Abstract
The emergence of infectious diseases promises to be one of the leading mortality factors in the healthcare sector. Although several drugs are available on the market, newly found microorganisms carrying multidrug resistance (MDR) against which existing drugs cannot function effectively, giving rise to escalated antibiotic dosage therapies and the need to develop novel drugs, which require time, money, and manpower. Thus, the exploitation of antimicrobials has led to the production of MDR bacteria, and their prevalence and growth are a major concern. Novel approaches to prevent antimicrobial drug resistance are in practice. Nanotechnology-based innovation provides physicians and patients the opportunity to overcome the crisis of drug resistance. Nanoparticles have promising potential in the healthcare sector. Recently, nanoparticles have been designed to address pathogenic microorganisms. A multitude of processes that can vary with various traits, including size, morphology, electrical charge, and surface coatings, allow researchers to develop novel composite antimicrobial substances for use in different applications performing antimicrobial activities. The antimicrobial activity of inorganic and carbon-based nanoparticles can be applied to various research, medical, and industrial uses in the future and offer a solution to the crisis of antimicrobial resistance to traditional approaches. Metal-based nanoparticles have also been extensively studied for many biomedical applications. In addition to reduced size and selectivity for bacteria, metal-based nanoparticles have proven effective against pathogens listed as a priority, according to the World Health Organization (WHO). Moreover, antimicrobial studies of nanoparticles were carried out not only in vitro but in vivo as well in order to investigate their efficacy. In addition, nanomaterials provide numerous opportunities for infection prevention, diagnosis, treatment, and biofilm control. This study emphasizes the antimicrobial effects of nanoparticles and contrasts nanoparticles’ with antibiotics’ role in the fight against pathogenic microorganisms. Future prospects revolve around developing new strategies and products to prevent, control, and treat microbial infections in humans and other animals, including viral infections seen in the current pandemic scenarios.
1. Introduction
Antibiotics are drugs that may either destroy microbes or suppress them. Based on their target population, antibiotics are graded as antibacterial, antifungal, and antiviral. The term antibiotic is generally used to refer to most antibacterial compounds [1]. Antibiotics have been used for generations to prevent and treat infections and aid with many medical therapies, varying from organ transplantation to chemical therapy. Inhibition of enzymes, DNA interference, RNA and protein synthesis, and destruction of the membrane structure are well-known antimicrobial pathways of antibiotics [2]. Depending on the formation and the mode of multi-drug resistance, many groups or classes of antibiotics have been developed. Thus, it is virtually impossible to imagine the future without antibiotics.
Unfortunately, due to the rise of antibiotic resistance in microorganisms, this may become a reality [3]. Emerging drug resistance in microorganisms relates directly to the exploitation of antibiotics, excessive doses of medications leading to increased toxicity, prolonged hospitalization, and growing fatality [4], their widespread use in agriculture, and the lack of development of new antibiotics [5]. Moreover, the modern convenience of mobility (of products and infected people) helps disseminate pathogens across the globe at a rate never seen before [6]. Apart from the adverse socio-economic consequences, the public health challenge of antibiotic resistance to pandemic infectious diseases is serious [6]. World Health Organization (WHO) has recognized antimicrobial resistance (AMR) as one of the major challenges to public health [7].
Feasible pathways known to prevent antibiotic resistance in microbes include decreasing the consumption of antimicrobial medicines and improving drug release, modifying antibiotic targets, developing medicines for degrading or modifying enzymes in microorganisms, developing a biofilm coating containing the bacteria, and avoiding antibiotic exposure [8]. Eventually, such developments will lead towards reduced drug accumulation in microbial cells or a brief intracellular residency of drugs that do not effectively reach therapeutic concentrations [9]. Currently, however, higher doses and repeated drug administration are prevalent, contributing significantly to adverse side effects on animals and humans. Resistance has been developed towards several forms of antibiotics widely used against pathogenic microorganisms [10]. Most significantly, no new kinds of antibiotics have been developed in recent years. Furthermore, the development and promotion of new antibiotics is a costly and time-consuming procedure, requiring new compounds and multiple clinical studies and licensing [11]. The possibility that bacterial resistance towards all-new antibiotics will develop promptly would lead to decreasing antibiotic usage and declining sales, further exacerbating public health and economic condition. Therefore, the failure of antibiotic advancement would eventually lead to an elevated mortality threat from infections [12]. Consequentially, modern therapies are critical for overcoming these challenges.
