Several studies demonstrated that the PK/PD index which correlates best with antibacterial effect depends on which dosing intervals are included in the analysis [46,61,62]. Also, higher inoculum sizes have been shown to require higher PK/PD target magnitudes to achieve the same absolute reduction in CFU counts [63]. This last point may be explained by the higher probabilities of pre-existing or emerging resistant mutants in large bacterial populations, or lower total (due to drug degradation or binding) or per-bacterium (due to larger population size) effective drug concentrations [64].
2.3. Plasma PK and Infection Site PD
It is important to realise that in most dose fractionation studies, plasma PK is linked to bacterial kill at the site of infection. This may limit the translational capacity of PK/PD targets in two additional ways.
First, drugs distribute unevenly between plasma and different tissues. The selected PK/PD target is thus dependent on the type of infection model used. The limitations associated with this are well illustrated in a study by Maglio et al., which found bactericidal effects of clarithromycin in the mouse lung model but not in the mouse thigh model, most likely resulting from intrapulmonary accumulation of the drug [65]. Therefore, basing clarithromycin PK/PD targets and susceptibility breakpoints on the mouse thigh model would result in overly pessimistic susceptibility breakpoints and an overestimation of the PK/PD target required in plasma to eradicate infections in the lung. The authors note that this might explain the lack of observed clinical failures with macrolides in the treatment of pneumonia, despite pathogens being classified as resistant [65].
Second, tissue distribution in humans may deviate from that in animal infection models. Therefore, even if a relevant infection model is used, the link between plasma PK and antimicrobial effect at the infection site, and thus the PK/PD target found in these studies, may not be directly translatable to humans. For example, a study by Rodvold et al. found that ceftobiprole penetration from plasma into ELF was much higher in mice than in humans. In other words, a given drug exposure in plasma results in much lower ELF exposure in humans than in mice for this drug, and using the PK/PD target based on mouse plasma PK to guide dosing strategies for treating pneumonia would likely result in insufficient lung exposure in patients [66].
For these reasons, it makes sense to characterise not only bacterial response but also PK at the infection site, so PK/PD targets are based on a direct link between PK and PD [38,67,68,69]. Several studies have linked drug exposure in ELF to bacterial kill in a murine lung model to establish PK/PD targets specific for the treatment of pneumonia [66,70,71]. Ideally more such site-specific PK/PD targets would be established, especially for antibiotics that distribute unequally across different tissues or for infections in sites to which distribution may be hampered or at which accumulation might occur, such as meningitis, pneumonia, osteomyelitis, or infections caused by intracellular pathogens [30,42,72,73]. Doing so adds to the complexity of the experimental procedures, but it increases the translational value of these targets, and the confidence we can have in outcomes of PTA analyses using human target site PK. However, for some infection types, such as lower urinary tract infections or complicated intra-abdominal infections, the animal infection models with which PK/PD targets can be established do not yet exist [41].
2.4. Informative Value of PD Endpoints
PK/PD targets are selected based on total bacterial counts measured at a single time point, generally after 24 h of drug exposure. A major limitation of this approach is that a single measurement provides no information on the time-course of effect, the rate and extent of kill, and the trend in bacterial population size at the time of measurement.
Moreover, potential resistance development is often ignored in dose fractionation studies, and the short study duration in combination with the single measurement of bacterial density means potential regrowth due to amplification of resistant subpopulations can often not be captured. However, drug exposure relates differently to the emergence of resistant subpopulations than to bacterial kill [63,74,75,76,77]. Drug exposures resulting in net bacterial kill during the first 24 h, and thus appearing effective, may actually provide the selection pressure that drives the development of resistance [74]. Higher PK/PD index magnitudes have been found to be required for resistance suppression than for reductions in bacterial count, with correspondingly lower target attainment rates for resistance suppression targets in PTA analyses [63,70]. It should be noted that the shape of the PK profile may also affect the development of resistance. In particular, slowly increasing antibiotic concentrations, as sometimes observed in peripheral tissues, may select for less susceptible subpopulations [78], promote the development of adaptive resistance [78,79] or the formation of persister cells [80].
2.5. Reliance on the MIC
All PK/PD indices include the MIC as a means to link PK to antimicrobial effect. However, the MIC is a measure with major limitations, including the static drug concentration under which it is measured, the poor precision and upward bias stemming from the twofold dilution steps used in its determination, and considerable assay variation [81,82]. Moreover, the endpoint on which the MIC is based, the presence of visible growth after overnight incubation, is also based on a single measurement and is additionally highly uninformative due to its dichotomous nature and the fact that growth media starts to become visibly turbid only at bacterial concentrations of 107–108 CFU/mL [82]. By relying heavily on the MIC, these limitations are propagated into the PK/PD indices and the PTA analyses based on PK/PD index targets.
3. Time–Kill Curve Approaches
Time–kill curve approaches represent alternatives to the PK/PD index approach to investigate antimicrobial PK/PD. In time–kill experiments, bacteria are inoculated in an experimental system and exposed to an antibiotic. Through periodic sampling time–kill curves can be established, depicting bacterial concentrations over time. In this section, we discuss how in vitro, in vivo, and in silico time–kill curve approaches can be used as a translational tool to investigate antimicrobial PK/PD, again with a focus on target site PK/PD.
3.1. Time-Course of Effect
A common advantage of time–kill approaches is that they allow for the evaluation of the complete PK/PD time-course, which overcomes many limitations related to the sequential workflow and dependence on summary endpoints of the PK/PD index approach [83,84,85,86,87,88]. Different antibiotics, pathogens, dosing regimens, or shapes of PK profiles can be compared in much more detail, even if these profiles have similar PK/PD index magnitudes and thus would be deemed equally effective in conventional PTA analyses. By plating samples on drug-containing agar plates, the emergence of less susceptible subpopulations can also be followed over time.
3.2. In Vitro Time–Kill Experiments
Most time–kill experiments are performed in vitro. A classification and detailed overview of the many different types of time–kill setups and their respective advantages and disadvantages has been published by Gloede et al. [89]. Broadly, they can be divided into static systems, in which drug concentrations are often assumed to be constant and no media exchange occurs, and dynamic systems, in which drug concentrations are made to change over time and fresh media is exchanged with or supplied to the culture.
3.2.1. Simulating Target Site PK
Data from static time–kill systems are useful in computational PK/PD modelling and can be used to answer specific pharmacodynamic questions, e.g., on the impact of specific pathophysiological conditions on PK/PD. However, they are not representative of the clinical situation from a PK perspective. In dynamic time–kill systems, on the other hand, any clinical PK profile can be precisely mimicked. This makes dynamic time–kill experiments suitable for simulating bacterial response to PK observed in specific tissues (Figure 1b). Indeed, setups of varying complexity have been used to simulate target site PK/PD of antimicrobials, ranging from simple stepwise dilution approaches [8,11,90,91,92,93,94,95,96,97], to more advanced continuous dilution methods [98,99,100,101,102,103,104,105,106,107,108,109], and setups in which drug and media diffuse through membranes to a peripheral compartment containing the bacteria [110,111,112,113,114,115,116,117], such as the hollow fibre infection model [118]. Various setups that aim to mimic specific target site PK characteristics have also been described [87,89]. For example, in vitro bladder infection models, pioneered by O’Grady et al., can be used to simulate the continuous filling and periodic emptying of the bladder [119,120,121,122].
Dynamic time–kill setups allow for flexibility in experimental design, not only in terms of the PK profile to simulate but also in terms of the initial inoculum size, for example. However, the number of experiments that can be feasibly performed is limited. Setups such as the hollow fibre infection model are particularly resource intensive and time consuming. So, especially when no subsequent computational PK/PD modelling is planned, the experimental conditions should be carefully selected. By far the most common approach in dynamic time–kill experiments is to simulate typical PK profiles. However, by doing so, the variability in PK between patients is ignored. This is particularly relevant when tissue PK is simulated, as tissue exposure often varies widely between patients. The lower end of the drug exposure distribution, in particular, deserves consideration in dynamic time–kill experiments, since we are generally more interested in assessing whether patients with low drug exposures face risk of treatment failure, and in vitro we cannot evaluate toxicity that might occur in patients with high tissue drug concentrations. For example, Hamada et al. simulated in a hollow-fibre infection model the 10th, 25th and 50th percentile of soft tissue exposure levels attained after a common dosage of vancomycin [106], an example of a drug for which in several different patient populations tissue exposure has been shown to be highly variable [13,25,123,124,125,126].
3.2.2. Translational Capacity of In Vitro Environment
The vast majority of in vitro PK/PD studies are performed in growth media such as Mueller–Hinton broth. Using standardised growth media enhances reproducibility, facilitates comparison between studies, and allows for assessing the interaction between drug and pathogen without potentially confounding factors. However, it has long been appreciated that the infection site environment plays a role in bacterial growth and antimicrobial activity [99]. Host factors such as physiological proteins and immune system components are absent in these growth media. Additionally, the pH value, which widely varies between different body fluids, may affect antimicrobial effect, and drug exposure may be different in homogeneous suspensions compared to the complex infection site matrix with anatomical barriers in vivo [89]. Moreover, bacteria grown in nutrient-rich growth media may be phenotypically different compared to bacteria grown in vivo [127], and growth rates are generally much higher in vitro [127,128,129]. Static time–kill systems have additional limitations. The lack of fresh media supply may result in nutrient depletion and (toxic) waste product accumulation, and the bacteria are exposed to non-dynamic and sometimes clinically irrelevant drug concentrations. These factors may impact bacterial response both phenotypically and genotypically [130]. Moreover, the duration of static time–kill experiments is usually restricted to 24 h, which may be too short to capture regrowth due to resistance development.
