Sub-Inhibitory Antibiotic Exposure and Virulence in Pseudomonas aeruginosa
Abstract
1. Introduction
2. Virulence Factors in Pseudomonas aeruginosa
3. Regulation of Virulence in P. aeruginosa
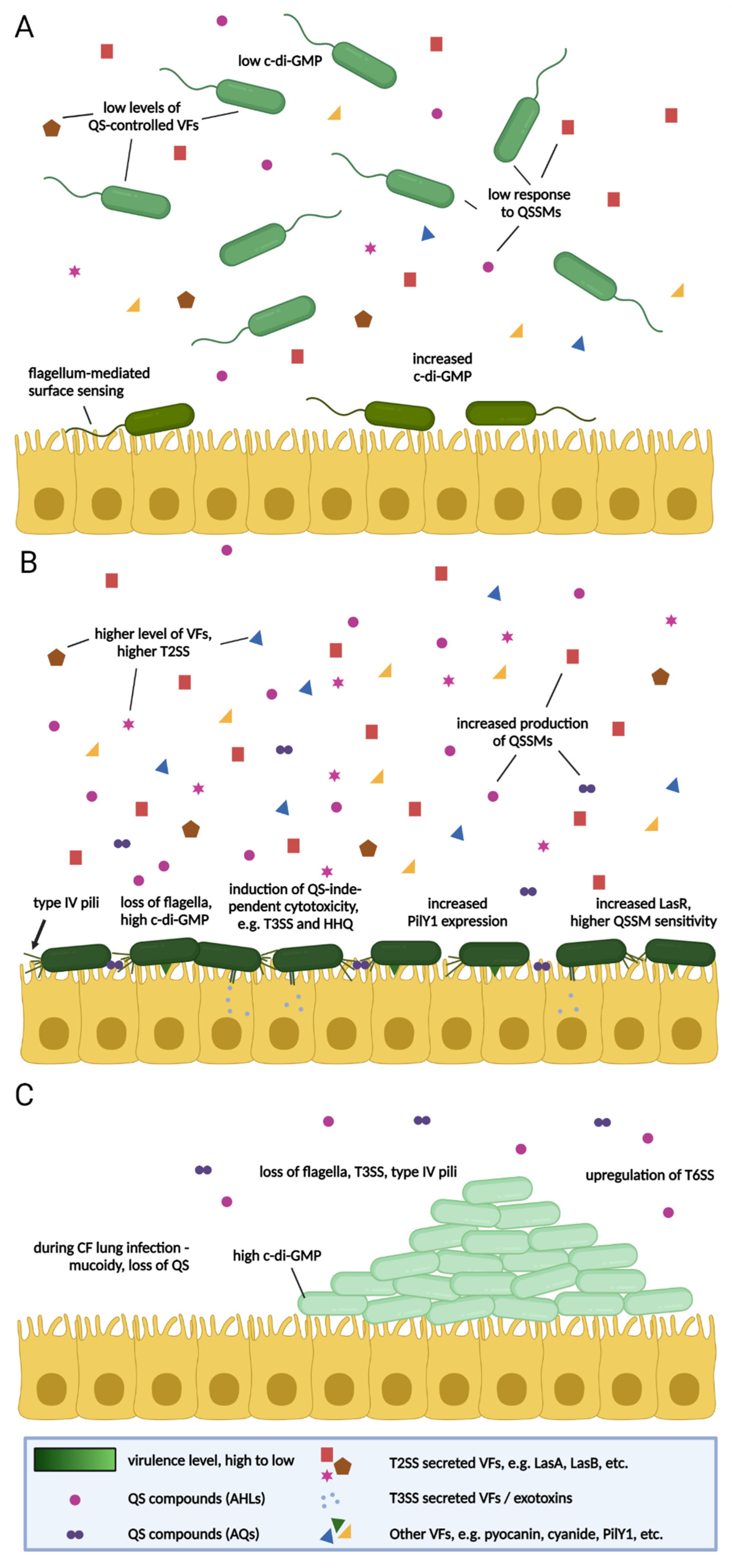
4. The Importance of Surface Attachment
5. Virulence in P. aeruginosa—Context and Time
| Pyocyanin | Pyochelin | Pyoverdine | Las A | Elastase | Swimming | Swarming | Twitching | Adherence | Biofilm | Vesicle form. | T3SS | T6SS | Alginate | Phospholipase C | Alk. Protease | Exotoxin A | 3oC12-HSL | C4-HSL | PQS family | Setting—A: Animal; B: Batch Culture; C: Cell culture; CI: Clinical | Length—S: Short (24 h); I: Intermediate (1–4 d); L: Long (>4 d) | Other Competition +/− Cooperation +/− | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycosides | |||||||||||||||||||||||
| Kanamycin | [114] | [114] [115] | [114] | [114] | [114] | [116] [114] | [116] | [116] | [115] | [114] |  |  | |||||||||||
| Gentamicin | [114,117,118] | [114,115,118] | [114] | [119] [120,121,122,123] | [114] | [114,124] [125] | [124] | [126,127,128] [122,126,129] | [116] [114,118,124,130] | [119] | [115,121,127,131,132] | [119][123,133] | [119][118,120,133] | [121,122,123] | [114,118,124,134] | [124,134] |  |  | Comp. [135,136] Cooperation [117] in vitro virul. [133] | ||||
| Amikacin | [114] | [114] | [114] | [114] | [114] | [137] | [114] | [132] | [114,134] | [134] |  |  | |||||||||||
| Netilmicin | [114] | [114] | [114] | [114] | [114] | [138] | [114] | [132] | [114] | [134] |  |  | |||||||||||
| Paromomycin | [114] | [114] | [114] | [114] | [114] | [114] | [114] |  |  | ||||||||||||||
| Neomycin B | [114] | [114] | [114] | [114] | [114] | [114] | [114] |  |  | ||||||||||||||
| Tobramycin | [139,140] [141] | [107] | [107,115] | [141] | [120,121,142,143,144] | [145] | [145] [140,141] | [127,129,146] [147] | [116,145,148] [116] [139,140,141] | [149] | [107,143,144] [145] | [149] | [107] [132] [115,121,127] | [143] | [141] [120,121,149] | [143] | [148] [142,149] | [148] [140] | [148] [107] |  |  | in vivo virul. [143,144,150] in vitro virul. [107,151] | |
| Streptomycin | [139] | [115,139] | [139] | [139] | [139] | [139] | [139] | [115,132,139] | [133,139] | [133] | [139] | [139] | 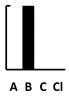 |  | Haemolysis [139] in vitro virul [133] | ||||||||
| Beta-Lactams | |||||||||||||||||||||||
| Penicillins | |||||||||||||||||||||||
| Carbenicillin | [115] | [121,122] | [122,129] | [115] | [121] [122] | 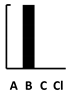 |  | Competition [135] [136] | |||||||||||||||
| Aztreonam | [132] |  | 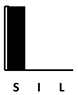 | ||||||||||||||||||||
| Ticarcillin | [129] | [132] | [133] | [133] | 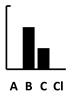 |  | in vitro virul. [133] | ||||||||||||||||
| Ampicillin | [152] | [152] | [152] | [152] | [152] | [152] | [152] | [152] |  |  | |||||||||||||
| Piperacillin/tazobactam | [153,154] | [153,154] | [153,154] | [154] |  |  | |||||||||||||||||
| Carbepenems | |||||||||||||||||||||||
| Meropenem | [129] |  |  | ||||||||||||||||||||
| Imipenem | [155] | [155] | [153] | [153] | [129,153] | [156] | [132][156] | [155] | [155] |  |  | ||||||||||||
| Cephalosporins | |||||||||||||||||||||||
| Ceftazidime | [84,155,157] | [115] | [123,143,155,157,158] [121,142,144] | [145,157,159] | [145,159,160] | [129,146] [137] | [145,157,159,160,161] | [143,144] | [132] [115] | [123,143] | [157,158] | [123,143] [121] | [142,145,155,157,158,159] | [142,145,155,157,158,159] | [157] |  |  | in vivo virul. [143,144,150,157] | |||||
| Cefotaxim | [161] | [133] | [133] |  |  | in vitro virul. [133] | |||||||||||||||||
| Ceftriaxone | [162] |  |  | ||||||||||||||||||||
| Cefepime | [155] | [155] | [147] | [155] | [155] |  |  | ||||||||||||||||
| Cefsulodin | [128] |  |  | ||||||||||||||||||||
| Macrolides | |||||||||||||||||||||||
| Azithromycin | [152] [163,164,165,166] | [166] | [167] [152] | [168][123,152,158,164,167,169,170,171] | [167,169,172,173] | [124,125,163,165,166,173] | [124,165,167,173] | [174,175] | [124,165,166,173,176,177,178] | [158,167] | [131,164,177,178] [152] | [123,172] | [158,165,170] [152] | [123] [152] | [124,158,164,168,171,173,176,179] | [124,158,165,171,173,176,179] |  |  | in vivo virul. [150,168] [164,172,173] in vitro virul. [167] | ||||
| Clarithromycin | [169] | [169] | [125] | [147] | [131] |  |  | in vivo virul. [150] | |||||||||||||||
| Erythromycin | [165] | [123,169,180] | [169] | [125,165] | [165] | [181] [182] | [183] [165,178] | [131,178] [132] | [181] [123] | [123,181] | [183] | [165] [183] |  |  | in vivo virul. [150] [181,184] | ||||||||
| Roxithromycin | [123] | [183] | [132] [185] | [123] | [123] | [183] | [183] |  |  | ||||||||||||||
| Tetracyclines | |||||||||||||||||||||||
| Tetracycline | [152] | [186] | [152] | [152,186] | [145] | [125,145] | [116,145] [139] | [145] [152] [186] | [152] | [152] [187] | [152][186] | [152] | 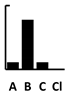 |  | in vivo virul. [145] | ||||||||
| Doxycycline | [188] | [188] | [188] | [188] | 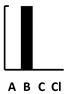 | 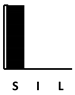 | in vivo virul. [188] | ||||||||||||||||
| Fluoroquinolones | |||||||||||||||||||||||
| Ciprofloxacin | [189] [190] | [191] | [115] | [143,144,158,191,192] | [193] [145,191] [194] | [194] [145,159,191,195] | [145,159,191,194,196] | [126,129,137,147,197] | [145] [159,192] [193] | [143,144] [145] | [191] [115,132] | [143,191] | [158,191] | [143] | [192,194,196] [145,158,159,191] [193] | [145,158,159,191] [193] | [193] |  | 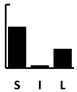 | in vivo virulence [143,144,198] | |||
| Enoxacin | [199] | [132] |  |  | |||||||||||||||||||
| Lomefloxacin | [199] |  |  | ||||||||||||||||||||
| Norfloxacin | [200] | [132] |  |  | |||||||||||||||||||
| Ofloxacin | [123] | [132] | [123] | [123] |  |  | in vivo virul. [150] | ||||||||||||||||
| Perfloxacin | [132] |  |  | ||||||||||||||||||||
| Cationic peptides | |||||||||||||||||||||||
| Polymyxin B | [128] |  | 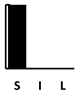 | competition [136] | |||||||||||||||||||
| Colistin | [201] [84] | [201] | [201] | [201] | [201] | [201] | [201] | [201] |  |  | |||||||||||||
| Others/Non-categorised | |||||||||||||||||||||||
| Trimethoprim | [197] | 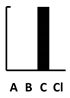 | 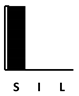 | ||||||||||||||||||||
| Sulfamethoxazole | [183] | [183] | [183] |  |  | ||||||||||||||||||
| Chloramphenicol | [125] | [128] | [131] | 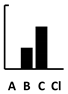 |  | ||||||||||||||||||
| Rifaximin (Ansamycin) | [159,190] |  |  | ||||||||||||||||||||
| Vancomycin(Glycopeptide) | [152] | [190] | [152] | [152] | [152] | [190] | [190] | [190] | [190] | [190] |  |  | |||||||||||
| Nalidixic acid(Quinolone) | [132] |  |  | ||||||||||||||||||||
| Clindamycin(Lincasamide) | [202] | [131] |  |  | |||||||||||||||||||
6. The Impact of Sub-MIC Antibiotics on Virulence in P. aeruginosa
6.1. Shorter-Term Studies
6.1.1. In Vitro Studies
Mode of Growth
Biofilm
Surface-Attached
Co-Operation between P. aeruginosa Strains
Competition between P. aeruginosa Strains and Other Bacteria
6.1.2. In Vivo Studies
6.2. Longer-Term Studies
6.2.1. In Vitro Studies
Planktonic
Biofilm
6.2.2. In Vivo Studies
Co-Operation between P. aeruginosa Strains
Acute Infections
Chronic Infections
7. The Translatability of In Vitro Virulence Assay Findings
8. Future Directions
8.1. What Is a Sub-MIC?
8.2. Can We Weaponise Environmental Composition?
8.3. How Do We Avoid Falling Prey to the Garbage in, Garbage out Principle?
9. Concluding Statement
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gooderham, W.J.; Hancock, R.E.W. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009, 33, 279–294. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Filloux, A. Fit and resistant is a worst case scenario with bacterial pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, 20360–20361. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Bush, K.; Harbarth, S.; Paul, M.; Rex, J.H.; Tacconelli, E.; Thwaites, G.E. Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 2020, 18, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Fourteen years in resistance. Int. J. Antimicrob. Agents 2012, 39, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. V The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef] [PubMed]