Medicine has been modernized with the introduction of nanotechnology, the most significant breakthrough in recent years. There is a steady rise in the market for nanotechnology products. Nanotechnology, a groundbreaking science, will influence our efforts to improve human health. The medical industry has studied the longevity, efficiency, durability, flexibility, and inimitable physicochemical characteristics of nanoparticles. They are being utilized in numerous therapeutic approaches, such as the targeted delivery of medications, prognostic visual monitoring of therapy, and even tumor identification [13,14]. Several conventional approaches have been used to synthesize nanoparticles, for example, effective techniques such as physical vapor deposition, laser ablation, sputtering, melt mixing, and chemical methods such as photo-reduction, sol—gel, thermolysis, and micro-emulsion. As a result of these techniques, nanoparticles can become unstable, harmful compounds can attach to nanoparticles’ surface, and hazardous by-products can develop.
The biogenic nanoparticle synthesis relies on green methodologies. Green synthesis advantages include the production of stable nanoparticles, the use of a biomass-based surface coating that provides extra active surface areas for biological interaction, the exclusion of dangerous formation of byproducts, and additional stabilizing or reducing factors that eventually make the procedure economical [15,16].However, constant human exposure to nanoparticles in the work environment may lead to unexpected danger to human health. Furthermore, secondary exposure to nanoparticles may occur through inhaling nanoparticles in the form of atmospheric toxins. Occasionally, these inhaled nanoparticles escape the immune system and are dispersed throughout the body, creating issues with systemic health.
In this study, we concentrated on antimicrobial drug resistance, nanoparticles, and their correlation with conventional antibiotics.
2. Microbes
Microbes are single-celled micro-organisms. They are so small that thousands of them can fit onto the point of a needle and can only be seen through a microscope. Microbes are the oldest forms of life on our planet [17], and microbial traces date back to over 3.5 billion years ago. The waste would not rot without microbial growth, and therefore less air would be accessible to breathe. Such microbes are present nearly everywhere–in food, soil, bone, water, humans, animals, and plants. Microbes are also considered microscopic creatures because they can exist everywhere around the environment. Some of them thrive in the cold, and others survive the extreme heat. Certain microbes require oxygen while others do not. Some microbes primarily induce certain forms of infectious conditions [17,18]. Fungi, parasites, bacteria, and viruses are all different classes of microbes. There are several common infections that are triggered by microbes. For example, Influenza can be caused by bacteria, fungi, protozoa, or viruses [19]. Inflammatory Bowel Disease is caused by bacteria [20]. Onychomycosis is caused by fungi [21]. Severe Acute Respiratory Syndrome (SARS) is caused by a virus [22]. Babesiosis is caused by Protozoa [23]. Protothecosis is caused by algae [24]. Gastrointestinal endogenous is caused by Archea [25,26] (Figure 1).
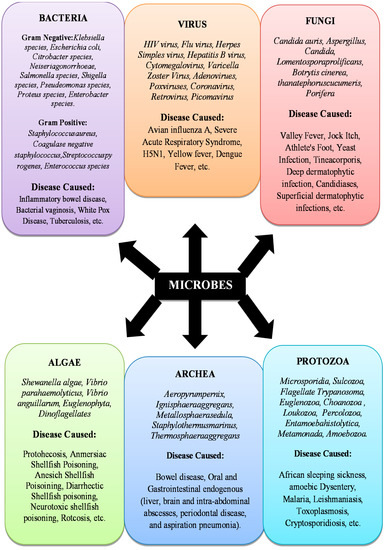
Figure 1.
Infectious microbial species.