Thus, despite the fact that in vitro dynamic time–kill studies can be used to simulate human PK, results should not be directly translated to humans. However, these studies are useful when comparing different drugs and dosing regimens, and evaluating the effect of antibiotic combination treatment at clinically relevant PK.
Performing time–kill studies in in vitro environments that mimic the infection site theoretically makes sense to improve their translational value. However, a recent review indicated that research on antimicrobial PD in body fluids (e.g., urine, bile fluid, or cerebrospinal fluid) or growth media supplemented with human material (e.g., serum or albumin) remains scarce [131]. This is most likely due to limitations related to variability and availability. The impact that substituting standardised growth media for adapted media or body fluids has on antimicrobial activity varies between strains, antimicrobial agents, and the fluids used [131]. Moreover, the composition of a given body fluid can vary significantly between individuals and within individuals at different time points, e.g., based on diet, which may affect bacterial growth and antimicrobial effect. For example, we recently found large variability in in vitro bacterial growth and ceftriaxone effect in individual ascitic fluids from ten different ascites patients [132]. There are practical limitations too, as obtaining body fluids may involve challenging or invasive procedures, and extracting it in sufficient quantities might not be feasible. This last point renders performing experiments with dynamic setups that require a considerable amount of media challenging or impossible with certain pure body fluids.
3.3. In Vivo Time–Kill Experiments
Time–kill experiments in animal infection models are a seemingly attractive alternative to their in vitro counterparts, as studying PK/PD in an in vivo infection site environment has obvious advantages. When it comes to translational value from a PK perspective, however, in vivo time–kill experiments suffer from similar limitations as the dose fractionation studies used to determine PK/PD targets.
Time–kill experiments in rodents follow the same workflow as the dose fractionation studies described in Section 2.1, with the difference that bacterial counts are measured at various time points, rather than only at the end of the experiment [54,129,133,134,135]. However, each individual animal can only contribute data for a single time point, as quantifying bacterial counts requires the infected tissue to be removed and the animal in question to be sacrificed. Moreover, since this means that the course of the infection cannot be followed within the same animal, the sacrifice of multiple animals per time point is necessary to account for the variability in PK and PD between animals. The frequent sampling that renders time–kill curves valuable therefore requires the sacrifice of large numbers of animals, which raises ethical concerns and explains in part why time–kill experiments in rodents are not often performed.
3.4. PK/PD Modelling of Time–Kill Data
Data from time–kill experiments can be used to inform PK/PD models, which provide a quantitative description of bacterial growth and drug effect. These models are usually semi-mechanistic or mechanism-based, which means that their structure can be based on knowledge of, for example, the mechanism of action of the antibiotic, or the various mechanisms by which a pathogen may acquire reduced susceptibility to it [83,86].
3.4.1. Simulating Target Site PK/PD
With drug effect and bacterial growth described in equations, a PK/PD model can be used to simulate bacterial response to untested scenarios. For example, PK/PD models based on in vitro static time–kill data have shown the ability to predict bacterial response under dynamic drug concentrations [136,137] and across strains with different drug susceptibilities, as well as the inoculum effect [138].
The flexibility and extrapolation capacities of computational PK/PD models render them useful for detailed comparisons of drugs and dosing regimens or informing the design of further preclinical experiments or clinical trials. It also makes them particularly useful to predict PK/PD at different infection sites, since there can be large differences between different tissues in terms of PK but also in terms of the typical bacterial burden of an infection [139]. Several studies have linked models describing PK in specific body compartments to PK/PD models in order to simulate bacterial response over time at the target site (Figure 1c) [97,140,141,142,143,144,145]. In this way, comparisons in terms of target site antimicrobial effect have been made between antibiotics [145], dosing regimens [140,141,143,145], drug formulations [141], and infection sites [142,143,144].
However, few target site-specific time–kill simulations have been performed with PK models that include a measure of variability around PK parameters [143,144]. As discussed, the between-patient variability in peripheral tissue PK is often significant and it is thus important that this is reflected in stochastic time–kill curve simulations. Besides variability in PK, the variability in bacterial response as observed in the time–kill experiments (e.g., within strains or between strains with the same MIC) can also be taken into account in the simulations [143,144]. This is an advantage over conventional PTA analyses using a PK/PD index target, in which only the variability in PK is considered.
PK/PD model simulations can be used to calculate time–kill curve-based PTAs and define breakpoints. For example, Iqbal et al. linked a PK model describing plasma, muscle, and subcutaneous PK in sepsis patients to a PK/PD model developed using in vitro static time–kill data to predict target site-specific breakpoints for stasis, 2-log10 kill, and resistance development suppression for moxifloxacin against E. coli and Staphylococcus aureus [143]. To allow the prediction of time–kill curves for strains with different susceptibilities, the model included the MIC as a covariate on drug effect parameters. Due to the relatively similar moxifloxacin PK observed in plasma, skin, and subcutis, the breakpoints for the different infection sites were generally the same. However, as the authors note, this may not be the case for drugs that display impaired or delayed distribution to the target site, or where target site PK is associated with high variability [143]. As this study illustrates, to inform clinical dosing strategies time–kill curve approaches also rely on easily obtainable susceptibility measures such as the MIC. In contrast to the PK/PD index approach, however, time–kill curve-based PTA analyses and breakpoint selection take into account the entire time-course of and variability in PK and PD simultaneously.
The PK input in simulations of bacterial response to human drug exposure often comes from population PK models developed using a mainly data-driven (‘top down’) approach; the optimal model structure and parameter estimates are based on the fit of the observed PK data. When it comes to predicting the efficacy of antibiotics in different tissues or when no clinical PK data is available, physiologically-based (PB) PK models offer a notable alternative. These models employ a more mechanistic (‘bottom up’) approach to predicting drug disposition to different tissues by using a compartmental structure that is based on anatomical and physiological characteristics of the body [146,147]. The utility of such models in predicting target site PK/PD was shown by Sadiq et al., who developed a whole-body PBPK model for ciprofloxacin based on plasma concentrations and plasma-to-tissue distribution coefficients from the literature to predict typical PK profiles in a variety of tissues [142]. This PBPK model was subsequently linked to a PK/PD model by Khan et al. [148] to predict the effect of ciprofloxacin against several E.coli isolates in the different tissues. Predicted drug exposures, and, accordingly, bacterial killing, varied widely between tissues. Such a modelling framework can be used to predict the time-courses of antimicrobial effect in different tissues and identify sites where an antibiotic may display strong activity, or lack thereof [142].
PK/PD modelling and simulation approaches also have great value in the translation of the effect of antibiotic combination therapies. Drug interaction effects depend on the concentrations of the individual antibiotics and the ratio between them, and consequently on the dosing amounts and intervals of each drug and the specific shape of the PK profiles [149]. Due to the many dosing designs that need to be considered and inter-species differences in PK, dose fractionation studies in animals are not well suited to investigate optimal dosing strategies of antibiotic combinations. Moreover, the summarised nature of PK/PD index targets fails to account for the PK profile-specific dynamic changes in the concentrations of the drugs and the ratio between them [83]. The hollow fibre infection model is often used to investigate drug combinations [150,151,152,153,154], but testing all potentially clinically relevant dosing strategies is impractical. In PK/PD models, in contrast, drug interactions can be described in equations based on limited experimental data and, through simulations, optimal dosing strategies can be identified [133,149,155,156,157,158]. Again, these simulations should be based on PK at the target site as this is where the drug interaction is relevant.
3.4.2. Translational PK/PD Modelling
Most PK/PD models of antimicrobials are based on in vitro static time–kill experiments in rich growth media. These experiments provide a cheap, practical, and relatively easy way to assess bacterial response to a wide range of drug concentrations, which is useful for estimating drug effect parameters in PK/PD models. However, they are poorly reflective of the infection site environment in vivo. It should be noted that despite this limitation, PK/PD models based on static time–kill experiments have shown the capacity to predict time–kill under dynamic drug concentrations [136,137], as well as PK/PD index targets derived from animal infection models [50,62,136,158,159]. The fact that PK/PD models can be based on simple static time–kill experiments requiring small volumes of growth media also presents the opportunity to develop translational PK/PD models based on experiments in human body fluids or adapted growth media.
Two important strengths of translational modelling and simulation are its ability to inform the optimal design of further preclinical or clinical studies, and its potential to integrate data from multiple sources. For example, simulations using PK/PD models developed based on static time–kill data can be used to select PK profiles to test in dynamic time–kill experiments, data from which can subsequently be used to validate the model structure or improve parameter estimation [83,137]. Efforts have also been made to include the effect of the immune system on bacterial kill in PK/PD models [142,160], based on data from murine infection models, for example [161,162,163]. To fully leverage these strengths, however, it is important that data are compatible (e.g., by using the same bacterial strains), and close coordination between research disciplines is therefore required.