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Williams, H.D.; Davies, J.C. Basic science for the chest physician: Pseudomonas aeruginosa and the cystic fibrosis airway. Thorax 2012, 67, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Byrd, M.S.; Pang, B.; Hong, W.; Waligora, E.; Juneau, R.; Armbruster, C.E.; Weimer, K.E.D.; Murrah, K.; Mann, E.E.; Lu, H.; et al. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect. Immun. 2011, 79, 3087–3095. [Google Scholar] [CrossRef]
- Tamber, S.; Ochs, M.M.; Hancock, R.E.W. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 45–54. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2019, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Mulat, M.; Pandita, A.; Khan, F. Medicinal Plant Compounds for Combating the Multi-drug Resistant Pathogenic Bacteria: A Review. Curr. Pharm. Biotechnol. 2019, 20, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Krzyżek, P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front. Microbiol. 2019, 10, 2473. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Wright, G.D.; Hung, D.T.; Helmann, J.D. How antibiotics kill bacteria: New models needed? Nat. Med. 2013, 19, 544–545. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zeng, J.; Wang, X.; Drlica, K.; Zhao, X. Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. USA 2019, 116, 10064–10071. [Google Scholar] [CrossRef]
- Lam, P.-L.; Wong, R.S.-M.; Lam, K.-H.; Hung, L.-K.; Wong, M.-M.; Yung, L.-H.; Ho, Y.-W.; Wong, W.-Y.; Hau, D.K.-P.; Gambari, R.; et al. The role of reactive oxygen species in the biological activity of antimicrobial agents: An updated mini review. Chem. Biol. Interact. 2020, 320, 109023. [Google Scholar] [CrossRef] [PubMed]
- Dolan, S.K. Current Knowledge and Future Directions in Developing Strategies to Combat Pseudomonas aeruginosa Infection. J. Mol. Biol. 2020, 432, 5509–5528. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J.B. Why is Pseudomonas aeruginosa a pathogen? F1000 Biol. Rep. 2010, 2, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Irazoqui, J.E.; Troemel, E.R.; Feinbaum, R.L.; Luhachack, L.G.; Cezairliyan, B.O.; Ausubel, F.M. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010, 6, e1000982. [Google Scholar] [CrossRef]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E.; et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef]
- Filloux, A. Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function. Front. Microbiol. 2011, 2, 155. [Google Scholar] [CrossRef] [PubMed]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.G.; Baumann, C.; Merdes, A.; Soscia, C.; Rattei, T.; Hachani, A.; Jones, C.; Bennett, K.L.; Filloux, A.; Superti-Furga, G.; et al. Internalization of Pseudomonas aeruginosa Strain PAO1 into Epithelial Cells Is Promoted by Interaction of a T6SS Effector with the Microtubule Network. mBio 2015, 6, e00712. [Google Scholar] [CrossRef]
- Jolly, A.L.; Takawira, D.; Oke, O.O.; Whiteside, S.A.; Chang, S.W.; Wen, E.R.; Quach, K.; Evans, D.J.; Fleiszig, S.M.J. Pseudomonas aeruginosa-induced bleb-niche formation in epithelial cells is independent of actinomyosin contraction and enhanced by loss of cystic fibrosis transmembrane-conductance regulator osmoregulatory function. mBio 2015, 6, e02533. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef]
- Gallique, M.; Bouteiller, M.; Merieau, A. The type VI secretion system: A dynamic system for bacterial communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef]
- Sana, T.G.; Berni, B.; Bleves, S. The T6SSs of Pseudomonas aeruginosa Strain PAO1 and Their Effectors: Beyond Bacterial-Cell Targeting. Front. Cell. Infect. Microbiol. 2016, 6, 61. [Google Scholar] [CrossRef]
- Ryall, B.; Davies, J.C.; Wilson, R.; Shoemark, A.; Williams, H.D. Pseudomonas aeruginosa, cyanide accumulation and lung function in CF and non-CF bronchiectasis patients. Eur. Respir. J. 2008, 32, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.D.; Zlosnik, J.E.A.; Ryall, B. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 2007, 52, 1–71. [Google Scholar] [CrossRef]
- Ugidos, A.; Morales, G.; Rial, E.; Williams, H.D.; Rojo, F. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ. Microbiol. 2008, 10, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lee, K.-M.; Go, J.; Ryu, J.-C.; Ryu, J.-H.; Yoon, S.S. The ferrichrome receptor A as a new target for Pseudomonas aeruginosa virulence attenuation. FEMS Microbiol. Lett. 2016, 363, fnw104. [Google Scholar] [CrossRef] [PubMed]
- Rada, B.; Leto, T.L. Redox warfare between airway epithelial cells and Pseudomonas: Dual oxidase versus pyocyanin. Immunol. Res. 2009, 43, 198–209. [Google Scholar] [CrossRef]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Price-Whelan, A.; Cornell, W.C.; Dietrich, L.E.P. Interdependency of respiratory metabolism and phenazine-associated physiology in Pseudomonas aeruginosa PA14. J. Bacteriol. 2020, 202, e00700-19. [Google Scholar] [CrossRef]
- Williams, P.; Cámara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 2010, 313, 96–102. [Google Scholar] [CrossRef]
- Bomberger, J.M.; Maceachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef]
- Cecil, J.D.; Sirisaengtaksin, N.; O’Brien-Simpson, N.M.; Krachler, A.M. Outer Membrane Vesicle-Host Cell Interactions. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- MacDonald, I.A.; Kuehna, M.J. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Bru, J.-L.; Rawson, B.; Trinh, C.; Whiteson, K.; Høyland-Kroghsbo, N.M.; Siryaporn, A. PQS Produced by the Pseudomonas aeruginosa Stress Response Repels Swarms Away from Bacteriophage and Antibiotics. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Bitto, N.J.; Chapman, R.; Pidot, S.; Costin, A.; Lo, C.; Choi, J.; D’Cruze, T.; Reynolds, E.C.; Dashper, S.G.; Turnbull, L.; et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017, 7, 7072. [Google Scholar] [CrossRef]
- Vrla, G.D.; Esposito, M.; Zhang, C.; Kang, Y.; Seyedsayamdost, M.R.; Gitai, Z. Cytotoxic alkyl-quinolones mediate surface-induced virulence in Pseudomonas aeruginosa. PLoS Pathog. 2020, 16, e1008867. [Google Scholar] [CrossRef]
- Mashburn, L.M.; Whiteley, M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 2005, 437, 422–425. [Google Scholar] [CrossRef]
- Mashburn-Warren, L.; Howe, J.; Garidel, P.; Richter, W.; Steiniger, F.; Roessle, M.; Brandenburg, K.; Whiteley, M. Interaction of quorum signals with outer membrane lipids: Insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 2008, 69, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26. [Google Scholar] [CrossRef]
- Hauser, A.R. Pseudomonas aeruginosa: So many virulence factors, so little time. Crit. Care Med. 2011, 39, 2193–2194. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Schneper, L.; Kumari, H.; Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013, 41, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Winzer, K.; Chan, W.C.; Cámara, M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007, 362, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef]
- Pesci, E.C. New signal molecules on the quorum-sensing block: Response. Trends Microbiol. 2000, 8, 103–104. [Google Scholar] [CrossRef]
- Dubern, J.-F.; Diggle, S.P. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 2008, 4, 882–888. [Google Scholar] [CrossRef]
- Heeb, S.; Fletcher, M.P.; Chhabra, S.R.; Diggle, S.P.; Williams, P.; Cámara, M. Quinolones: From antibiotics to autoinducers. FEMS Microbiol. Rev. 2011, 35, 247–274. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhou, L.; Jin, K.; Jiang, H.; He, Y.-W. Quorum sensing systems differentially regulate the production of phenazine-1-carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Sci. Rep. 2016, 6, 30352. [Google Scholar] [CrossRef]