Infectious Microbial Species
The development and effects of a disease on the human body depend on the form and strain of the pathogen [27]. The immune system generally provides an excellent defense against infectious agents. However, microbes may overtake the capacity of the immune system to fend them off. In such cases, an infection is harmful. Some microbes have no negative effect, while others generate toxins or toxic agents that cause adverse body responses. Due to this difference, some infections are harmless and hardly detectable, while others may be severe and life-threatening. Infection can spread in various ways, partly due to the differences in microbes themselves; fungi, parasites, bacteria, and viruses are all different varieties of microbes. These differences result from the mechanism of action on the body and the genetic content, function, shape, and size of the microbe.
3. Conventional Antibiotics
With a historical context of human disease, an exceedingly high percentage of infectious agents has been involved in infectious diseases. Microbes have been considered responsible for many infectious diseases during the second half of the nineteenth century. As a result, antimicrobial chemotherapy was introduced as the principal therapeutic technique against the pathogenic species.
Penicillin was discovered in 1928 by Fleming. In a region containing an infected blue mold (a fungus of the penicillium genus), the development of Staphylococcus aureus in cultivation dishes was inhibited, leading to the discovery that a microbe is developing compounds that may prevent other microbial growth. Subsequently, the antibiotic penicillin came into clinical usage in the 1940s. In the period of antimicrobial chemotherapy, penicillin was an excellent safety and efficacy agent, and during the Second World War, it saved the lives of many wounded troops. Penicillin had become the first antibiotic and marked the beginning of modern antibiotic production [5] (Table 1).

Table 1.
History of antibiotics, their discoveries, and events that have occurred.
3.1. Mode of Action of Antibiotics
Antibiotics can be classified according to their mode of action, the spectrum of the action, or their chemical structure. Bactericidal or bacteriostatic antibiotics may be commonly present across the Gram-negative and Gram-positive spectrum [156]. Depending on molecular structure, they can be classed as macrolides, β lactam, aminoglycosides, glycopeptides, tetracycline, and quinolones. The bacteria are either bacteriological or bacteriostatic; the targets can be of a wide variety (Gram-negatives or Gram-positive bacteria) [5].
β-Lactam inhibits the growth of bacterial cell walls by binding penicillin-binding enzymes (PBPs). PBPs perform the function of connecting the peptide units in the peptidoglycan sheet. After β- lactams are connected to PBPs, cell lysis occurs. The lactam antibiotics are also divided into monobactams, cephalosporins, carbapenems, carbacephems, and penicillins. Penicillin-resistant bacteria were reported to appear in the late 1960s. The enzymes β-lactamases, which could degrade β-lactam antibiotics, were synthesized by these bacteria. However, the emergence of carbapenem, the new class of β-lactams, solved this problem as carbapenem is not sensitive to the β-lactamases. Carbapenems exhibit the broadest spectrum of activity of all of the recognized lactams [5]. However, some bacterial species showed resistance to carbapenem [157].
Glycopeptides also attack the synthesis of bacterial cell walls, and additionally, they block the PBPs and inhibit peptidoglycan synthesis [5]. By attacking protein synthesis in the cell’s tetracycline, macrolides, oxazolidinones, and aminoglycosides block bacterial growth. The binding of macrolides to the 50S ribosomal subunit causes the inhibition of mRNA elongation during translation [5]. Oxazolidinones are also connected to the 50S subunit; however, they inhibit protein synthesis by inhibiting the development of a 70S initiation complex similar to macrolides [158]. These two groups combined make up the 50S category of blockers. Aminoglycosides and tetracycline attach to the ribosomal 30S subunit that prevents the utilization of aminoacyl-tRNA to the ribosome, thus inhibiting the synthesis of proteins. Tetracycline and macrolides are typically bacteriostatic, while the mode of action of aminoglycosides is broadly bactericidal [5].
Nucleic acid synthesis is necessary for cell survival. Quinolones inhibit bacterial growth by inhibiting the operation of helicases in DNA which, just before reproduction or repair of DNA, are crucial for relaxing DNA double-helical structure. Quinolones also interfere with bacterial functions of topoisomerase II and topoisomerase IV, which negatively impact RNA polymerase and thus inhibit the synthesis of RNA [5].