Integrating in vivo time–kill or efficacy data in the model development process not only increases the translational power of model predictions, but also maximises the value of sparse animal data as inter-species differences in PK are accounted for, which is desirable both from a scientific and ethical perspective. A recent work by Sou et al. provides an example of how sparse in vivo efficacy data in combination with PK/PD modelling can provide a translational step between in vitro PK/PD and predicting clinical efficacy. The authors developed a semi-mechanistic PK/PD model for apramycin, a novel aminoglycoside antibiotic, against E. coli based on in vitro static time–kill data of four strains. Data obtained in a murine thigh infection model with the same strains were subsequently used to correct for the slower growth rate observed in vivo, and final model parameters were re-estimated using both in vitro and in vivo data. This final PK/PD model was then coupled to a human apramycin PK model to calculate a human efficacious dose [61]. As with the PK/PD index methodology, for certain infection types or antibiotics it is important to measure drug concentrations at the infection site to optimise the translational value of in vivo data in PK/PD models. For example, Lin et al. developed an in vivo semi-mechanistic PK/PD model, the structure of which was based on a model developed with in vitro static time–kill data, for aerosolised colistin against Pseudomonas aeruginosa using time–kill data from a murine lung infection model and a population PK model describing ELF and plasma PK in mice [134]. The PK/PD model was subsequently linked to a PK model describing colistin concentrations in ELF of critically ill patients [164] in order to predict an optimal clinical dosing regimen [134].
4. Conclusions
We have discussed the capacity of different translational approaches to predicting antimicrobial efficacy at the target site. It is clear that each approach has both strengths and drawbacks (Table 1). PK/PD index targets are based on antibacterial effect in vivo, and PTA analyses using these targets allow for estimating efficacy across MICs in an economic way. However, PK/PD targets are often based on plasma PK in rodents, and differences in drug distribution and PK profile shape between species and tissues are ignored. Moreover, they are fixed summary endpoints that provide no detailed information on antimicrobial effect and often do not consider resistance development. Time–kill experiments, in contrast, allow the evaluation of the complete PK/PD time-course, including resistance development. On their own, however, these approaches lack translational power due to the experimental environment (in case of in vitro dynamic time–kill experiments) or inter-species differences in PK (in case of in vivo time–kill experiments). Additionally, a limited number of conditions can be investigated. PK/PD models can optimally bridge translational gaps in both PK and PD through integrating different sources of data and allow for simulating untested scenarios. We believe that combining PK/PD models based on both in vitro and in vivo (time–kill) data with target site PK models is the most powerful approach to optimise antibiotic dosing regimens.

Table 1.
Strengths and limitations of translational approaches for antibiotic PK/PD.
Ultimately, however, the choice of translational approach should be based on the primary objective of the analysis and the available information on PK and PD. For example, if rapid equilibration between plasma and target site concentrations is observed for a time-dependent antibiotic, current dosing strategies are likely appropriate to treat infections at the site in question (provided that plasma concentrations of this drug are known to be efficacious). In such cases, PTA analyses are valuable if large variability in target site concentrations between individuals is observed, but laborious in vitro or in silico simulations of target site PK/PD may have limited added value. The value of time–kill approaches increases, however, if the shape of the target site PK profile is very different to that in plasma or other tissues, or if impaired distribution to or accumulation at the target site is observed. When it comes to novel antibiotics, model-based translation should play a central role in de-risking and accelerating the development process. PK/PD models quantify concentration–effect relationships early on and can thereby inform the design of further translational research. These models can continuously be improved as data from increasingly complex systems become available, and ultimately guide the design of optimal dosing strategies [83]. Moreover, in the absence of clinical PK data, model-based approaches can be used to predict target site concentrations, e.g., through PBPK modelling. Such predictions may serve as the basis for studies in hollow-fibre infection models. If the antibiotic in development belongs to a well-known class of antibiotics, reasonable predictions of PK/PD relationships and tissue distribution in animals and humans can be made. In such cases, available models of related compounds can play a role in reducing the amount of preclinical (target site) PK and PK/PD work needed [61] and streamline the development of the new compound.
In conclusion, in order to make the best use of available and newly developed antibiotics, improve patient outcomes, and limit resistance development, we should move away from plasma PK-based dosing strategies. Instead, clinical target site PK data and the experimental and computational tools available to us should be leveraged to tailor antibiotic treatment strategies to the site of infection.
Author Contributions
Conceptualization, W.v.O. and M.Z.; writing—original draft preparation, W.v.O.; writing—review and editing, W.v.O. and M.Z.; supervision, M.Z.; funding acquisition, M.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 861323.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017; Available online: https://apps.who.int/iris/handle/10665/311820 (accessed on 27 October 2021).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Review of Antimicrobial Resistance: London, UK, 2016; Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 27 October 2021).
- European Centre for Disease Prevention and Control; European Medicines Agency. The bacterial challenge: Time to react. In A Call to Narrow the Gap between Multidrug-Resistant Bacteria in the EU and Development of New Antibacterial Agents; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2009; Available online: https://www.ecdc.europa.eu/en/publications-data/ecdcemea-joint-technical-report-bacterial-challenge-time-react (accessed on 27 October 2021).
- World Health Organization. 2020 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2021; Available online: https://www.who.int/publications/i/item/9789240021303 (accessed on 24 November 2021).
- Liu, P.; Müller, M.; Derendorf, H. Rational dosing of antibiotics: The use of plasma concentrations versus tissue concentrations. Int. J. Antimicrob. Agents 2002, 19, 285–290. [Google Scholar] [CrossRef]
- Lonsdale, D.O.; Udy, A.A.; Roberts, J.A.; Lipman, J. Antibacterial therapeutic drug monitoring in cerebrospinal fluid: Difficulty in achieving adequate drug concentrations. J. Neurosurg. 2013, 118, 297–301. [Google Scholar] [CrossRef]
- Zeitlinger, M.A.; Dehghanyar, P.; Mayer, B.X.; Schenk, B.S.; Neckel, U.; Heinz, G.; Georgopoulos, A.; Müller, M.; Joukhadar, C. Relevance of Soft-Tissue Penetration by Levofloxacin for Target Site Bacterial Killing in Patients with Sepsis. Antimicrob. Agents Chemother. 2003, 47, 3548–3553. [Google Scholar] [CrossRef] [PubMed]
- Joukhadar, C.; Frossard, M.; Mayer, B.X.; Brunner, M.; Klein, N.; Siostrzonek, P.; Eichler, H.G.; Müller, M. Impaired target site penetration of beta-lactams may account for therapeutic failure in patients with septic shock. Crit. Care Med. 2001, 29, 385–391. [Google Scholar] [CrossRef]
- Joukhadar, C.; Klein, N.; Mayer, B.X.; Kreischitz, N.; Delle-Karth, G.; Palkovits, P.; Heinz, G.; Müller, M. Plasma and tissue pharmacokinetics of cefpirome in patients with sepsis. Crit. Care Med. 2002, 30, 1478–1482. [Google Scholar] [CrossRef]