- Cornelis, P. Putting an end to the Pseudomonas aeruginosa IQS controversy. Microbiologyopen 2020, 9, e962. [Google Scholar] [CrossRef]
- Trottmann, F.; Franke, J.; Ishida, K.; García-Altares, M.; Hertweck, C. A Pair of Bacterial Siderophores Releases and Traps an Intercellular Signal Molecule: An Unusual Case of Natural Nitrone Bioconjugation. Angew. Chem. Int. Ed. Engl. 2019, 58, 200–204. [Google Scholar] [CrossRef]
- Schuster, M.; Greenberg, E.P. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Dees, J.L.; Ibberson, C.B.; Huse, H.K.; Mathiesen, I.H.; Kirketerp-Møller, K.; Wolcott, R.D.; Rumbaugh, K.P.; Bjarnsholt, T.; Whiteley, M. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. USA 2018, 115, E5125–E5134. [Google Scholar] [CrossRef]
- Laventie, B.-J.J.; Sangermani, M.; Estermann, F.; Manfredi, P.; Planes, R.; Hug, I.; Jaeger, T.; Meunier, E.; Broz, P.; Jenal, U. A Surface-Induced Asymmetric Program Promotes Tissue Colonization by Pseudomonas aeruginosa. Cell Host Microbe 2019, 25, 140–152. [Google Scholar] [CrossRef]
- Siryaporn, A.; Kuchma, S.L.; O’Toole, G.A.; Gitai, Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl. Acad. Sci. USA 2014, 111, 16860–16865. [Google Scholar] [CrossRef]
- Chuang, S.K.; Vrla, G.D.; Fröhlich, K.S.; Gitai, Z. Surface association sensitizes Pseudomonas aeruginosa to quorum sensing. Nat. Commun. 2019, 10, 4118. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Y.; Sugiman-Marangos, S.; Harvey, H.; Bell, S.D.; Charlton, C.L.; Junop, M.S.; Burrows, L.L. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 2015, 290, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Persat, A.; Inclan, Y.F.; Engel, J.N.; Stone, H.A.; Gitai, Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 7563–7568. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Gonzalez, D.; Mavridou, D.A.; Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 2018, 41, 15–20. [Google Scholar] [CrossRef]
- Kang, P.J.; Hauser, A.R.; Apodaca, G.; Fleiszig, S.M.; Wiener-Kronish, J.; Mostov, K.; Engel, J.N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 1997, 24, 1249–1262. [Google Scholar] [CrossRef]
- Huse, H.K.; Kwon, T.; Zlosnik, J.E.A.; Speert, D.P.; Marcotte, E.M.; Whiteley, M. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio 2010, 1, e00199-10–e00199-17. [Google Scholar] [CrossRef]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Hoboth, C.; Hoffmann, R.; Eichner, A.; Henke, C.; Schmoldt, S.; Imhof, A.; Heesemann, J.; Hogardt, M. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 2009, 200, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Ryall, B.; Zlosnik, J.E.A.E.A.; Speert, D.P.P.; Bundy, J.G.G.; Williams, H.D.D. Metabolic adaptations of Pseudomonas aeruginosa during cystic fibrosis chronic lung infections. Environ. Microbiol. 2013, 15, 398–408. [Google Scholar] [CrossRef]
- Yang, L.; Jelsbak, L.; Marvig, R.L.; Damkiær, S.; Workman, C.T.; Rau, M.H.; Hansen, S.K.; Folkesson, A.; Johansen, H.K.; Ciofu, O.; et al. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. USA 2011, 108, 7481–7486. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 2010, 300, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Heesemann, J. Microevolution of pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr. Top. Microbiol. Immunol. 2013, 91–118. [Google Scholar] [CrossRef]
- Cullen, L.; Weiser, R.; Olszak, T.; Maldonado, R.F.; Moreira, A.S.; Slachmuylders, L.; Brackman, G.; Paunova-Krasteva, T.S.; Zarnowiec, P.; Czerwonka, G.; et al. Phenotypic characterisation of an international Pseudomonas aeruginosa reference panel: Strains of cystic fibrosis origin show less in vivo virulence than non-CF strains. Microbiology 2015. [Google Scholar] [CrossRef]
- Ryall, B.; Carrara, M.; Zlosnik, J.E.A.; Behrends, V.; Lee, X.; Wong, Z.; Lougheed, K.E.; Williams, H.D. The Mucoid Switch in Pseudomonas aeruginosa Represses Quorum Sensing Systems and Leads to Complex Changes to Stationary Phase Virulence Factor Regulation. PLoS ONE 2014, 9, e96166. [Google Scholar] [CrossRef]
- Deretic, V.; Martin, D.W.; Schurr, M.J.; Mudd, M.H.; Hibler, N.S.; Curcic, R.; Boucher, J.C. Conversion to mucoidy in Pseudomonas aeruginosa. Nat. Biotechnol. 1993, 11, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.F.; Ohman, D.E. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 2009, 72, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Ryall, B.; Wang, X.; Bundy, J.G.; Williams, H.D. Metabolic profiling of Pseudomonas aeruginosa demonstrates that the anti-sigma factor MucA modulates osmotic stress tolerance. Mol. Biosyst. 2010, 6, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.D.; Behrends, V.; Bundy, J.G.; Ryall, B.; Zlosnik, J.E.A. Hypertonic Saline Therapy in Cystic Fibrosis: Do Population Shifts Caused by the Osmotic Sensitivity of Infecting Bacteria Explain the Effectiveness of this Treatment? Front. Microbiol. 2010, 1, 120. [Google Scholar] [CrossRef] [PubMed]
- Michon, A.-L.; Jumas-Bilak, E.; Chiron, R.; Lamy, B.; Marchandin, H. Advances toward the elucidation of hypertonic saline effects on Pseudomonas aeruginosa from cystic fibrosis patients. PLoS ONE 2014, 9, e90164. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.A.; Fothergill, J.L.; Paterson, S.; Brockhurst, M.A.; Winstanley, C. Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium. BMC Microbiol. 2013, 13, 170. [Google Scholar] [CrossRef]
- Mowat, E.; Paterson, S.; Fothergill, J.L.; Wright, E.; Ledson, M.J.; Walshaw, M.J.; Brockhurst, M.; Winstanley, C. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 2011, 183, 1674–1679. [Google Scholar] [CrossRef]
- Long, J.; Zaborina, O.; Holbrook, C.; Zaborin, A.; Alverdy, J. Depletion of intestinal phosphate after operative injury activates the virulence of P aeruginosa causing lethal gut-derived sepsis. Surgery 2008, 144, 189–197. [Google Scholar] [CrossRef]
- Line, L.; Alhede, M.; Kolpen, M.; Kühl, M.; Ciofu, O.; Bjarnsholt, T.; Moser, C.; Toyofuku, M.; Nomura, N.; Høiby, N.; et al. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 2014, 5, 554. [Google Scholar] [CrossRef]
- Palmer, K.L.; Brown, S.; Whiteley, M. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 4449–4455. [Google Scholar] [CrossRef]
- Schobert, M.; Tielen, P. Contribution of oxygen-limiting conditions to persistent infection of Pseudomonas aeruginosa. Future Microbiol. 2010, 49, 603–621. [Google Scholar] [CrossRef]
- Schobert, M.; Jahn, D. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 2010, 300, 549–556. [Google Scholar] [CrossRef]
- Schreiber, K.; Boes, N.; Eschbach, M.; Jaensch, L.; Wehland, J.; Bjarnsholt, T.; Givskov, M.; Hentzer, M.; Schobert, M. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 2006, 188, 659–668. [Google Scholar] [CrossRef]
- Eschbach, M.; Schreiber, K.; Trunk, K.; Buer, J.; Jahn, D.; Schobert, M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 2004, 186, 4596–4604. [Google Scholar] [CrossRef]
- Sabra, W.; Lünsdorf, H.; Zeng, A.-P. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology 2003, 149, 2789–2795. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Fujii, T.; Takaya, N.; Maseda, H.; Sawada, I.; Nakajima, T.; Uchiyama, H. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2007, 189, 4969–4972. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.H.; Davies, D.G.; Gilbert, P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.S.; Hennigan, R.F.; Hilliard, G.M.; Ochsner, U.A.; Parvatiyar, K.; Kamani, M.C.; Allen, H.L.; DeKievit, T.R.; Gardner, P.R.; Schwab, U.; et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: Relationships to cystic fibrosis pathogenesis. Dev. Cell 2002, 3, 593–603. [Google Scholar] [CrossRef]
- Van Alst, N.E.; Wellington, M.; Clark, V.L.; Haidaris, C.G.; Iglewski, B.H. Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect. Immun. 2009, 77, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Van Alst, N.E.; Picardo, K.F.; Iglewski, B.H.; Haidaris, C.G. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect. Immun. 2007, 75, 3780–3790. [Google Scholar] [CrossRef] [PubMed]