The para-aminobenzoic acid (PABA), a substrate for synthesizing folic acid used in bacterial cells, is imitated by sulfonamides structurally. Sulfonamide avoids cell division and causes the inhibition of the growth of bacteria, and it is necessary for the synthesis of nucleic acid. Regrettably, these modern antibiotics have been detected in bacteria which makes it difficult to treat infections due to these bacteria [159] (Figure 2).
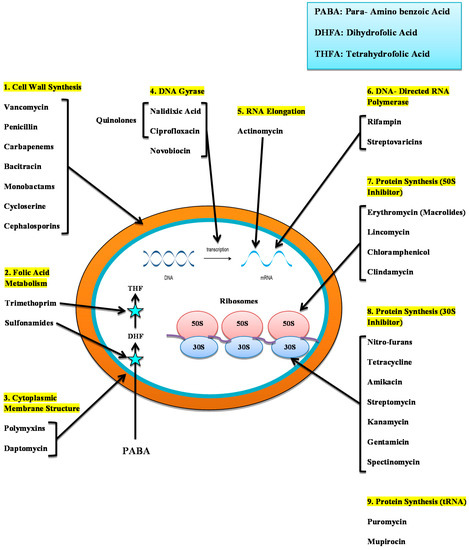
Figure 2.
Diagrammatic representation of the mode of action of antibiotics. 1. Cell wall synthesis Inhibition. 2. Folic acid metabolism Inhibition. 3. Disruption of Cell Membranes. 4. DNA Gyrase. 5. Inhibition of RNA elongation. 6. RNA synthesis inhibitors. 7. Protein Synthesis Inhibitors (50S inhibitor). 8. Protein Synthesis Inhibitors (30S Inhibitor). 9. Inhibition of Protein Synthesis (tRNA).
3.2. Origin of Antibiotic Resistance
Antibiotic resistance is considered present when a drug begins to lose its ability to successfully suppress bacteria growth. In the presence of active antibiotics, bacteria are “immune” and continue to divide. Bacteria are called resistant bacteria when they replicate even in the presence of antibiotics [160]. If the microbes are less susceptible or resistant, an effect greater than the usual concentration of the same drug is needed. Antimicrobial resistance has been found immediately after the launch of new antimicrobial compounds [161]. The mechanism of natural selection, in which evolution allows all bacteria to have a degree of low resistance, may partly explain antibiotic resistance [162].For instance, one study confirmed that sulfamethoxazole and trimethoprim (TMP-SMZ), ampicillin, and tetracycline, commonly used in earlier years, now no longer play a role in Thailand’s treatment of non-cholera diarrhea [163]. However, a study in Bangladesh demonstrated the effective therapeutic use of the same drugs [164]. Even before the use of antibiotics in infection control, resistance has been documented [165].
Agricultural antibiotics are typically identical and adjacent to commonly used antibiotic compounds [166], which can also encourage drug resistance. The food chain may be viewed as the primary route of spreading antibiotic-resistant bacteria between the animal and human populations. Livestock obtains antibiotics from food, water, or parents who may bear microbial resistance to a specific antibiotic [166]. Antibiotic resistance in livestock feed increases with the use of antibiotics as growth promoters [167]. According to the study of the rural villages in Barcelona, a fecal carrier of Quinolone-resistant Escherichia coli was reported in one-fourth of the babies, the possible source of which could be poultry or swine. These children have been exposed to quinolones [168] (Table 2).

Table 2.
Multidrug-Resistant (MDR) species.
3.3. Development of Antibiotic Resistance
As suggested above, bacteria seem to have a natural mechanism that promotes resistance. The resistance mechanism arises by mutations at the gene stage [180]. Selective pressure is caused by antibiotics, and even the genes function in accordance with selective pressure [181]. Bacteria possess the ability to transfer genetic material directly between themselves by transferring plasmids, meaning that natural selection may not be the only process through which resistance develops. The bacteria in a colony thus may mutate, resulting in resistance [182]. Broad-spectrum antibiotic drugs used in the treatment of nosocomial infections in health centers may, in fact, improve microbial resistance since large colonies of mutated bacteria are often found in such places [183]. Increased association between antibiotic-resistant infections and antibiotic use has been demonstrated [184]. Resistance development can also arise in cases where patients fail to complete the course of their prescribed medication. In such cases, the bacteria remain unaffected and become much more immune to the action of antibiotics [181]. Thus, bacteria can acquire multiple resistance characteristics over time and become immune to many antibiotic classes [185,186,187]. Some FDA-approved antibiotics for the treatment of microbial infections and their resistant microbes are listed in Table 3.