- Sauermann, R.; Delle-Karth, G.; Marsik, C.; Steiner, I.; Zeitlinger, M.; Mayer-Helm, B.X.; Georgopoulos, A.; Müller, M.; Joukhadar, C. Pharmacokinetics and pharmacodynamics of cefpirome in subcutaneous adipose tissue of septic patients. Antimicrob. Agents Chemother. 2005, 49, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Buerger, C.; Plock, N.; Dehghanyar, P.; Joukhadar, C.; Kloft, C. Pharmacokinetics of unbound linezolid in plasma and tissue interstitium of critically ill patients after multiple dosing using microdialysis. Antimicrob. Agents Chemother. 2006, 50, 2455–2463. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.; Sinnollareddy, M.G.; Roberts, M.S.; Williams, P.; Peake, S.L.; Lipman, J.; Roberts, J.A. Plasma and interstitial fluid population pharmacokinetics of vancomycin in critically ill patients with sepsis. Int. J. Antimicrob. Agents 2019, 53, 137–142. [Google Scholar] [CrossRef]
- Minichmayr, I.K.; Schaeftlein, A.; Kuti, J.L.; Zeitlinger, M.; Kloft, C. Clinical Determinants of Target Non-Attainment of Linezolid in Plasma and Interstitial Space Fluid: A Pooled Population Pharmacokinetic Analysis with Focus on Critically Ill Patients. Clin. Pharmacokinet. 2017, 56, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Sinnollareddy, M.G.; Roberts, M.S.; Lipman, J.; Lassig-Smith, M.; Starr, T.; Robertson, T.; Peake, S.L.; Roberts, J.A. In Vivo Microdialysis To Determine Subcutaneous Interstitial Fluid Penetration and Pharmacokinetics of Fluconazole in Intensive Care Unit Patients with Sepsis. Antimicrob. Agents Chemother. 2016, 60, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, I.; Schmidtko, A.; Bräutigam, L.; Kirschbaum, A.; Geisslinger, G.; Lötsch, J. Tissue distribution of imipenem in critically ill patients. Clin. Pharmacol. Ther. 2002, 71, 325–333. [Google Scholar] [CrossRef]
- Dahyot-Fizelier, C.; Lefeuvre, S.; Laksiri, L.; Marchand, S.; Sawchuk, R.J.; Couet, W.; Mimoz, O. Kinetics of imipenem distribution into the peritoneal fluid of patients with severe peritonitis studied by microdialysis. Clin. Pharmacokinet. 2010, 49, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, U.M.; Brunner, M.; Schmid, R.; Müller, M. Soft tissue concentrations of ciprofloxacin in obese and lean subjects following weight-adjusted dosing. Int. J. Obes. Relat. Metab. Disord. 2001, 25, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, L.; Simon, P.; Busse, D.; Petroff, D.; Dorn, C.; Huisinga, W.; Dietrich, A.; Zeitlinger, M.; Wrigge, H.; Kloft, C. Risk of target non-attainment in obese compared to non-obese patients in calculated linezolid therapy. Clin. Microbiol. Infect. 2020, 26, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Wittau, M.; Paschke, S.; Kurlbaum, M.; Scheele, J.; Ly, N.S.; Hemper, E.; Kornmann, M.; Henne-Bruns, D.; Bulitta, J.B. Population Pharmacokinetics and Target Attainment of Ertapenem in Plasma and Tissue Assessed via Microdialysis in Morbidly Obese Patients after Laparoscopic Visceral Surgery. Antimicrob. Agents Chemother. 2017, 61, e00952-16. [Google Scholar] [CrossRef]
- Wittau, M.; Scheele, J.; Kurlbaum, M.; Brockschmidt, C.; Wolf, A.M.; Hemper, E.; Henne-Bruns, D.; Bulitta, J.B. Population Pharmacokinetics and Target Attainment of Meropenem in Plasma and Tissue of Morbidly Obese Patients after Laparoscopic Intraperitoneal Surgery. Antimicrob. Agents Chemother. 2015, 59, 6241–6247. [Google Scholar] [CrossRef]
- Brill, M.J.E.; Houwink, A.P.I.; Schmidt, S.; Van Dongen, E.P.A.; Hazebroek, E.J.; van Ramshorst, B.; Deneer, V.H.; Mouton, J.W.; Knibbe, C.A.J. Reduced subcutaneous tissue distribution of cefazolin in morbidly obese versus non-obese patients determined using clinical microdialysis. J. Antimicrob. Chemother. 2014, 69, 715–723. [Google Scholar] [CrossRef]
- Toma, O.; Suntrup, P.; Stefanescu, A.; London, A.; Mutch, M.; Kharasch, E. Pharmacokinetics and tissue penetration of cefoxitin in obesity: Implications for risk of surgical site infection. Anesth. Analg. 2011, 113, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.; Schmidt, S.; Rout, W.R.; Ben-David, K.; Burkhardt, O.; Derendorf, H. Soft tissue penetration of cefuroxime determined by clinical microdialysis in morbidly obese patients undergoing abdominal surgery. Int. J. Antimicrob. Agents 2009, 34, 231–235. [Google Scholar] [CrossRef]
- Skhirtladze, K.; Hutschala, D.; Fleck, T.; Thalhammer, F.; Ehrlich, M.; Vukovich, T.; Müller, M.; Tschernko, E.M. Impaired target site penetration of vancomycin in diabetic patients following cardiac surgery. Antimicrob. Agents Chemother. 2006, 50, 1372–1375. [Google Scholar] [CrossRef] [PubMed]
- Joukhadar, C.; Stass, H.; Müller-Zellenberg, U.; Lackner, E.; Kovar, F.; Minar, E.; Müller, M. Penetration of moxifloxacin into healthy and inflamed subcutaneous adipose tissues in humans. Antimicrob. Agents Chemother. 2003, 47, 3099–3103. [Google Scholar] [CrossRef]
- Kim, A.; Suecof, L.A.; Sutherland, C.A.; Gao, L.; Kuti, J.L.; Nicolau, D.P. In vivo microdialysis study of the penetration of daptomycin into soft tissues in diabetic versus healthy volunteers. Antimicrob. Agents Chemother. 2008, 52, 3941–3946. [Google Scholar] [CrossRef] [PubMed]
- Brunner, M.; Pernerstorfer, T.; Mayer, B.X.; Eichler, H.G.; Müller, M. Surgery and intensive care procedures affect the target site distribution of piperacillin. Crit. Care Med. 2000, 28, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. Antibacterial Therapies for Patients with an Unmet Medical Need for the Treatment of Serious Bacterial Diseases. Guidance for Industry. Available online: https://www.fda.gov/media/86250/download (accessed on 27 October 2021).
- European Medicines Agency. Guideline on the Use of Pharmacokinetics and Pharmacodynamics in the Development of Antimicrobial Medicinal Products. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-use-pharmacokinetics-pharmacodynamics-development-antimicrobial-medicinal-products_en.pdf (accessed on 27 October 2021).
- Gaynes, R.; Edwards, J.R. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 2005, 41, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Stamey, T.A.; Fair, W.R.; Timothy, M.M.; Millar, M.A.; Mihara, G.; Lowery, Y.C. Serum versus Urinary Antimicrobial Concentrations in Cure of Urinary-Tract Infections. N. Engl. J. Med. 1974, 291, 1159–1163. [Google Scholar] [CrossRef]
- Frimodt-Møller, N. Correlation between pharmacokinetic/pharmacodynamic parameters and efficacy for antibiotics in the treatment of urinary tract infection. Int. J. Antimicrob. Agents 2002, 19, 546–553. [Google Scholar] [CrossRef]
- Müller, M. Microdialysis in clinical drug delivery studies. Adv. Drug Deliv. Rev. 2000, 45, 255–269. [Google Scholar] [CrossRef]
- de la Peña, A.; Liu, P.; Derendorf, H. Microdialysis in peripheral tissues. Adv. Drug Deliv. Rev. 2000, 45, 189–216. [Google Scholar] [CrossRef]
- Joukhadar, C.; Müller, M. Microdialysis: Current applications in clinical pharmacokinetic studies and its potential role in the future. Clin. Pharmacokinet. 2005, 44, 895–913. [Google Scholar] [CrossRef]
- Joukhadar, C.; Derendorf, H.; Müller, M. Microdialysis. A novel tool for clinical studies of anti-infective agents. Eur. J. Clin. Pharmacol. 2001, 57, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, K.A.; George, J.M.; Yoo, L. Penetration of anti-infective agents into pulmonary epithelial lining fluid: Focus on antibacterial agents. Clin. Pharmacokinet. 2011, 50, 637–664. [Google Scholar] [CrossRef] [PubMed]
- Mouton, J.W.; Theuretzbacher, U.; Craig, W.A.; Tulkens, P.M.; Derendorf, H.; Cars, O. Tissue concentrations: Do we ever learn? J. Antimicrob. Chemother. 2008, 61, 235–237. [Google Scholar] [CrossRef]
- Andes, D.; Craig, W.A. Animal model pharmacokinetics and pharmacodynamics: A critical review. Int. J. Antimicrob. Agents 2002, 19, 261–268. [Google Scholar] [CrossRef]
- Bulitta, J.B.; Hope, W.W.; Eakin, A.E.; Guina, T.; Tam, V.H.; Louie, A.; Drusano, G.L.; Hoover, J.L. Generating Robust and Informative Nonclinical In Vitro and In Vivo Bacterial Infection Model Efficacy Data to Support Translation to Humans. Antimicrob. Agents Chemother. 2019, 63, e02307-18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lepak, A.J.; Andes, D.R. Animal models in the pharmacokinetic/pharmacodynamic evaluation of antimicrobial agents. Bioorg. Med. Chem. 2016, 24, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Bergen, P.J.; Lora-Tamayo, J.; Landersdorfer, C.B.; Li, J. PK/PD models in antibacterial development. Curr. Opin. Microbiol. 2013, 16, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Craig, W.A. Pharmacokinetic/pharmacodynamic parameters: Rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 1998, 26, 1–10. [Google Scholar] [CrossRef]