- Behrends, V.; Geier, B.; Williams, H.D.; Bundy, J.G. Direct assessment of metabolite utilization by Pseudomonas aeruginosa during growth on artificial sputum medium. Appl. Environ. Microbiol. 2013, 79, 2467–2470. [Google Scholar] [CrossRef][Green Version]
- Sonnleitner, E.; Bläsi, U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 2014, 10, e1004440. [Google Scholar] [CrossRef]
- Behrends, V.; Bell, T.J.; Liebeke, M.; Cordes-Blauert, A.; Ashraf, S.N.; Nair, C.; Zlosnik, J.E.A.; Williams, H.D.; Bundy, J.G. Metabolite profiling to characterize disease-related bacteria: Gluconate excretion by Pseudomonas aeruginosa mutants and clinical isolates from cystic fibrosis patients. J. Biol. Chem. 2013, 288, 15098–15109. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Moreno, R.; Fajardo, A.; Martínez-Solano, L.; Escalante, R.; Rojo, F.; Martínez, J.L. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 2010, 12, 3196–3212. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.-H.; Zhang, X.-F.; Zhang, L.-H. The global regulator Crc plays a multifaceted role in modulation of type III secretion system in Pseudomonas aeruginosa. Microbiologyopen 2013, 2, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Luckett, J.C.A.; Darch, O.; Watters, C.; Abuoun, M.; Wright, V.; Paredes-Osses, E.; Ward, J.; Goto, H.; Heeb, S.; Pommier, S.; et al. A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog. 2012, 8, e1002854. [Google Scholar] [CrossRef]
- Everett, J.; Turner, K.; Cai, Q.; Gordon, V.; Whiteley, M.; Rumbaugh, K. Arginine Is a Critical Substrate for the Pathogenesis of Pseudomonas aeruginosa in Burn Wound Infections. mBio 2017, 8, e02160-16. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.-G.; Merritt, J.H.; Hampton, T.H.; Hodgkinson, J.T.; Janecek, M.; Spring, D.R.; Welch, M.; O’Toole, G.A. 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J. Bacteriol. 2011, 193, 6770–6780. [Google Scholar] [CrossRef]
- Anderson, G.G.; Moreau-Marquis, S.; Stanton, B.A.; O’Toole, G.A. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 2008, 76, 1423–1433. [Google Scholar] [CrossRef]
- Gödeke, J.; Pustelny, C.; Häussler, S. Recycling of Peptidyl-tRNAs by Peptidyl-tRNA Hydrolase Counteracts Azithromycin-Mediated Effects on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 1617. [Google Scholar] [CrossRef]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; de Lorenzo, V. Pseudomonas putida KT2440 Strain Metabolizes Glucose through a Cycle Formed by Enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and Pentose Phosphate Pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef] [PubMed]
- Kohlstedt, M.; Wittmann, C. GC-MS-based 13 C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 2019, 54, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Meylan, S.; Porter, C.B.M.M.; Yang, J.H.; Belenky, P.; Gutierrez, A.; Lobritz, M.A.; Park, J.; Kim, S.H.; Moskowitz, S.M.; Collins, J.J. Carbon Sources Tune Antibiotic Susceptibility in Pseudomonas aeruginosa via Tricarboxylic Acid Cycle Control. Cell Chem. Biol. 2017, 24, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Su, Y.-B.; Li, H.; Han, Y.; Guo, C.; Tian, Y.; Peng, X.-X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Chen, S.; Sysoeva, T.A.; You, L. Universal antibiotic tolerance arising from antibiotic-triggered accumulation of pyocyanin in Pseudomonas aeruginosa. PLoS Biol. 2019, 17, e3000573. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Lee, J.-W.; Javaid, A.; Park, S.-K.; Kim, Y.-M. Inhibition of biofilm and virulence properties of Pseudomonas aeruginosa by sub-inhibitory concentrations of aminoglycosides. Microb. Pathog. 2020, 146, 104249. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Brown, M.R. Novel modes of action of aminoglycoside antibiotics against Pseudomonas aeruginosa. Lancet 1988, 1, 1359–1361. [Google Scholar] [CrossRef]
- Jones, C.; Allsopp, L.; Horlick, J.; Kulasekara, H.; Filloux, A. Subinhibitory concentration of kanamycin induces the Pseudomonas aeruginosa type VI secretion system. PLoS ONE 2013, 8, e81132. [Google Scholar] [CrossRef]
- Vasse, M.; Noble, R.J.; Akhmetzhanov, A.R.; Torres-Barceló, C.; Gurney, J.; Benateau, S.; Gougat-Barbera, C.; Kaltz, O.; Hochberg, M.E. Antibiotic stress selects against cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, 546–551. [Google Scholar] [CrossRef]
- Gupta, P.; Sarkar, A.; Sandhu, P.; Daware, A.; Das, M.C.; Akhter, Y.; Bhattacharjee, S. Potentiation of antibiotic against Pseudomonas aeruginosa biofilm: A study with plumbagin and gentamicin. J. Appl. Microbiol. 2017, 123, 246–261. [Google Scholar] [CrossRef] [PubMed]
- Kadurugamuwa, J.L.; Beveridge, T.J. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 1997, 40, 615–621. [Google Scholar] [CrossRef]
- Warren, R.L.; Baker, N.R.; Johnson, J.; Stapleton, M.J. Selective inhibition of the accumulation of extracellular proteases of Pseudomonas aeruginosa by gentamicin and tobramycin. Antimicrob. Agents Chemother. 1985, 27, 468–472. [Google Scholar] [CrossRef]
- Geers, T.A.; Baker, N.R. The effect of sublethal levels of antibiotics on the pathogenicity of Pseudomonas aeruginosa for tracheal tissue. J. Antimicrob. Chemother. 1987, 19, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Ogaard, A.R.; Bjøro, K.; Bukholm, G.; Berdal, B.P. Pseudomonas aeruginosa virulence factors: Modifications by sub-inhibitory concentrations of carbenicillin or gentamicin. Acta Pathol. Microbiol. Immunol. Scand. B 1986, 94, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Mizukane, R.; Hirakata, Y.; Kaku, M.; Ishii, Y.; Furuya, N.; Ishida, K.; Koga, H.; Kohno, S.; Yamaguchi, K. Comparative in vitro exoenzyme-suppressing activities of azithromycin and other macrolide antibiotics against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1994, 38, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Bahari, S.; Zeighami, H.; Mirshahabi, H.; Roudashti, S.; Haghi, F. Inhibition of Pseudomonas aeruginosa quorum sensing by subinhibitory concentrations of curcumin with gentamicin and azithromycin. J. Glob. Antimicrob. Resist. 2017, 10, 21–28. [Google Scholar] [CrossRef]
- Kawamura-Sato, K.; Iinuma, Y.; Hasegawa, T.; Horii, T.; Yamashino, T.; Ohta, M. Effect of subinhibitory concentrations of macrolides on expression of flagellin in Pseudomonas aeruginosa and Proteus mirabilis. Antimicrob. Agents Chemother. 2000, 44, 2869–2872. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Kim, S.O.; Davidson, R.J.; Hoban, D.J.; Nicolle, L.E. Effect of subinhibitory concentrations of Ciprofloxacin and gentamicin on the adherence of Pseudomonas aeruginosa to Vero cells and voided uroepithelial cells. Chemotherapy 1993, 39, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Geers, T.A.; Baker, N.R. The effect of sublethal concentrations of aminoglycosides on adherence of Pseudomonas aeruginosa to hamster tracheal epithelium. J. Antimicrob. Chemother. 1987, 19, 561–568. [Google Scholar] [CrossRef]
- Di Martino, P.; Rebière-Huët, J.; Hulen, C. Effects of Antibiotics on Adherence of Pseudomonas aeruginosa and Pseudomonas fluorescens to A549 Pneumocyte Cells. Chemotherapy 2000, 46, 129–134. [Google Scholar] [CrossRef]
- Wolter, J.M.; McCormack, J.G. The effect of subinhibitory concentrations of antibiotics on adherence of Pseudomonas aeruginosa to cystic fibrosis (CF) and non-CF-affected tracheal epithelial cells. J. Infect. 1998, 37, 217–223. [Google Scholar] [CrossRef]
- Marr, A.K.; Overhage, J.; Bains, M.; Hancock, R.E.W. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 2007, 153, 474–482. [Google Scholar] [CrossRef]
- Hosoyama, T.; Yuge, T.; Takahashi, S.; Suzuki, T.; Okubo, T.; Iyobe, S.; Nakagawa, A. Inhibitory effect of antimicrobial agents on alginate production in Pseudomonas aeruginosa. J. Antibiot. 1999, 52, 65–67. [Google Scholar] [CrossRef][Green Version]
- Majtán, V.; Hybenová, D. Inhibition of Pseudomonas aeruginosa alginate expression by subinhibitory concentrations of antibiotics. Folia Microbiol. 1996, 41, 61–64. [Google Scholar] [CrossRef]