Table 3.
FDA (Food and Drug and Administration)-approved antibiotics for the treatment of Microbial infections [188].
The Antibiotic resistance in bacteria develops through the mechanisms shown in Figure 3:
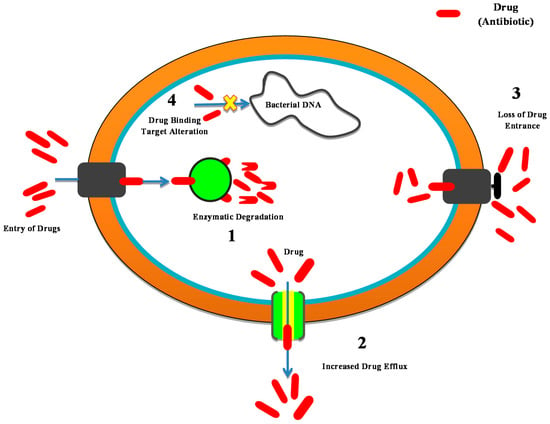
Figure 3.
Development of the mechanism of resistance in bacteria. 1. Antibiotic enzyme inactivation/degradation. 2. Excretion of the drug through the use of efflux pumps. 3. Reduced absorption by variations in the external membrane permeability. 4. Drug target modifications.
- Antibiotic enzyme inactivation/degradation; an endogenous cellular enzyme is modified to interact with that of the antibiotic in a manner in which the bacteria are no longer affected. B-lactamase enzymes are among the most important examples; they hydrolyze most commonly administered antibiotics, i.e., b-lactams (cephalosporin and penicillin), and are the most widespread source of antibiotic resistance in Gram-negative bacteria.
- The excretion of the drug through efflux pumps; Bacteria are triggered to eliminate the antibiotic by stimulating the proteins that can eradicate an extensive range of substances from the periplasm to the outside cell. This is a mechanism of resistance especially essential for P. aeruginosa and Acinetobacter spp.
- Reduced absorption by variations in the external membrane permeability; these changes inhibit the successful entry of antibiotics.
- Drug target modifications to weaken or demolish the antibiotic binding efficacy and thereby minimize its potential.
3.4. Availability of Antibiotics
Antibiotic production is no longer deemed an economically viable strategy for pharmaceutical companies [11]. Because antibiotics are widely used and are primarily curative, they are less effective in managing conditions related to diabetes, psychological disease, asthma, or gastrointestinal complications [11,189].
An annual cost—benefit study by the Office of Health Economics in London reports that the current net value of the new antibiotic is $50 million compared with about 1 billion dollars in neuromuscular condition treatment drugs [11]. The expense of modern antibiotics is typically USD 1000–USD 3000 a year, while USD 100–USD 1000 is spent on cancer chemotherapy [11,190,191]. The effectiveness, usability, and generally lower prices of antibiotics have, on occasion, resulted in poor evaluation from investors and the general public [190].
Furthermore, restricted use of antibiotics has been recommended by microbiologists and infectious disease specialists [190]. In some cases, healthcare professionals limit the use of the latest medication, believing it might promote resistance, and instead, continue administering current antibiotics that have shown equivalent effectiveness prior to the introduction of a new antibiotic [190]. New drugs are therefore often used as the “last line” of defense medicine in the fight against severe diseases. Such a method limits the usage of modern drugs and reduces investor returns [190]. Many pharmaceutical companies fear that the million-dollar investments required to develop new antibiotics may provide inadequate returns [189,190,191]. Graph 1 shows a relative decrease in the manufacturing of new antibiotics over a period of time (Figure 4).

Figure 4.
The number of approvals of new antibiotic drugs relative to year interval.