- Craig, W.A. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagn. Microbiol. Infect. Dis. 1995, 22, 89–96. [Google Scholar] [CrossRef]
- Vogelman, B.; Gudmundsson, S.; Leggett, J.; Turnidge, J.; Ebert, S.; Craig, W.A. Correlation of antimicrobial pharmacokinetic parameters with therapeutic efficacy in an animal model. J. Infect. Dis. 1988, 158, 831–847. [Google Scholar] [CrossRef]
- Ambrose, P.G.; Bhavnani, S.M.; Rubino, C.M.; Louie, A.; Gumbo, T.; Forrest, A.; Drusano, G.L. Pharmacokinetics-Pharmacodynamics of Antimicrobial Therapy: It’s Not Just for Mice Anymore. Clin. Infect. Dis. 2007, 44, 79–86. [Google Scholar] [CrossRef]
- Drusano, G.L.; Preston, S.L.; Hardalo, C.; Hare, R.; Banfield, C.; Andes, D.; Vesga, O.; Craig, W.A. Use of preclinical data for selection of a phase II/III dose for evernimicin and identification of a preclinical MIC breakpoint. Antimicrob. Agents Chemother. 2001, 45, 13–22. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bulik, C.; Bhavnani, S.; Hammel, J.; Forrest, A.; Dudley, M.; Ellis-Grosse, E.; Drusano, G.; Ambrose, P. Relationship between regulatory approval and pharmacokinetic-pharmacodynamic target attainment: Focus on community-and hospital-acquired pneumonia. In Proceedings of the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Denver, CO, USA, 10–13 September 2013. [Google Scholar]
- Nielsen, E.I.; Otto, C.; Friberg, L.E. Pharmacokinetic/Pharmacodynamic (PK/PD) Indices of Antibiotics Predicted by a Semimechanistic PKPD Model: A Step toward Model-Based Dose Optimization. Antimicrob. Agents Chemother. 2011, 55, 4619–4630. [Google Scholar] [CrossRef]
- Dhaese, S.; Heffernan, A.; Liu, D.; Abdul-Aziz, M.H.; Stove, V.; Tam, V.H.; Lipman, J.; Roberts, J.A.; De Waele, J.J. Prolonged Versus Intermittent Infusion of β-Lactam Antibiotics: A Systematic Review and Meta-Regression of Bacterial Killing in Preclinical Infection Models. Clin. Pharmacokinet. 2020, 59, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.U.; Brugger, H.-P.; Feller, C.; Stritzko, T.; Stalder, B. Antibiotic Therapy of Infections Due to Pseudomonas aeruginosa in Normal and Granulocytopenic Mice: Comparison of Murine and Human Pharmacokinetics. J. Infect. Dis. 1986, 153, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.J.; Holford, N.H.G. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 303–332. [Google Scholar] [CrossRef] [PubMed]
- Craig, W.A.; Redington, J.; Ebert, S.C. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J. Antimicrob. Chemother. 1991, 27, 29–40. [Google Scholar] [CrossRef]
- Andes, D.; Craig, W.A. In vivo activities of amoxicillin and amoxicillin-clavulanate against Streptococcus pneumoniae: Application to breakpoint determinations. Antimicrob. Agents Chemother. 1998, 42, 2375–2379. [Google Scholar] [CrossRef]
- Crandon, J.L.; Nicolau, D.P. Human simulated studies of aztreonam and aztreonam-avibactam to evaluate activity against challenging gram-negative organisms, including metallo-β-lactamase producers. Antimicrob. Agents Chemother. 2013, 57, 3299–3306. [Google Scholar] [CrossRef] [PubMed]
- Keel, R.A.; Crandon, J.L.; Nicolau, D.P. Efficacy of human simulated exposures of ceftaroline administered at 600 milligrams every 12 hours against phenotypically diverse Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011, 55, 4028–4032. [Google Scholar] [CrossRef][Green Version]
- Blatter, M.; Fluckiger, U.; Entenza, J.; Glauser, M.P.; Francioli, P. Simulated human serum profiles of one daily dose of ceftriaxone plus netilmicin in treatment of experimental streptococcal endocarditis. Antimicrob. Agents Chemother. 1993, 37, 1971–1976. [Google Scholar] [CrossRef] [PubMed]
- Piroth, L.; Martin, L.; Coulon, A.; Lequeu, C.; Duong, M.; Buisson, M.; Portier, H.; Chavanet, P. Development of a new experimental model of penicillin-resistant Streptococcus pneumoniae pneumonia and amoxicillin treatment by reproducing human pharmacokinetics. Antimicrob. Agents Chemother. 1999, 43, 2484–2492. [Google Scholar] [CrossRef] [PubMed]
- Robaux, M.A.; Dube, L.; Caillon, J.; Bugnon, D.; Kergueris, M.F.; Navas, D.; Le Conte, P.; Baron, D.; Potel, G. In vivo efficacy of continuous infusion versus intermittent dosing of ceftazidime alone or in combination with amikacin relative to human kinetic profiles in a Pseudomonas aeruginosa rabbit endocarditis model. J. Antimicrob. Chemother. 2001, 47, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Sou, T.; Hansen, J.; Liepinsh, E.; Backlund, M.; Ercan, O.; Grinberga, S.; Cao, S.; Giachou, P.; Petersson, A.; Tomczak, M.; et al. Model-Informed Drug Development for Antimicrobials: Translational PK and PK/PD Modeling to Predict an Efficacious Human Dose for Apramycin. Clin. Pharmacol. Ther. 2021, 109, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersson, A.N.; David-Pierson, P.; Parrott, N.J.; Kuhlmann, O.; Lave, T.; Friberg, L.E.; Nielsen, E.I. Simulation-Based Evaluation of PK/PD Indices for Meropenem Across Patient Groups and Experimental Designs. Pharm. Res. 2016, 33, 1115–1125. [Google Scholar] [CrossRef]
- Jumbe, N.; Louie, A.; Leary, R.; Liu, W.; Deziel, M.R.; Tam, V.H.; Bachhawat, R.; Freeman, C.; Kahn, J.B.; Bush, K.; et al. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J. Clin. Investig. 2003, 112, 275–285. [Google Scholar] [CrossRef]
- Udekwu, K.I.; Parrish, N.; Ankomah, P.; Baquero, F.; Levin, B.R. Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 2009, 63, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Maglio, D.; Capitano, B.; Banevicius, M.A.; Geng, Q.; Nightingale, C.H.; Nicolau, D.P. Differential Efficacy of Clarithromycin in Lung versus Thigh Infection Models. Chemotherapy 2004, 50, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, K.A.; Nicolau, D.P.; Lodise, T.P.; Khashab, M.; Noel, G.J.; Kahn, J.B.; Gotfried, M.; Murray, S.A.; Nicholson, S.; Laohavaleeson, S.; et al. Identifying exposure targets for treatment of staphylococcal pneumonia with ceftobiprole. Antimicrob. Agents Chemother. 2009, 53, 3294–3301. [Google Scholar] [CrossRef]
- Trang, M.; Dudley, M.N.; Bhavnani, S.M. Use of Monte Carlo simulation and considerations for PK-PD targets to support antibacterial dose selection. Curr. Opin. Pharmacol. 2017, 36, 107–113. [Google Scholar] [CrossRef]
- Ambrose, P.G. Antibacterial drug development program successes and failures: A pharmacometric explanation. Curr. Opin. Pharmacol. 2017, 36, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, P.G.; Bhavnani, S.M.; Ellis-Grosse, E.J.; Drusano, G.L. Pharmacokinetic-Pharmacodynamic Considerations in the Design of Hospital-Acquired or Ventilator-Associated Bacterial Pneumonia Studies: Look before You Leap! Clin. Infect. Dis. 2010, 51, S103–S110. [Google Scholar] [CrossRef] [PubMed]
- Drusano, G.L.; Lodise, T.P.; Melnick, D.; Liu, W.; Oliver, A.; Mena, A.; VanScoy, B.; Louie, A. Meropenem penetration into epithelial lining fluid in mice and humans and delineation of exposure targets. Antimicrob. Agents Chemother. 2011, 55, 3406–3412. [Google Scholar] [CrossRef] [PubMed]
- Louie, A.; Fregeau, C.; Liu, W.; Kulawy, R.; Drusano, G.L. Pharmacodynamics of levofloxacin in a murine pneumonia model of Pseudomonas aeruginosa infection: Determination of epithelial lining fluid targets. Antimicrob. Agents Chemother. 2009, 53, 3325–3330. [Google Scholar] [CrossRef] [PubMed]
- Landersdorfer, C.B.; Bulitta, J.B.; Kinzig, M.; Holzgrabe, U.; Sörgel, F. Penetration of antibacterials into bone: Pharmacokinetic, pharmacodynamic and bioanalytical considerations. Clin. Pharmacokinet. 2009, 48, 89–124. [Google Scholar] [CrossRef] [PubMed]
- Nau, R.; Sörgel, F.; Eiffert, H. Penetration of Drugs through the Blood-Cerebrospinal Fluid/Blood-Brain Barrier for Treatment of Central Nervous System Infections. Clin. Microbiol. Rev. 2010, 23, 858–883. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.H.; Louie, A.; Deziel, M.R.; Liu, W.; Drusano, G.L. The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: A new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob. Agents Chemother. 2007, 51, 744–747. [Google Scholar] [CrossRef]
- Tam, V.H.; Louie, A.; Deziel, M.R.; Liu, W.; Leary, R.; Drusano, G.L. Bacterial-Population Responses to Drug-Selective Pressure: Examination of Garenoxacin’s Effect on Pseudomonas aeruginosa. J. Infect. Dis. 2005, 192, 420–428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tam, V.H.; Louie, A.; Fritsche, T.R.; Deziel, M.; Liu, W.; Brown, D.L.; Deshpande, L.; Leary, R.; Jones, R.N.; Drusano, G.L. Impact of drug-exposure intensity and duration of therapy on the emergence of Staphylococcus aureus resistance to a quinolone antimicrobial. J. Infect. Dis. 2007, 195, 1818–1827. [Google Scholar] [CrossRef] [PubMed]