- Hostacká, A.; Majtán, V. Alterations in Pseudomonas aeruginosa exoproducts by sub-MICs of some antibiotics. Folia Microbiol. 1993, 38, 349–352. [Google Scholar] [CrossRef]
- Baskin, H.; Bayrakal, V.; Bahar, İ.H. Effects of Gentamicin, Amikacin and Netilmicin on the Pathogenic Factors and Two Extracellular Quorum Sensing Systems of Pseudomonas aeruginosa Strains. Turkiye Klin. J. Med. Sci. 2012, 32, 1319–1326. [Google Scholar] [CrossRef][Green Version]
- Al-Ani, F.Y.; Al-Shibib, A.S. In vitro pyocin activity of Pseudomonas aeruginosa strains pretreated with antibiotics. Folia Microbiol. 1985, 30, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Stevens, G.S.; Ottolenghi, A.C. Pyocin sensitivity of pseudomonas aeruginosa pretreated with antibiotics. Can. J. Microbiol. 1977, 23, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.; Pascual, A.; Perea, E.J. Effect of preincubation of Pseudomonas aeruginosa in subinhibitory concentrations of amikacin, ceftazidime and ciprofloxacin on adherence to plastic catheters. Chemotherapy 1991, 37, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Furneri, P.M.; Garozzo, A.; Musumarra, M.P.; Scuderi, A.C.; Russo, A.; Bonfiglio, G. Effects on adhesiveness and hydrophobicity of sub-inhibitory concentrations of netilmicin. Int. J. Antimicrob. Agents 2003, 22, 164–167. [Google Scholar] [CrossRef]
- Khan, F.; Lee, J.-W.; Pham, D.T.N.; Lee, J.-H.; Kim, H.-W.; Kim, Y.-K.; Kim, Y.-M. Streptomycin mediated biofilm inhibition and suppression of virulence properties in Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 2020, 104, 799–816. [Google Scholar] [CrossRef]
- Babić, F.; Venturi, V.; Maravić-Vlahovicek, G. Tobramycin at subinhibitory concentration inhibits the RhlI/R quorum sensing system in a Pseudomonas aeruginosa environmental isolate. BMC Infect. Dis. 2010, 10, 148. [Google Scholar] [CrossRef]
- Chanda, W.; Joseph, T.P.; Padhiar, A.A.; Guo, X.; Min, L.; Wang, W.; Lolokote, S.; Ning, A.; Cao, J.; Huang, M.; et al. Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp. Ther. Med. 2017, 14, 4328–4338. [Google Scholar] [CrossRef] [PubMed]
- Garske, L.A.; Beatson, S.A.; Leech, A.J.; Walsh, S.L.; Bell, S.C. Sub-inhibitory concentrations of ceftazidime and tobramycin reduce the quorum sensing signals of Pseudomonas aeruginosa. Pathology 2004, 36, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Grimwood, K.; To, M.; Rabin, H.R.; Woods, D.E. Inhibition of Pseudomonas aeruginosa exoenzyme expression by subinhibitory antibiotic concentrations. Antimicrob. Agents Chemother. 1989, 33, 41–47. [Google Scholar] [CrossRef]
- Grimwood, K.; To, M.; Rabin, H.R.; Woods, D.E. Subinhibitory antibiotics reduce Pseudomonas aeruginosa tissue injury in the rat lung model. J. Antimicrob. Chemother. 1989, 24, 937–945. [Google Scholar] [CrossRef]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [CrossRef]
- Vishwanath, S.; Guay, C.M.; Ramphal, R. Effects of subminimal inhibitory concentrations of antibiotics on the adherence of Pseudomonas aeruginosa to tracheobronchial mucin. J. Antimicrob. Chemother. 1987, 19, 579–583. [Google Scholar] [CrossRef]
- Ferrara, A.; Dos Santos, C.; Lupi, A. Effect of different antibacterial agents and surfactant protein-A (SP-A) on adherence of some respiratory pathogens to bronchial epithelial cells. Int. J. Antimicrob. Agents 2001, 17, 401–405. [Google Scholar] [CrossRef]
- Tahrioui, A.; Duchesne, R.; Bouffartigues, E.; Rodrigues, S.; Maillot, O.; Tortuel, D.; Hardouin, J.; Taupin, L.; Groleau, M.-C.; Dufour, A.; et al. Extracellular DNA release, quorum sensing, and PrrF1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin-enhanced biofilm formation. NPJ Biofilms Microbiomes 2019, 5, 15. [Google Scholar] [CrossRef]
- Koeppen, K.; Barnaby, R.; Jackson, A.A.; Gerber, S.A.; Hogan, D.A.; Stanton, B.A. Tobramycin reduces key virulence determinants in the proteome of Pseudomonas aeruginosa outer membrane vesicles. PLoS ONE 2019, 14, e0211290. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Tateda, K.; Matsumoto, T.; Miyazaki, S.; Watanabe, A.; Nukiwa, T.; Yamaguchi, K. Macrolide-treated Pseudomonas aeruginosa induces paradoxical host responses in the lungs of mice and a high mortality rate. J. Antimicrob. Chemother. 2002, 50, 59–66. [Google Scholar] [CrossRef]
- Anderson, G.G.; Kenney, T.F.; Macleod, D.L.; Henig, N.R.; O’Toole, G.A. Eradication of Pseudomonas aeruginosa biofilms on cultured airway cells by a fosfomycin/tobramycin antibiotic combination. Pathog. Dis. 2013, 67, 39–45. [Google Scholar] [CrossRef]
- Shen, L.; Shi, Y.; Zhang, D.; Wei, J.; Surette, M.G.; Duan, K. Modulation of secreted virulence factor genes by subinhibitory concentrations of antibiotics in Pseudomonas aeruginosa. J. Microbiol. 2008, 46, 441–447. [Google Scholar] [CrossRef]
- Fonseca, A.P.; Sousa, J.C. Effect of antibiotic-induced morphological changes on surface properties, motility and adhesion of nosocomial Pseudomonas aeruginosa strains under different physiological states. J. Appl. Microbiol. 2007, 103, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.P.; Extremina, C.; Fonseca, A.F.; Sousa, J.C. Effect of subinhibitory concentration of piperacillin/tazobactam on Pseudomonas aeruginosa. J. Med. Microbiol. 2004, 53, 903–910. [Google Scholar] [CrossRef] [PubMed]
- El-Mowafy, S.A.; Abd El Galil, K.H.; Habib, E.-S.E.; Shaaban, M.I. Quorum sensing inhibitory activity of sub-inhibitory concentrations of β-lactams. Afr. Health Sci. 2017, 17, 199–207. [Google Scholar] [CrossRef]
- Bagge, N.; Schuster, M.; Hentzer, M.; Ciofu, O.; Givskov, M.; Greenberg, E.P.; Høiby, N. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 2004, 48, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.M.; Ahmad, I.; Baig, M.H.; Khan, M.S.; Khan, M.S.; Hassan, I.; Al-Shabib, N.A. Broad-spectrum inhibition of AHL-regulated virulence factors and biofilms by sub-inhibitory concentrations of ceftazidime. RSC Adv. 2016, 6, 27952–27962. [Google Scholar] [CrossRef]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Roudashti, S.; Zeighami, H.; Mirshahabi, H.; Bahari, S.; Soltani, A.; Haghi, F. Synergistic activity of sub-inhibitory concentrations of curcumin with ceftazidime and ciprofloxacin against Pseudomonas aeruginosa quorum sensing related genes and virulence traits. World J. Microbiol. Biotechnol. 2017, 33, 50. [Google Scholar] [CrossRef] [PubMed]
- Otani, S.; Hiramatsu, K.; Hashinaga, K.; Komiya, K.; Umeki, K.; Kishi, K.; Kadota, J.-I. Sub-minimum inhibitory concentrations of ceftazidime inhibit Pseudomonas aeruginosa biofilm formation. J. Infect. Chemother. 2018, 24, 428–433. [Google Scholar] [CrossRef]
- Bassaris, H.P.; Lianou, P.E.; Votta, E.G.; Papavassiliou, J.T. Effects of subinhibitory concentrations of cefotaxime on adhesion and polymorphonuclear leukocyte function with gram-negative bacteria. J. Antimicrob. Chemother. 1984, 14 (Suppl. B), 91–96. [Google Scholar] [CrossRef] [PubMed]
- Onaolapo, J.A.; Salami, J.O. Effect of subminimum inhibitory concentration of ceftriaxone on adherence of Pseudomonas aeruginosa to inert surfaces in an experimental model. Afr. J. Med. Med. Sci. 1995, 24, 275–281. [Google Scholar] [PubMed]
- Pérez-Martínez, I.; Haas, D. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3399–3405. [Google Scholar] [CrossRef]
- Hoffmann, N.; Lee, B.; Hentzer, M.; Rasmussen, T.B.; Song, Z.; Johansen, H.K.; Givskov, M.; Høiby, N. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr(-/-) mice. Antimicrob. Agents Chemother. 2007, 51, 3677–3687. [Google Scholar] [CrossRef] [PubMed]
- Seleem, N.M.; El Latif, H.K.A.; Shaldam, M.A.; El-Ganiny, A. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1687–1702. [Google Scholar] [CrossRef]
- Stellari, F.; Bergamini, G.; Sandri, A.; Donofrio, G.; Sorio, C.; Ruscitti, F.; Villetti, G.; Assael, B.M.; Melotti, P.; Lleo, M.M. In vivo imaging of the lung inflammatory response to Pseudomonas aeruginosa and its modulation by azithromycin. J. Transl. Med. 2015, 13, 251. [Google Scholar] [CrossRef]
- Nalca, Y.; Jänsch, L.; Bredenbruch, F.; Geffers, R.; Buer, J.; Häussler, S. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: A global approach. Antimicrob. Agents Chemother. 2006, 50, 1680–1688. [Google Scholar] [CrossRef]