4. Nanomaterials
Nanotechnology-based approaches rang from the engineering and material sciences to biology and medicine. Materials with dimensions between 1–100 nm are commonly referred to as nanomaterial. They vary in shapes and sizes to help give characteristic features for a large spectrum of uses. The properties of the material undergo significant changes when limited to a rather small scale. Health sciences have shown considerable interest in various applications of nanotechnology. Nanomaterials are usually metal and metal oxides or their composite, carbon-based, and emulsion-based. They are cost-effective and can fight antibiotic-resistant bacteria. Approximately 10 million deaths by 2050 are predicted due to the growing risk of antibiotic resistance [192]. Nano-based technologies can offer a long-term and effective approach to managing drug resistance.
4.1. History and Development of Nanomaterials
The protection of ceramic matrixes has been in use for over 4500 years, including the use of organic asbestos nanofibers [193]. More than 4000 years ago, the ancient Egyptians used NMs for synthesizing five nm diameter lead sulfide (PbS) nanoparticles (NPs) for hair dye [194]. NMs were based on the synthetic chemical process. “Egyptian blue” was the first synthetic stain produced and utilized by the ancient Egyptians in the third century BC with a synthetic combination in the range of nanometer-sized quartz and glass [195].
The synthesis by chemical methodologies of the metallic NPs dated back to the 14th century BC when Mesopotamians and Egyptians began the manufacture of glass from metals that could be referred to as the beginning of the metal nanoparticles era [196]. These materials may be the first experimental cases of synthetic NMs. In Frattesina di Rovigo (Italy), a red glass colored by Cu nanoparticles surface excitation was discovered during the late Bronze Age (1200–1000 BC) [197]. Likewise, it is recorded that Cu NPs and cuprous oxide (cuprite Cu2O) were found in Celtic red enamels from 400–100 BC [198]. However, the most notable example of prehistoric metallic NPs is a Roman glass workpiece [199]. The Mesopotamians began to use glazed ceramics for metallic decorations in the 9th century [193]. Moreover, clay minerals with a size of a few nanometers are the most ancient example of nanomaterial. In 5000 BC, in Cyprus, clay was used to bleach clothes and wool [200]. The synthesis of an AuNP colloidal solution, which was the very first scientific effort in NP preparation, was documented in 1857 by Michael Faraday. This was apparently among the first publications that detected and explored quantum-size effects. The cause of the particular colors of metal colloids was later explained by Mie in 1908 [201]. In the 1940s, SiO2 NPs were developed to provide rubber insulation to replace carbon black [202]. Today, manufactured nanomaterials can significantly enhance the properties of bulk materials with respect to their strength, conductivity, resilience, and lightness and can offer beneficial applications and serve as structural and detecting materials for protection. Despite other potential applications, taking advantage of the favorable shape and size to enhance the appearance of materials remains the primary use of NPs.
In 2003, Samsung launched the antibacterial technology Silver NanomicTM from ionic AgNPs in their air conditioners, washing machines, air purifiers, and cooling systems [203]. In auto production, NPs and nanostructured materials (NSMs) are commonly used as tire fillers to improve adherence to the surface, car body fillers to improve stability, and translucent layers for heating, mist, and ice-free window panes [204]. By the end of 2003, Mercedes-Benz launched an NP base coat for metallic and nonmetallic surfaces in series production. The coating improves the rubbing resistance and strengthens the gloss. The latest studies concentrated on the production of specialized earth-based astronomical telescopes with adaptive optics and magnetic mirrors with ferrofluid shape-shifting potential [205]. In solar cells with dye sensitization potential, TiO NPs are widely used [206]. In 2012, Logitech launched the first large commercial usage of dye-sensitized solar cells, an external light-powered iPad keypad. AbraxaneTM was developed in 2005, sold, and launched on the pharmaceutical market as a human serum-albumin NP substance comprising paclitaxel [207].