- Louie, A.; Maynard, M.; Duncanson, B.; Nole, J.; Vicchiarelli, M.; Drusano, G.L. Determination of the Dynamically Linked Indices of Fosfomycin for Pseudomonas aeruginosa in the Hollow Fiber Infection Model. Antimicrob. Agents Chemother. 2018, 62, e02627-17. [Google Scholar] [CrossRef]
- Cheah, S.E.; Li, J.; Tsuji, B.T.; Forrest, A.; Bulitta, J.B.; Nation, R.L. Colistin and Polymyxin B Dosage Regimens against Acinetobacter baumannii: Differences in Activity and the Emergence of Resistance. Antimicrob. Agents Chemother. 2016, 60, 3921–3933. [Google Scholar] [CrossRef]
- Sandoval-Motta, S.; Aldana, M. Adaptive resistance to antibiotics in bacteria: A systems biology perspective. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; He, X.; Zou, X.; Wang, G.; Li, S.; Wu, Y. Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 3267–3272. [Google Scholar] [CrossRef][Green Version]
- Mouton, J.W.; Meletiadis, J.; Voss, A.; Turnidge, J. Variation of MIC measurements: The contribution of strain and laboratory variability to measurement precision. J. Antimicrob. Chemother. 2018, 73, 2374–2379. [Google Scholar] [CrossRef]
- Mouton, J.W.; Muller, A.E.; Canton, R.; Giske, C.G.; Kahlmeter, G.; Turnidge, J. MIC-based dose adjustment: Facts and fables. J. Antimicrob. Chemother. 2017, 73, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Friberg, L.E. Pivotal Role of Translation in Anti-Infective Development. Clin. Pharm. Therap. 2021, 109, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.; Ly, N.; Diep, J.K.; Rao, G.G. Moving From Point-Based Analysis to Systems-Based Modeling: Integration of Knowledge to Address Antimicrobial Resistance Against MDR Bacteria. Clin. Pharmacol. Ther. 2021, 110, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.G.; Landersdorfer, C.B. Antibiotic pharmacokinetic/pharmacodynamic modelling: MIC, pharmacodynamic indices and beyond. Int. J. Antimicrob. Agents 2021, 58, 106368. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.I.; Friberg, L.E. Pharmacokinetic-Pharmacodynamic Modeling of Antibacterial Drugs. Pharmacol. Rev. 2013, 65, 1053–1090. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; de la Peña, A.; Derendorf, H. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: Kill curves versus MIC. Antimicrob. Agents Chemother. 2004, 48, 369–377. [Google Scholar] [CrossRef]
- Rayner, C.R.; Smith, P.F.; Andes, D.; Andrews, K.; Derendorf, H.; Friberg, L.E.; Hanna, D.; Lepak, A.; Mills, E.; Polasek, T.M.; et al. Model-Informed Drug Development for Anti-Infectives: State of the Art and Future. Clin. Pharmacol. Ther. 2021, 109, 867–891. [Google Scholar] [CrossRef] [PubMed]
- Gloede, J.; Scheerans, C.; Derendorf, H.; Kloft, C. In vitro pharmacodynamic models to determine the effect of antibacterial drugs. J. Antimicrob. Chemother. 2010, 65, 186–201. [Google Scholar] [CrossRef]
- Brunner, M.; Hollenstein, U.; Delacher, S.; Jäger, D.; Schmid, R.; Lackner, E.; Georgopoulos, A.; Eichler, H.G.; Müller, M. Distribution and antimicrobial activity of ciprofloxacin in human soft tissues. Antimicrob. Agents Chemother. 1999, 43, 1307–1309. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frossard, M.; Joukhadar, C.; Erovic, B.M.; Dittrich, P.; Mrass, P.E.; Van Houte, M.; Burgmann, H.; Georgopoulos, A.; Müller, M. Distribution and antimicrobial activity of fosfomycin in the interstitial fluid of human soft tissues. Antimicrob. Agents Chemother. 2000, 44, 2728–2732. [Google Scholar] [CrossRef]
- Zeitlinger, M.A.; Marsik, C.; Georgopoulos, A.; Müller, M.; Heinz, G.; Joukhadar, C. Target site bacterial killing of cefpirome and fosfomycin in critically ill patients. Int. J. Antimicrob. Agents 2003, 21, 562–567. [Google Scholar] [CrossRef]
- Joukhadar, C.; Klein, N.; Dittrich, P.; Zeitlinger, M.; Geppert, A.; Skhirtladze, K.; Frossard, M.; Heinz, G.; Müller, M. Target site penetration of fosfomycin in critically ill patients. J. Antimicrob. Chemother. 2003, 51, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Sauermann, R.; Zeitlinger, M.; Erovic, B.M.; Marsik, C.; Georgopoulos, A.; Müller, M.; Brunner, M.; Joukhadar, C. Pharmacodynamics of piperacillin in severely ill patients evaluated by using a PK/PD model. Int. J. Antimicrob. Agents 2003, 22, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Zeitlinger, M.A.; Erovic, B.M.; Sauermann, R.; Georgopoulos, A.; Müller, M.; Joukhadar, C. Plasma concentrations might lead to overestimation of target site activity of piperacillin in patients with sepsis. J. Antimicrob. Chemother. 2005, 56, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Joukhadar, C.; Dehghanyar, P.; Traunmüller, F.; Sauermann, R.; Mayer-Helm, B.; Georgopoulos, A.; Müller, M. Increase of microcirculatory blood flow enhances penetration of ciprofloxacin into soft tissue. Antimicrob. Agents Chemother. 2005, 49, 4149–4153. [Google Scholar] [CrossRef] [PubMed]
- Delacher, S.; Derendorf, H.; Hollenstein, U.; Brunner, M.; Joukhadar, C.; Hofmann, S.; Georgopoulos, A.; Eichler, H.G.; Müller, M. A combined in vivo pharmacokinetic-in vitro pharmacodynamic approach to simulate target site pharmacodynamics of antibiotics in humans. J. Antimicrob. Chemother. 2000, 46, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, Y.; Bulitta, J.B.; Forrest, A.; Sakoulas, G.; Lesse, A.J.; Mylotte, J.M.; Tsuji, B.T. Pharmacodynamics of vancomycin at simulated epithelial lining fluid concentrations against methicillin-resistant Staphylococcus aureus (MRSA): Implications for dosing in MRSA pneumonia. Antimicrob. Agents Chemother. 2009, 53, 3894–3901. [Google Scholar] [CrossRef] [PubMed]
- Satta, G.; Cornaglia, G.; Foddis, G.; Pompei, R. Evaluation of ceftriaxone and other antibiotics against Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae under in vitro conditions simulating those of serious infections. Antimicrob. Agents Chemother. 1988, 32, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Fan, H.W.; Sutherland, C.; DeRyke, A.C.; Nicolau, D.P. Antibacterial effects of moxifloxacin and levofloxacin simulating epithelial lining fluid concentrations against community-acquired methicillin-resistant Staphylococcus aureus. Drugs R D 2007, 8, 69–77. [Google Scholar] [CrossRef]
- Deryke, C.A.; Du, X.; Nicolau, D.P. Evaluation of bacterial kill when modelling the bronchopulmonary pharmacokinetic profile of moxifloxacin and levofloxacin against parC-containing isolates of Streptococcus pneumoniae. J. Antimicrob. Chemother. 2006, 58, 601–609. [Google Scholar] [CrossRef][Green Version]
- Florea, N.R.; Tessier, P.R.; Zhang, C.; Nightingale, C.H.; Nicolau, D.P. Pharmacodynamics of moxifloxacin and levofloxacin at simulated epithelial lining fluid drug concentrations against Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 1215–1221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jain, J.G.; Housman, S.T.; Nicolau, D.P. Humanized tissue pharmacodynamics of cefazolin against commonly isolated pathogens in skin and skin structure infections. J. Antimicrob. Chemother. 2014, 69, 2443–2447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- MacVane, S.H.; So, W.; Nicolau, D.P.; Kuti, J.L. In vitro activity of human-simulated epithelial lining fluid exposures of ceftaroline, ceftriaxone, and vancomycin against methicillin-susceptible and -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 7520–7526. [Google Scholar] [CrossRef]
- So, W.; Crandon, J.L.; Hamada, Y.; Nicolau, D.P. Antibacterial activity of achievable epithelial lining fluid exposures of Amikacin Inhale with or without meropenem. J. Antimicrob. Chemother. 2016, 71, 428–437. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hamada, Y.; Kuti, J.L.; Nicolau, D.P. In Vitro Pharmacodynamics of Vancomycin against Methicillin-Susceptible and -Resistant Staphylococcus aureus: Considering the Variability in Observed Tissue Exposure. Antimicrob. Agents Chemother. 2015, 60, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, I.M.; Grupper, M.; Nicolau, D.P. Anti-staphylococcal activity resulting from epithelial lining fluid (ELF) concentrations of amikacin inhale administered via the pulmonary drug delivery system. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, I.M.; Grupper, M.; Nicolau, D.P. Antibacterial activity of human simulated epithelial lining fluid concentrations of amikacin inhale alone and in combination with meropenem against Acinetobacter baumannii. Infect. Dis. 2017, 49, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Almarzoky Abuhussain, S.S.; Kuti, J.L.; Nicolau, D.P. Antibacterial Activity of Human Simulated Epithelial Lining Fluid Concentrations of Ceftazidime-Avibactam Alone or in Combination with Amikacin Inhale (BAY41-6551) against Carbapenem-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2018, 62, e00113-18. [Google Scholar] [CrossRef] [PubMed]