- Köhler, T.; Perron, G.G.; Buckling, A.; van Delden, C. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 2010, 6, e1000883. [Google Scholar] [CrossRef]
- Molinari, G.; Guzmán, C.A.; Pesce, A.; Schito, G.C. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 1993, 31, 681–688. [Google Scholar] [CrossRef]
- Swatton, J.E.; Davenport, P.W.; Maunders, E.A.; Griffin, J.L.; Lilley, K.S.; Welch, M. Impact of Azithromycin on the Quorum Sensing-Controlled Proteome of Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0147698. [Google Scholar] [CrossRef]
- Tateda, K.; Comte, R.; Pechere, J.C.; Köhler, T.; Yamaguchi, K.; Van Delden, C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001, 45, 1930–1933. [Google Scholar] [CrossRef]
- Nguyen, D.; Emond, M.J.; Mayer-Hamblett, N.; Saiman, L.; Marshall, B.C.; Burns, J.L. Clinical response to azithromycin in cystic fibrosis correlates with in vitro effects on Pseudomonas aeruginosa phenotypes. Pediatr. Pulmonol. 2007, 42, 533–541. [Google Scholar] [CrossRef]
- Bala, A.; Kumar, R.; Harjai, K. Inhibition of quorum sensing in Pseudomonas aeruginosa by azithromycin and its effectiveness in urinary tract infections. J. Med. Microbiol. 2011, 60, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Carfartan, G.; Gerardin, P.; Turck, D.; Husson, M.-O. Effect of subinhibitory concentrations of azithromycin on adherence of Pseudomonas aeruginosa to bronchial mucins collected from cystic fibrosis patients. J. Antimicrob. Chemother. 2004, 53, 686–688. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vranes, J. Effect of subminimal inhibitory concentrations of azithromycin on adherence of Pseudomonas aeruginosa to polystyrene. J. Chemother. 2000, 12, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Favre-Bonté, S.; Köhler, T.; Van Delden, C. Biofilm formation by Pseudomonas aeruginosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 2003, 52, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Ichimiya, T.; Takeoka, K.; Hiramatsu, K.; Hirai, K.; Yamasaki, T.; Nasu, M. The Influence of Azithromycin on the Biofilm Formation of Pseudomonas aeruginosa in vitro. Chemotherapy 1996, 42, 186–191. [Google Scholar] [CrossRef]
- Nagino, K.; Kobayashi, H. Influence of macrolides on mucoid alginate biosynthetic enzyme from Pseudomonas aeruginosa. Clin. Microbiol. Infect. 1997, 3, 432–439. [Google Scholar] [CrossRef][Green Version]
- Kai, T.; Tateda, K.; Kimura, S.; Ishii, Y.; Ito, H.; Yoshida, H.; Kimura, T.; Yamaguchi, K. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009, 22, 483–486. [Google Scholar] [CrossRef]
- Kita, E.; Sawaki, M.; Oku, D.; Hamuro, A.; Mikasa, K.; Konishi, M.; Emoto, M.; Takeuchi, S.; Narita, N.; Kashiba, S. Suppression of virulence factors of Pseudomonas aeruginosa by erythromycin. J. Antimicrob. Chemother. 1991, 27, 273–284. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kaku, M.; Mizukane, R.; Ishida, K.; Furuya, N.; Matsumoto, T.; Tateda, K.; Yamaguchi, K. Potential effects of erythromycin on host defense systems and virulence of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1992, 36, 1922–1927. [Google Scholar] [CrossRef][Green Version]
- Tsang, K.W.; Ng, P.; Ho, P.L.; Chan, S.; Tipoe, G.; Leung, R.; Sun, J.; Ho, J.C.; Ip, M.S.; Lam, W.K. Effects of erythromycin on Pseudomonas aeruginosa adherence to collagen and morphology in vitro. Eur. Respir. J. 2003, 21, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Bruchmann, J.; Kirchen, S.; Schwartz, T. Sub-inhibitory concentrations of antibiotics and wastewater influencing biofilm formation and gene expression of multi-resistant Pseudomonas aeruginosa wastewater isolates. Environ. Sci. Pollut. Res. Int. 2013, 20, 3539–3549. [Google Scholar] [CrossRef]
- Sofer, D.; Gilboa-Garber, N.; Belz, A.; Garber, N.C. ‘Subinhibitory’ Erythromycin Represses Production of Pseudomonas aeruginosa Lectins, Autoinducer and Virulence Factors. Chemotherapy 1999, 45, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Dupont, M.J.; Lapointe, J.R. Effect on Pseudomonas aeruginosa alginate expression of direct plating and culture of fresh cystic fibrosis sputum on to pseudomonas isolation agar containing subinhibitory concentrations of roxithromycin and rifampicin. J. Antimicrob. Chemother. 1995, 36, 231–236. [Google Scholar] [CrossRef] [PubMed]
- LeVatte, M.A.; Woods, D.E.; Shahrabadi, M.S.; Semple, R.; Sokol, P.A. Subinhibitory concentrations of tetracycline inhibit surface expression of the Pseudomonas aeruginosa ferripyochelin binding protein in vivo. J. Antimicrob. Chemother. 1990, 26, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Shibl, A.M.; Al-Sowaygh, I.A. Antibiotic inhibition of protease production by Pseudomonas aeruginosa. J. Med. Microbiol. 1980, 13, 345–348. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I. Doxycycline interferes with quorum sensing-mediated virulence factors and biofilm formation in gram-negative bacteria. World J. Microbiol. Biotechnol. 2013, 29, 949–957. [Google Scholar] [CrossRef]
- Hossain, M.A.; Sattenapally, N.; Parikh, H.I.; Li, W.; Rumbaugh, K.P.; German, N.A. Design, synthesis, and evaluation of compounds capable of reducing Pseudomonas aeruginosa virulence. Eur. J. Med. Chem. 2020, 185, 111800. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Coppo, E.; Barbieri, R.; Debbia, E.A.; Marchese, A. The effect of sub-inhibitory concentrations of rifaximin on urease production and on other virulence factors expressed by Klebsiella pneumoniae, Proteus mirabilis, Pseudomonas aeruginosa and Staphylococcus aureus. J. Chemother. 2017, 29, 67–73. [Google Scholar] [CrossRef]
- Gupta, P.; Chhibber, S.; Harjai, K. Subinhibitory concentration of ciprofloxacin targets quorum sensing system of Pseudomonas aeruginosa causing inhibition of biofilm formation & reduction of virulence. Indian J. Med. Res. 2016, 143, 643–651. [Google Scholar] [CrossRef]
- Wassermann, T.; Meinike Jørgensen, K.; Ivanyshyn, K.; Bjarnsholt, T.; Khademi, S.M.H.; Jelsbak, L.; Høiby, N.; Ciofu, O. The phenotypic evolution of Pseudomonas aeruginosa populations changes in the presence of subinhibitory concentrations of ciprofloxacin. Microbiology 2016, 162, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Gao, Y.; Yin, D.; Song, Y.; Kang, J.; Li, X.; Zhang, Z.; Feng, X.; Duan, J. The effect of the sub-minimal inhibitory concentration and the concentrations within resistant mutation window of ciprofloxacin on MIC, swimming motility and biofilm formation of Pseudomonas aeruginosa. Microb. Pathog. 2019, 137, 103765. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Abdelsamad, A.; Wassermann, T.; Porse, A.; Becker, J.; Sommer, M.O.A.A.; Høiby, N.; Ciofu, O. The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: Beyond selection of antibiotic resistance. NPJ Biofilms Microbiomes 2020, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Jedrey, H.; Lilley, K.S.; Welch, M. Ciprofloxacin binding to GyrA causes global changes in the proteome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2018, 365, 134. [Google Scholar] [CrossRef]
- Ahmed, M.N.; Porse, A.; Sommer, M.O.A.; Høiby, N.; Ciofu, O. Evolution of Antibiotic Resistance in Biofilm and Planktonic Pseudomonas aeruginosa Populations Exposed to Subinhibitory Levels of Ciprofloxacin. Antimicrob. Agents Chemother. 2018, 62, e00320-18. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.R.; Beumer, H.; Hoepelman, A.I.; Rozenberg-Arska, M.; Verhoef, J. Changes in adherence of respiratory pathogens to HEp-2 cells induced by subinhibitory concentrations of sparfloxacin, ciprofloxacin, and trimethoprim. Antimicrob. Agents Chemother. 1993, 37, 885–888. [Google Scholar] [CrossRef][Green Version]
- Ravizzola, G.; Pirali, F.; Paolucci, A.; Terlenghi, L.; Peroni, L.; Colombi, A.; Turano, A. Reduced virulence in ciprofloxacin-resistant variants of Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 1987, 20, 825–829. [Google Scholar] [CrossRef]