4.2. Nanoparticles Act as Antimicrobials
In the 1980s, the advance in nanotechnology allowed Nano-scale construction by the tracking of atomic particles. Subsequently, nanotechnology has become important in diverse areas like biomaterials, organic chemistry, medicine, and others. In the medical science and healthcare industry, nanomedicine and nano-scale particles [208] are used for therapeutic purposes, as biomaterials, and diagnostic tools [209]. Thus nanomaterials and future molecular nanotechnology are widespread medical applications [210,211]. Nanotechnology has made many difficult diagnoses possible and has expanded the knowledge of disease pathogenesis. Many medications and procedures are seriously impacted because of their poorer efficacy, usefulness, and adverse effects. As the scale of nanoparticles is close to biological molecules and forms, in vivo and in vitro therapeutic strategies and higher specificities are useful [208]. Higher specific activity medications can increase effectiveness and decrease adverse effects. Smaller-sized nanoparticles may have treatment application in the high-risk areas, reducing the potential harm and delivering the exact medication dosage required.
Despite their past therapeutic successes, the use of antibiotics became problematic at the beginning of the 20th century [212], with the development of drug-resistant bacteria brought on partly through their extensive use [213,214]. Increased pathogen antibiotic resistance to colistin [215], carbapenems [216,217], and tigecycline [218], and stalled production of new antibiotics made it impossible to cure the contagious disease caused by harmful pathogens. Even though a new antibiotic may be found, it could not provide a guarantee of its effectiveness against all multi-drug-resistant infections [219]. Nonetheless, the threat to public health is real since both the Gram-negative and Gram-positive multiple-drug-resistance bacteria are developing faster than ever [220]. Thus, technological developments must aid the efforts against these dangerous pathogens and help meet the long-term demand for successful treatment of drug-resistant bacterial infectious disease [221].
The latest developments in the production of medicinal nanomaterials may benefit the purpose of antibiotics. Theoretically, nanoparticles are a modern class of bacterial antimicrobials and synthetic pathogens. Nanoparticles as antibiotics can hopefully minimize resistance and support the efficient delivery of antibiotics [222]. Recent research suggests that certain antimicrobial-activated metal nano constructs have indeed helped combat infectious diseases [223]. Such constructs benefit from lower toxicity, lower cost, and better pharmacokinetic factors while helping to eliminate drug-resistant bacteria. Their most significant benefit is that they maintain their efficacy longer than traditional antibiotics, which is extremely valuable in the long-term sustained therapeutic effect [224].
4.3. Classification of Nanomaterials
The nanomaterials are classified into four categories:
- Carbon-based nanomaterial
- Inorganic nanomaterial
- Organic-based nanomaterial
- Composite-based nanomaterial
4.3.1. Carbon-Based Nanomaterial
These nanomaterials typically have carbon and are found in spheres, ellipsoids, and hollow tubes. The types of carbon-based NMs include Fullerenes (C60), Carbon Nanotubes (CNT), Carbon nanofibers, Carbon Black, Graphene (Gr) [225] (Table 4).
Carbon Nanotubes (CNTs)
CNTs reported by 1991 are cylindrical informs connected with covalent bonds [226]. Multiwalled nanotubes and single-walled (SW) nanotubes have various types of single pipe CNTs 1–5 nm in diameter, and multiple nested tubes of lengths ranging from 100 nm to micrometers, respectively [227]. The cytotoxic influence of CNTs has been shown in both in vivo and alveolar macrophages [228,229]. The antimicrobial property of SWNTs indicates that their effective antibacterial and antiviral properties are based on the low aqueous dispersion of pure CNTs. The aqueous dispersion of CNTs has recently been demonstrated and could further be improved utilizing other surfactants or polymers. The best antimicrobial carbon-based nanomaterials identified are SWNTs. Initial interaction with bacteria, membrane dysfunction, and membrane oxidations are part of the SWNTs’ action in preventing microbial growth [230]. The use of CNTs for purifying water, E. coli inactivation, and poliovirus are being gradually studied, and MS2 phage elimination is also being investigated [231]. Thus far, CTTs offer relevant materials that can be used as antimicrobial agents.
Fullerenes
Fullerenes are ball clusters made up of atoms of carbon [232]. Antimicrobial activity was observed in fullerenes against many bacteria, including Salmonella, Streptococcus spp, and E. coli [232]. It Is suggested that their antibacterial activity was induced by energy metabolism inhibition after nanoparticles were internalized into the bacteria [233,234]. It was proposed that fullerene compounds prevent bacterial growth by destroying the respiratory chain [235].