- Noreddin, A.M.; Roberts, D.; Nichol, K.; Wierzbowski, A.; Hoban, D.J.; Zhanel, G.G. Pharmacodynamic modeling of clarithromycin against macrolide-resistant [PCR-positive mef(A) or erm(B)] Streptococcus pneumoniae simulating clinically achievable serum and epithelial lining fluid free-drug concentrations. Antimicrob. Agents Chemother. 2002, 46, 4029–4034. [Google Scholar] [CrossRef][Green Version]
- Sevillano, D.; Alou, L.; Aguilar, L.; Echevarría, O.; Giménez, M.J.; Prieto, J. Azithromycin iv pharmacodynamic parameters predicting Streptococcus pneumoniae killing in epithelial lining fluid versus serum: An in vitro pharmacodynamic simulation. J. Antimicrob. Chemother. 2006, 57, 1128–1133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alou, L.; Giménez, M.J.; Sevillano, D.; Aguilar, L.; Cafini, F.; Echeverría, O.; Pérez-Trallero, E.; Prieto, J. A pharmacodynamic approach to antimicrobial activity in serum and epithelial lining fluid against in vivo-selected Streptococcus pneumoniae mutants and association with clinical failure in pneumonia. J. Antimicrob. Chemother. 2006, 58, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Sevillano, D.; Alou, L.; Cafini, F.; Gimenez, M.J.; Gomez-Lus, M.L.; Prieto, J.; Aguilar, L. Influence of the MBC/MIC ratio on the antibacterial activity of vancomycin versus linezolid against methicillin-resistant Staphylococcus aureus isolates in a pharmacodynamic model simulating serum and soft tissue interstitial fluid concentrations reported in diabetic patients. J. Antimicrob. Chemother. 2013, 68, 2291–2295. [Google Scholar] [CrossRef] [PubMed]
- Sime, F.B.; Johnson, A.; Whalley, S.; Santoyo-Castelazo, A.; Montgomery, A.B.; Walters, K.A.; Lipman, J.; Hope, W.W.; Roberts, J.A. Pharmacodynamics of Aerosolized Fosfomycin and Amikacin against Resistant Clinical Isolates of Pseudomonas aeruginosa and Klebsiella pneumoniae in a Hollow-Fiber Infection Model: Experimental Basis for Combination Therapy. Antimicrob. Agents Chemother. 2016, 61, e01763-16. [Google Scholar] [CrossRef]
- Zhanel, G.G.; DeCorby, M.; Noreddin, A.; Mendoza, C.; Cumming, A.; Nichol, K.; Wierzbowski, A.; Hoban, D.J. Pharmacodynamic activity of azithromycin against macrolide-susceptible and -resistant Streptococcus pneumoniae simulating clinically achievable free serum, epithelial lining fluid and middle ear fluid concentrations. J. Antimicrob. Chemother. 2003, 52, 83–88. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Johanson, C.; Hisanaga, T.; Mendoza, C.; Laing, N.; Noreddin, A.; Wierzbowski, A.; Hoban, D.J. Pharmacodynamic activity of telithromycin against macrolide-susceptible and macrolide-resistant Streptococcus pneumoniae simulating clinically achievable free serum and epithelial lining fluid concentrations. J. Antimicrob. Chemother. 2004, 54, 1072–1077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhanel, G.G.; Johanson, C.; Laing, N.; Hisanaga, T.; Wierzbowski, A.; Hoban, D.J. Pharmacodynamic activity of telithromycin at simulated clinically achievable free-drug concentrations in serum and epithelial lining fluid against efflux (mefE)-producing macrolide-resistant Streptococcus pneumoniae for which telithromycin MICs vary. Antimicrob. Agents Chemother. 2005, 49, 1943–1948. [Google Scholar] [CrossRef][Green Version]
- Zinner, S.H.; Husson, M.; Klastersky, J. An artificial capillary in vitro kinetic model of antibiotic bactericidal activity. J. Infect. Dis. 1981, 144, 583–587. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, F.; Pennington, J.H. Bacterial growth in an in vitro system simulating conditions in the urinary bladder. Br. J. Exp. Pathol. 1966, 47, 152–157. [Google Scholar]
- Greenwood, D.; O’Grady, F. An in vitro model of the urinary bladder. J. Antimicrob. Chemother. 1978, 4, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, D. An in-vitro model simulating the hydrokinetic aspects of the treatment of bacterial cystitis. J. Antimicrob. Chemother. 1985, 15, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Abbott, I.J.; Meletiadis, J.; Belghanch, I.; Wijma, R.A.; Kanioura, L.; Roberts, J.A.; Peleg, A.Y.; Mouton, J.W. Fosfomycin efficacy and emergence of resistance among Enterobacteriaceae in an in vitro dynamic bladder infection model. J. Antimicrob. Chemother. 2018, 73, 709–719. [Google Scholar] [CrossRef]
- Payne, C.J.; Thomson, A.H.; Stearns, A.T.; Watson, D.G.; Zhang, T.; Kingsmore, D.B.; Byrne, D.S.; Binning, A.S. Pharmacokinetics and tissue penetration of vancomycin continuous infusion as prophylaxis for vascular surgery. J. Antimicrob. Chemother. 2011, 66, 2624–2627. [Google Scholar] [CrossRef] [PubMed]
- Housman, S.T.; Bhalodi, A.A.; Shepard, A.; Nugent, J.; Nicolau, D.P. Vancomycin Tissue Pharmacokinetics in Patients with Lower-Limb Infections via In Vivo Microdialysis. J. Am. Podiatr. Med. Assoc. 2015, 105, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Kuti, J.L.; Nicolau, D.P. Vancomycin serum concentrations do not adequately predict tissue exposure in diabetic patients with mild to moderate limb infections. J. Antimicrob. Chemother. 2015, 70, 2064–2067. [Google Scholar] [CrossRef]
- Bue, M.; Tøttrup, M.; Hanberg, P.; Langhoff, O.; Birke-Sørensen, H.; Thillemann, T.M.; Andersson, T.L.; Søballe, K. Bone and subcutaneous adipose tissue pharmacokinetics of vancomycin in total knee replacement patients. Acta Orthop. 2018, 89, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Dalhoff, A. Differences between bacteria grown in vitro and in vivo. J. Antimicrob. Chemother. 1985, 15 (Suppl. A), 175–195. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, C.R.; Friedrich, L.V.; Bosso, J.A.; White, R.L. Determination of antibiotic effect in an in vitro pharmacodynamic model: Comparison with an established animal model of infection. Antimicrob. Agents Chemother. 2002, 46, 3574–3579. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, B.V.; Diniz, A.; Palma, E.C.; Buffé, C.; Dalla Costa, T. PK-PD modeling of β-lactam antibiotics: In vitro or in vivo models? J. Antibiot. 2011, 64, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Aulin, L.B.S.; Koumans, C.I.M.; Haakman, Y.; van Os, W.; Kraakman, M.E.M.; Gooskens, J.; Smits, W.K.; Liakopoulos, A.; van Hasselt, J.G.C. Distinct evolution of colistin resistance associated with experimental resistance evolution models in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2020, 76, 533–535. [Google Scholar] [CrossRef]
- Nussbaumer-Pröll, A.; Zeitlinger, M. Use of Supplemented or Human Material to Simulate PD Behavior of Antibiotics at the Target Site In Vitro. Pharmaceutics 2020, 12, 773. [Google Scholar] [CrossRef]
- van Os, W.; Wulkersdorfer, B.; Eberl, S.; Oesterreicher, Z.; Paternostro, R.; Schwabl, P.; Willinger, B.; Weber, M.; Reiberger, T.; Zeitlinger, M. Impact of individual ascitic fluids on bacterial growth and antimicrobial effect of ceftriaxone. In Proceedings of the European Congress of Clinical Microbiology and Infectious Diseases, Online. 9–12 July 2021. Abstract 03105. [Google Scholar]
- Yadav, R.; Bulitta, J.B.; Wang, J.; Nation, R.L.; Landersdorfer, C.B. Evaluation of Pharmacokinetic/Pharmacodynamic Model-Based Optimized Combination Regimens against Multidrug-Resistant Pseudomonas aeruginosa in a Murine Thigh Infection Model by Using Humanized Dosing Schemes. Antimicrob. Agents Chemother. 2017, 61, e01268-17. [Google Scholar] [CrossRef]
- Lin, Y.W.; Zhou, Q.T.; Han, M.L.; Onufrak, N.J.; Chen, K.; Wang, J.; Forrest, A.; Chan, H.K.; Li, J. Mechanism-Based Pharmacokinetic/Pharmacodynamic Modeling of Aerosolized Colistin in a Mouse Lung Infection Model. Antimicrob. Agents Chemother. 2018, 62, e01965-17. [Google Scholar] [CrossRef] [PubMed]