- Sonstein, S.A.; Burnham, J.C. Effect of low concentrations of quinolone antibiotics on bacterial virulence mechanisms. Diagn. Microbiol. Infect. Dis. 1993, 16, 277–289. [Google Scholar] [CrossRef]
- Kumar, A.; Ting, Y.-P. Effect of sub-inhibitory antibacterial stress on bacterial surface properties and biofilm formation. Colloids Surf. B Biointerfaces 2013, 111, 747–754. [Google Scholar] [CrossRef]
- Cummins, J.; Reen, F.J.; Baysse, C.; Mooij, M.J.; O’Gara, F. Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology 2009, 155, 2826–2837. [Google Scholar] [CrossRef]
- Lianou, P.E.; Bassaris, H.P.; Votta, E.G.; Papavassiliou, J.T. Interaction of subminimal inhibitory concentrations of clindamycin and gram-negative aerobic organisms: Effects on adhesion and polymorphonuclear leukocyte function. J. Antimicrob. Chemother. 1985, 15, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Siehnel, R.J.; Garudathri, J.; Staudinger, B.J.; Hisert, K.B.; Ozer, E.A.; Hauser, A.R.; Eng, J.K.; Manoil, C.; Singh, P.K.; et al. In vivo Proteome of Pseudomonas aeruginosa in Airways of Cystic Fibrosis Patients. J. Proteome Res. 2019, 18, 2601–2612. [Google Scholar] [CrossRef]
- Crousilles, A.; Maunders, E.; Bartlett, S.; Fan, C.; Ukor, E.-F.; Abdelhamid, Y.; Baker, Y.; Floto, A.; Spring, D.R.; Welch, M. Which microbial factors really are important in Pseudomonas aeruginosa infections? Future Microbiol. 2015, 10, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.H.; Everett, J.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014, 10, e1004518. [Google Scholar] [CrossRef]
- Chung, J.C.S.; Rzhepishevska, O.; Ramstedt, M.; Welch, M. Type III secretion system expression in oxygen-limited Pseudomonas aeruginosa cultures is stimulated by isocitrate lyase activity. Open Biol. 2013, 3, 120131. [Google Scholar] [CrossRef] [PubMed]
- Perinbam, K.; Chacko, J.V.; Kannan, A.; Digman, M.A.; Siryaporn, A. A Shift in Central Metabolism Accompanies Virulence Activation in Pseudomonas aeruginosa. mBio 2020, 11, e02730-18. [Google Scholar] [CrossRef]
- Hogardt, M.; Schubert, S.; Adler, K.; Götzfried, M.; Heesemann, J. Sequence variability and functional analysis of MutS of hypermutable Pseudomonas aeruginosa cystic fibrosis isolates. Int. J. Med. Microbiol. 2006, 296, 313–320. [Google Scholar] [CrossRef]
- Rogers, G.B.; van der Gast, C.J.; Serisier, D.J. Predominant pathogen competition and core microbiota divergence in chronic airway infection. ISME J. 2015, 9, 217–225. [Google Scholar] [CrossRef]
- Trejo-Hernández, A.; Andrade-Domínguez, A.; Hernández, M.; Encarnación, S. Interspecies competition triggers virulence and mutability in Candida albicans-Pseudomonas aeruginosa mixed biofilms. ISME J. 2014, 8, 1974–1988. [Google Scholar] [CrossRef]
- Yassien, M.; Khardori, N.; Ahmedy, A.; Toama, M. Modulation of biofilms of Pseudomonas aeruginosa by quinolones. Antimicrob. Agents Chemother. 1995, 39, 2262–2268. [Google Scholar] [CrossRef]
- She, P.; Luo, Z.; Chen, L.; Wu, Y. Efficacy of levofloxacin against biofilms of Pseudomonas aeruginosa isolated from patients with respiratory tract infections in vitro. Microbiologyopen 2019, 8, e00720. [Google Scholar] [CrossRef]
- Okuda, J.; Yamane, S.; Nagata, S.; Kunikata, C.; Suezawa, C.; Yasuda, M. The Pseudomonas aeruginosa dnaK gene is involved in bacterial translocation across the intestinal epithelial cell barrier. Microbiology 2017, 163, 1208–1216. [Google Scholar] [CrossRef]
- Anderson, G.G.; Yahr, T.L.; Lovewell, R.R.; O’Toole, G.A. The Pseudomonas aeruginosa magnesium transporter MgtE inhibits transcription of the type III secretion system. Infect. Immun. 2010, 78, 1239–1249. [Google Scholar] [CrossRef]
- Wilder, C.N.; Diggle, S.P.; Schuster, M. Cooperation and cheating in Pseudomonas aeruginosa: The roles of the las, rhl and pqs quorum-sensing systems. ISME J. 2011, 5, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Hotterbeekx, A.; Kumar-Singh, S.; Goossens, H.; Malhotra-Kumar, S. In vivo and In vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 2017, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Price, K.E.; Naimie, A.A.; Griffin, E.F.; Bay, C.; O’Toole, G.A. Tobramycin-Treated Pseudomonas aeruginosa PA14 Enhances Streptococcus constellatus 7155 Biofilm Formation in a Cystic Fibrosis Model System. J. Bacteriol. 2016, 198, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Dehbashi, S.; Alikhani, M.Y.; Tahmasebi, H.; Arabestani, M.R. The inhibitory effects of Staphylococcus aureus on the antibiotic susceptibility and virulence factors of Pseudomonas aeruginosa: A549 cell line model. AMB Express 2021, 11, 50. [Google Scholar] [CrossRef]
- Lenhard, J.R.; Smith, N.M.; Quach, C.D.; Nguyen, T.Q.; Doan, L.H.; Chau, J. Bacterial brothers in arms: Cooperation of Staphylococcus aureus and Pseudomonas aeruginosa during antimicrobial exposure. J. Antimicrob. Chemother. 2019, 74, 2657–2665. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.M.; Martinez-Garcia, E.; Xavier, J.; Durham, W.M.; Kolter, R.; Kim, W.; Foster, K.R. Biofilm Formation As a Response to Ecological Competition. PLoS Biol. 2015, 13, e1002191. [Google Scholar] [CrossRef] [PubMed]
- Neves, P.R.; McCulloch, J.A.; Mamizuka, E.M.; Lincopan, N. PSEUDOMONAS | Pseudomonas aeruginosa. Encycl. Food Microbiol. Sec. Ed. 2014, 253–260. [Google Scholar] [CrossRef]
- Michel-Briand, Y.; Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef]
- Brazas, M.D.; Hancock, R.E.W. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 3222–3227. [Google Scholar] [CrossRef]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 2020, 2894. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.; Van Gennip, M.; Bjarnsholt, T.; Jensen, P.Ø.; Lee, B.; Hougen, H.P.; Calum, H.; Ciofu, O.; Givskov, M.; Molin, S.; et al. Novel experimental Pseudomonas aeruginosa lung infection model mimicking long-term host-pathogen interactions in cystic fibrosis. APMIS 2009, 117, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Hershenson, M.B.; Zhou, Y.; Sajjan, U. Azithromycin increases survival and reduces lung inflammation in cystic fibrosis mice. Inflamm. Res. 2009, 58, 491–501. [Google Scholar] [CrossRef]
- Tsai, W.C.; Rodriguez, M.L.; Young, K.S.; Deng, J.C.; Thannickal, V.J.; Tateda, K.; Hershenson, M.B.; Standiford, T.J. Azithromycin blocks neutrophil recruitment in Pseudomonas endobronchial infection. Am. J. Respir. Crit. Care Med. 2004, 170, 1331–1339. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Leal, T.; Bergamini, G.; Huaux, F.; Panin, N.; Noel, S.; Dhooghe, B.; Haaf, J.B.; Mauri, P.; Motta, S.; Di Silvestre, D.; et al. Azithromycin Attenuates Pseudomonas-Induced Lung Inflammation by Targeting Bacterial Proteins Secreted in the Cultured Medium. Front. Immunol. 2016, 7, 15. [Google Scholar] [CrossRef]
- Parnham, M.J. Immunomodulatory effects of antimicrobials in the therapy of respiratory tract infections. Curr. Opin. Infect. Dis. 2005, 18, 125–131. [Google Scholar] [CrossRef]
- Idris, S.F.; Chilvers, E.R.; Haworth, C.; McKeon, D.; Condliffe, A.M. Azithromycin therapy for neutrophilic airways disease: Myth or magic? Thorax 2009, 64, 186–189. [Google Scholar] [CrossRef]
- Torres-Barceló, C.; Kojadinovic, M.; Moxon, R.; MacLean, R.C. The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic. Proc. Biol. Sci. 2015, 282, 20150885. [Google Scholar] [CrossRef]
- Jørgensen, K.M.; Wassermann, T.; Jensen, P.Ø.; Hengzuang, W.; Molin, S.; Høiby, N.; Ciofu, O. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Morero, N.R.; Monti, M.R.; Argaraña, C.E. Effect of ciprofloxacin concentration on the frequency and nature of resistant mutants selected from Pseudomonas aeruginosa mutS and mutT hypermutators. Antimicrob. Agents Chemother. 2011, 55, 3668–3676. [Google Scholar] [CrossRef]
- Migliorini, L.B.; Brüggemann, H.; de Sales, R.O.; Koga, P.C.M.; de Souza, A.V.; Martino, M.D.V.; Galhardo, R.S.; Severino, P. Mutagenesis Induced by Sub-Lethal Doses of Ciprofloxacin: Genotypic and Phenotypic Differences Between the Pseudomonas aeruginosa Strain PA14 and Clinical Isolates. Front. Microbiol. 2019, 10, 1553. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Larson Freeman, T.J.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef]
- García-Contreras, R.; Nuñez-López, L.; Jasso-Chávez, R.; Kwan, B.W.; Belmont, J.A.; Rangel-Vega, A.; Maeda, T.; Wood, T.K. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2015, 9, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.N.; Porse, A.; Abdelsamad, A.; Sommer, M.; Høiby, N.; Ciofu, O. Lack of the Major Multifunctional Catalase KatA in Pseudomonas aeruginosa Accelerates Evolution of Antibiotic Resistance in Ciprofloxacin-Treated Biofilms. Antimicrob. Agents Chemother. 2019, 63, e00766-19. [Google Scholar] [CrossRef]
- Köhler, T.; Buckling, A.; van Delden, C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. USA 2009, 106, 6339–6344. [Google Scholar] [CrossRef]
- Kumar, M.; Rao, M.; Mathur, T.; Barman, T.K.; Joshi, V.; Chaira, T.; Singhal, S.; Pandya, M.; Al Khodor, S.; Upadhyay, D.J.; et al. Azithromycin Exhibits Activity Against Pseudomonas aeruginosa in Chronic Rat Lung Infection Model. Front. Microbiol. 2021, 12, 603151. [Google Scholar] [CrossRef]
- Nagata, T.; Mukae, H.; Kadota, J.; Hayashi, T.; Fujii, T.; Kuroki, M.; Shirai, R.; Yanagihara, K.; Tomono, K.; Koji, T.; et al. Effect of erythromycin on chronic respiratory infection caused by Pseudomonas aeruginosa with biofilm formation in an experimental murine model. Antimicrob. Agents Chemother. 2004, 48, 2251–2259. [Google Scholar] [CrossRef]
- Fujii, T.; Kadota, J.; Kawakami, K.; Iida, K.; Shirai, R.; Kaseda, M.; Kawamoto, S.; Kohno, S. Long term effect of erythromycin therapy in patients with chronic Pseudomonas aeruginosa infection. Thorax 1995, 50, 1246–1252. [Google Scholar] [CrossRef]
- Nagai, H.; Shishido, H.; Yoneda, R.; Yamaguchi, E.; Tamura, A.; Kurashima, A. Long-term low-dose administration of erythromycin to patients with diffuse panbronchiolitis. Respiration 1991, 58, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Kudoh, S.; Azuma, A.; Yamamoto, M.; Izumi, T.; Ando, M. Improvement of survival in patients with diffuse panbronchiolitis treated with low-dose erythromycin. Am. J. Respir. Crit. Care Med. 1998, 157, 1829–1832. [Google Scholar] [CrossRef]
- Burr, L.D.; Rogers, G.B.; Chen, A.C.-H.; Hamilton, B.R.; Pool, G.F.; Taylor, S.L.; Venter, D.; Bowler, S.D.; Biga, S.; McGuckin, M.A. Macrolide Treatment Inhibits Pseudomonas aeruginosa Quorum Sensing in Non-Cystic Fibrosis Bronchiectasis. An Analysis from the Bronchiectasis and Low-Dose Erythromycin Study Trial. Ann. Am. Thorac. Soc. 2016, 13, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Saiman, L.; Marshall, B.C.; Mayer-Hamblett, N.; Burns, J.L.; Quittner, A.L.; Cibene, D.A.; Coquillette, S.; Fieberg, A.Y.; Accurso, F.J.; Campbell, P.W.; et al. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: A randomized controlled trial. JAMA 2003, 290, 1749–1756. [Google Scholar] [CrossRef]
- Wolter, J.; Seeney, S.; Bell, S.; Bowler, S.; Masel, P.; McCormack, J. Effect of long term treatment with azithromycin on disease parameters in cystic fibrosis: A randomised trial. Thorax 2002, 57, 212–216. [Google Scholar] [CrossRef]
- van Delden, C.; Köhler, T.; Brunner-Ferber, F.; François, B.; Carlet, J.; Pechère, J.-C. Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: A randomized controlled trial. Intensive Care Med. 2012, 38, 1118–1125. [Google Scholar] [CrossRef]
- Pirzada, O.M.; McGaw, J.; Taylor, C.J.; Everard, M.L. Improved lung function and body mass index associated with long-term use of Macrolide antibiotics. J. Cyst. Fibros. 2003, 2, 69–71. [Google Scholar] [CrossRef]
- Hansen, C.R.; Pressler, T.; Koch, C.; Høiby, N. Long-term azitromycin treatment of cystic fibrosis patients with chronic Pseudomonas aeruginosa infection; an observational cohort study. J. Cyst. Fibros. 2005, 4, 35–40. [Google Scholar] [CrossRef]
- Ratjen, F.; Saiman, L.; Mayer-Hamblett, N.; Lands, L.C.; Kloster, M.; Thompson, V.; Emmett, P.; Marshall, B.; Accurso, F.; Sagel, S.; et al. Effect of azithromycin on systemic markers of inflammation in patients with cystic fibrosis uninfected with Pseudomonas aeruginosa. Chest 2012, 142, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.D.; Assoufi, B.K.; Hodson, M.E. Regular three monthly oral ciprofloxacin in adult cystic fibrosis patients infected with Pseudomonas aeruginosa. Respir. Med. 1993, 87, 587–593. [Google Scholar] [CrossRef]
- Littlewood, J.M.; Bevan, A.; Connett, G.; Conway, S.; Govan, J.; Hodson, M. Antibiotic Treatment for Cystic Fibrosis: Report of the UK Cystic Fibrosis Trust Antibiotic Group; Cystic Fibrosis Trust: London, UK, 2009. [Google Scholar]
- Neve, R.L.; Carrillo, B.D.; Phelan, V.V. Commercial porcine gastric mucin contributes to variation in production of small molecule virulence factors by Pseudomonas aeruginosa when cultured in different formulations of artificial sputum medium. bioRxiv 2021. [Google Scholar] [CrossRef]
- Phan, J.; Ranjbar, S.; Kagawa, M.; Gargus, M.; Hochbaum, A.I.; Whiteson, K.L. Thriving Under Stress: Pseudomonas aeruginosa Outcompetes the Background Polymicrobial Community Under Treatment Conditions in a Novel Chronic Wound Model. Front. Cell. Infect. Microbiol. 2020, 10, 569685. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Whiteley, M.; Rumbaugh, K.P.; Stewart, P.S.; Jensen, P.Ø.; Frimodt-Møller, N. The importance of understanding the infectious microenvironment. Lancet. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Belanger, C.R.; Lee, A.H.-Y.; Pletzer, D.; Dhillon, B.K.; Falsafi, R.; Hancock, R.E.W. Identification of novel targets of azithromycin activity against Pseudomonas aeruginosa grown in physiologically relevant media. Proc. Natl. Acad. Sci. USA 2020, 117, 33519–33529. [Google Scholar] [CrossRef]
- Buyck, J.M.; Plésiat, P.; Traore, H.; Vanderbist, F.; Tulkens, P.M.; Van Bambeke, F. Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin. Infect. Dis. 2012, 55, 534–542. [Google Scholar] [CrossRef][Green Version]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef]
- Sriramulu, D.D.; Lünsdorf, H.; Lam, J.S.; Römling, U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005, 54, 667–676. [Google Scholar] [CrossRef]
- Lozano, C.; López, M.; Rojo-Bezares, B.; Sáenz, Y. Antimicrobial Susceptibility Testing in Pseudomonas aeruginosa Biofilms: One Step Closer to a Standardized Method. Antibiot. 2020, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.R.; Geers, T. Protection of tracheal explants infected with Pseudomonas aeruginosa by subinhibitory concentrations of aminoglycosides. In The Influence of Antibiotics on the Host-Parasite Relationship II.; Springer: Berlin/Heidelberg, Germany, 1985; pp. 277–288. [Google Scholar]
- Mailloux, R.J.; Bériault, R.; Lemire, J.; Singh, R.; Chénier, D.R.; Hamel, R.D.; Appanna, V.D. The Tricarboxylic Acid Cycle, an Ancient Metabolic Network with a Novel Twist. PLoS ONE 2007, 2, e690. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.D.; Yuan, J.; Kimball, E.H.; Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar] [CrossRef]
- Schumacher, J.; Behrends, V.; Pan, Z.; Brown, D.R.; Heydenreich, F.; Lewis, M.R.; Bennett, M.H.; Razzaghi, B.; Komorowski, M.; Barahona, M.; et al. Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. mBio 2013, 4, e00881-13. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nolan, C.; Behrends, V. Sub-Inhibitory Antibiotic Exposure and Virulence in Pseudomonas aeruginosa. Antibiotics 2021, 10, 1393. https://doi.org/10.3390/antibiotics10111393
Nolan C, Behrends V. Sub-Inhibitory Antibiotic Exposure and Virulence in Pseudomonas aeruginosa. Antibiotics. 2021; 10(11):1393. https://doi.org/10.3390/antibiotics10111393
Chicago/Turabian StyleNolan, Charlotte, and Volker Behrends. 2021. "Sub-Inhibitory Antibiotic Exposure and Virulence in Pseudomonas aeruginosa" Antibiotics 10, no. 11: 1393. https://doi.org/10.3390/antibiotics10111393
APA StyleNolan, C., & Behrends, V. (2021). Sub-Inhibitory Antibiotic Exposure and Virulence in Pseudomonas aeruginosa. Antibiotics, 10(11), 1393. https://doi.org/10.3390/antibiotics10111393