- Lepak, A.J.; Marchillo, K.; Craig, W.A.; Andes, D.R. In vivo pharmacokinetics and pharmacodynamics of the lantibiotic NAI-107 in a neutropenic murine thigh infection model. Antimicrob. Agents Chemother. 2015, 59, 1258–1264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wicha, S.G.; Clewe, O.; Svensson, R.J.; Gillespie, S.H.; Hu, Y.; Coates, A.R.M.; Simonsson, U.S.H. Forecasting Clinical Dose-Response From Preclinical Studies in Tuberculosis Research: Translational Predictions With Rifampicin. Clin. Pharmacol. Ther. 2018, 104, 1208–1218. [Google Scholar] [CrossRef]
- Nielsen, E.I.; Cars, O.; Friberg, L.E. Predicting in vitro antibacterial efficacy across experimental designs with a semimechanistic pharmacokinetic-pharmacodynamic model. Antimicrob. Agents Chemother. 2011, 55, 1571–1579. [Google Scholar] [CrossRef]
- Nielsen, E.I.; Khan, D.D.; Cao, S.; Lustig, U.; Hughes, D.; Andersson, D.I.; Friberg, L.E. Can a pharmacokinetic/pharmacodynamic (PKPD) model be predictive across bacterial densities and strains? External evaluation of a PKPD model describing longitudinal in vitro data. J. Antimicrob. Chemother. 2017, 72, 3108–3116. [Google Scholar] [CrossRef]
- Martinez, M.N.; Papich, M.G.; Drusano, G.L. Dosing regimen matters: The importance of early intervention and rapid attainment of the pharmacokinetic/pharmacodynamic target. Antimicrob. Agents Chemother. 2012, 56, 2795–2805. [Google Scholar] [CrossRef] [PubMed]
- Nolting, A.; Dalla Costa, T.; Rand, K.H.; Derendorf, H. Pharmacokinetic-pharmacodynamic modeling of the antibiotic effect of piperacillin in vitro. Pharm. Res. 1996, 13, 91–96. [Google Scholar] [CrossRef]
- de la Peña, A.; Gräbe, A.; Rand, K.H.; Rehak, E.; Gross, J.; Thyroff-Friesinger, U.; Müller, M.; Derendorf, H. PK-PD modelling of the effect of cefaclor on four different bacterial strains. Int. J. Antimicrob. Agents 2004, 23, 218–225. [Google Scholar] [CrossRef]
- Sadiq, M.W.; Nielsen, E.I.; Khachman, D.; Conil, J.M.; Georges, B.; Houin, G.; Laffont, C.M.; Karlsson, M.O.; Friberg, L.E. A whole-body physiologically based pharmacokinetic (WB-PBPK) model of ciprofloxacin: A step towards predicting bacterial killing at sites of infection. J. Pharmacokinet. Pharmacodyn. 2017, 44, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Broeker, A.; Nowak, H.; Rahmel, T.; Nussbaumer-Pröll, A.; Österreicher, Z.; Zeitlinger, M.; Wicha, S.G. A pharmacometric approach to define target site-specific breakpoints for bacterial killing and resistance suppression integrating microdialysis, time-kill curves and heteroresistance data: A case study with moxifloxacin. Clin. Microbiol. Infect. 2020, 26, 1255.e1–1255.e8. [Google Scholar] [CrossRef]
- Barbour, A.M.; Schmidt, S.; Zhuang, L.; Rand, K.; Derendorf, H. Application of pharmacokinetic/pharmacodynamic modelling and simulation for the prediction of target attainment of ceftobiprole against meticillin-resistant Staphylococcus aureus using minimum inhibitory concentration and time-kill curve based approaches. Int. J. Antimicrob. Agents 2014, 43, 60–67. [Google Scholar] [CrossRef]
- Liu, P.; Rand, K.H.; Obermann, B.; Derendorf, H. Pharmacokinetic-pharmacodynamic modelling of antibacterial activity of cefpodoxime and cefixime in in vitro kinetic models. Int. J. Antimicrob. Agents 2005, 25, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Nestorov, I. Whole body pharmacokinetic models. Clin. Pharmacokinet. 2003, 42, 883–908. [Google Scholar] [CrossRef] [PubMed]
- Theil, F.P.; Guentert, T.W.; Haddad, S.; Poulin, P. Utility of physiologically based pharmacokinetic models to drug development and rational drug discovery candidate selection. Toxicol. Lett. 2003, 138, 29–49. [Google Scholar] [CrossRef]
- Khan, D.D.; Lagerbäck, P.; Cao, S.; Lustig, U.; Nielsen, E.I.; Cars, O.; Hughes, D.; Andersson, D.I.; Friberg, L.E. A mechanism-based pharmacokinetic/pharmacodynamic model allows prediction of antibiotic killing from MIC values for WT and mutants. J. Antimicrob. Chemother. 2015, 70, 3051–3060. [Google Scholar] [CrossRef] [PubMed]
- Brill, M.J.E.; Kristoffersson, A.N.; Zhao, C.; Nielsen, E.I.; Friberg, L.E. Semi-mechanistic pharmacokinetic–pharmacodynamic modelling of antibiotic drug combinations. Clin. Microbiol. Infect. 2018, 24, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, E.B.; Ledesma, K.R.; Chang, K.T.; Schwartz, M.S.; Motyl, M.R.; Tam, V.H. In vitro activity of MK-7655, a novel β-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2012, 56, 3753–3757. [Google Scholar] [CrossRef] [PubMed]
- Ly, N.S.; Bulitta, J.B.; Rao, G.G.; Landersdorfer, C.B.; Holden, P.N.; Forrest, A.; Bergen, P.J.; Nation, R.L.; Li, J.; Tsuji, B.T. Colistin and doripenem combinations against Pseudomonas aeruginosa: Profiling the time course of synergistic killing and prevention of resistance. J. Antimicrob. Chemother. 2015, 70, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- VanScoy, B.D.; Trang, M.; McCauley, J.; Conde, H.; Bhavnani, S.M.; Friedrich, L.V.; Alexander, D.C.; Ambrose, P.G. Pharmacokinetics-Pharmacodynamics of a Novel β-Lactamase Inhibitor, CB-618, in Combination with Meropenem in an In Vitro Infection Model. Antimicrob. Agents Chemother. 2016, 60, 3891–3896. [Google Scholar] [CrossRef]
- Zhao, M.; Bulman, Z.P.; Lenhard, J.R.; Satlin, M.J.; Kreiswirth, B.N.; Walsh, T.J.; Marrocco, A.; Bergen, P.J.; Nation, R.L.; Li, J.; et al. Pharmacodynamics of colistin and fosfomycin: A ‘treasure trove’ combination combats KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2017, 72, 1985–1990. [Google Scholar] [CrossRef] [PubMed]
- Sadouki, Z.; McHugh, T.D.; Aarnoutse, R.; Ortiz Canseco, J.; Darlow, C.; Hope, W.; van Ingen, J.; Longshaw, C.; Manissero, D.; Mead, A.; et al. Application of the hollow fibre infection model (HFIM) in antimicrobial development: A systematic review and recommendations of reporting. J. Antimicrob. Chemother. 2021, 76, 2252–2259. [Google Scholar] [CrossRef]
- Sy, S.K.B.; Zhuang, L.; Xia, H.; Beaudoin, M.-E.; Schuck, V.J.; Nichols, W.W.; Derendorf, H. A mathematical model-based analysis of the time–kill kinetics of ceftazidime/avibactam against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2018, 73, 1295–1304. [Google Scholar] [CrossRef]
- Kristoffersson, A.N.; Bissantz, C.; Okujava, R.; Haldimann, A.; Walter, I.; Shi, T.; Zampaloni, C.; Nielsen, E.I. A novel mechanism-based pharmacokinetic–pharmacodynamic (PKPD) model describing ceftazidime/avibactam efficacy against β-lactamase-producing Gram-negative bacteria. J. Antimicrob. Chemother. 2019, 75, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.K.B.; Zhuang, L.; Xia, H.; Schuck, V.J.; Nichols, W.W.; Derendorf, H. A model-based analysis of pharmacokinetic-pharmacodynamic (PK/PD) indices of avibactam against Pseudomonas aeruginosa. Clin. Microbiol. Infect. 2019, 25, 904.e9–904.e16. [Google Scholar] [CrossRef]
- Wicha, S.G.; Huisinga, W.; Kloft, C. Translational Pharmacometric Evaluation of Typical Antibiotic Broad-Spectrum Combination Therapies Against Staphylococcus Aureus Exploiting In Vitro Information. CPT Pharmacom. Syst. Pharmacol. 2017, 6, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.D.; Friberg, L.E.; Nielsen, E.I. A pharmacokinetic-pharmacodynamic (PKPD) model based on in vitro time-kill data predicts the in vivo PK/PD index of colistin. J. Antimicrob. Chemother. 2016, 71, 1881–1884. [Google Scholar] [CrossRef] [PubMed]
- Thorsted, A.; Nielsen, E.I.; Friberg, L.E. Pharmacodynamics of immune response biomarkers of interest for evaluation of treatment effects in bacterial infections. Int. J. Antimicrob. Agents 2020, 56, 106059. [Google Scholar] [CrossRef] [PubMed]
- Drusano, G.L.; Fregeau, C.; Liu, W.; Brown, D.L.; Louie, A. Impact of burden on granulocyte clearance of bacteria in a mouse thigh infection model. Antimicrob. Agents Chemother. 2010, 54, 4368–4372. [Google Scholar] [CrossRef]
- Drusano, G.L.; Vanscoy, B.; Liu, W.; Fikes, S.; Brown, D.; Louie, A. Saturability of granulocyte kill of Pseudomonas aeruginosa in a murine model of pneumonia. Antimicrob. Agents Chemother. 2011, 55, 2693–2695. [Google Scholar] [CrossRef]
- Guo, B.; Abdelraouf, K.; Ledesma, K.R.; Chang, K.T.; Nikolaou, M.; Tam, V.H. Quantitative impact of neutrophils on bacterial clearance in a murine pneumonia model. Antimicrob. Agents Chemother. 2011, 55, 4601–4605. [Google Scholar] [CrossRef] [PubMed]
- Boisson, M.; Jacobs, M.; Grégoire, N.; Gobin, P.; Marchand, S.; Couet, W.; Mimoz, O. Comparison of intrapulmonary and systemic pharmacokinetics of colistin methanesulfonate (CMS) and colistin after aerosol delivery and intravenous administration of CMS in critically ill patients. Antimicrob. Agents Chemother. 2014, 58, 7331–7339. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).