The antibacterial efficacy of a sulfobutyl fullerene derivative in environmental bacteria was assessed by Yu et al. (2005). They observed that after photoirradiation, the used derivatives could inhibit environmental bacteria [236]. Mizuno et al. (2011) have also stated that cationic replacement fullerene derivatives are highly successful in destroying a large variety of microbial cells following the white light irradiation. A new generation of synthetic fullerenes was tested against Gram-negative and Gram-positive bacteria and whether they carry basic or quaternary amino groups.
Graphene Oxide (GO)
Graphene is generally referred to as a monolayer of carbon atoms, closely enclosed within a 2-dimensional crystal [237]. The GO Nanosheets are created by the chemical modification of graphene with suspended epoxyl, hydroxyl, and carboxyl groups that can be dispersed rapidly into water. The membrane stress is known to constitute the critical antimicrobial function of GO through direct interaction with sharp Nanosheets [238]. The inhibitory effects on E. coli growth were demonstrated by both graphene and GO. Testing antimicrobial effects on graphene sheets was performed by Akhavan and Ghaderi (2010), confirming that the direct relationship of extremely sharp edges and bacteria has resulted in RNA effluxes by impaired cellular membranes of both Gram-positive and Gram-negative bacterial membranes. The antimicrobial effects of the two active GO nanostructures (graphene oxide-chlorophyllin and graphene oxide-chlorophyllin-Zn) on E. coli were reported by Azimi et al. (2014). The authors suggested that E. coli was affected by the functional GO.
4.3.2. Inorganic Nanomaterial
Silver Nanoparticles (AgNPs)
Silver has been used as an antibacterial agent for many years. Applications of silver as metal and metal oxide nanoparticles are included in nanomaterials. These nanomaterials can be synthesized into metals such as Ag or Au, metal oxides such as ZnO and TIO2 NP, and semiconductors such as ceramics and silicon. A few of these demonstrate high antimicrobial activity towards certain bacteria, viruses, and other microbes. Specific nanomaterials with strong antibacterial efficacy are thought to have high volume to surface ratios as well as a unique chemical—physical property [239] (Table 4).
Silver as an antibacterial agent has been used in various ways; as silver nitrate, silver sulfadiazine, and powdered silver for infectious diseases, dental research, catheters, and burned wound treatment [240]. The development of antibiotics in the 1940s limited the use of silver as an antibacterial agent. However, the emergence of antibiotic-resistant bacteria and diminished antibiotic efficaciousness has resurrected the therapeutic use of silver [241]. AgNPs are found suitable against almost all viruses, bacteria, and other eukaryotic microorganisms in various types of metal and metal oxides [242,243]. The effectiveness of AgNPs as antimicrobial agents depends entirely on their size and shape [244]. Mechanism of action of AgNP targets the cell division and respiratory chain, eventually destroying the cell [245]. Increased synergistic antimicrobial actions against Gram-Positive and Gram-Negative bacteria have been reported, resulting from the action of AgNPs in conjunction with conventional antibiotics such as vancomycin, erythromycin amoxicillin, and penicillin G [246].
Further studies suggested that Ag+ ion, which has a sulfur and nitrogen affinity, can suppress and destroy the protein structure by binding it to thiol and amino groups [247]. Lastly, silver NMs have been reported as photocatalytic [248] and capable of triggering reactive oxygen species ROS [249,250,251]. Some have denied that this effect is cell-type dependence, at least in eukaryotic cells [242]. Silver has a broad spectrum of applications, from injury dressing to surgical device coating and manufacturing applications. Often there are negative health consequences associated with the use of metallic silver [252]. These adverse effects include permanent skin pigmentation (argyria) and eye loss, organ destruction, inflammation, and changes in the blood cell count [253]. However, amounts of AgNPs commonly used, and the concentration-dependent toxicity that influences the mitochondrial activity (possibly responsible for AgNPs toxicity) is still below observation [254].
Gold Nanoparticles
Gold coating on carbon nanotubes improves drug production. Gold nanoparticles have been used to identify bacteria by adjusting colors to facilitate cytometry flow responsiveness. Gold nanoparticles’ action as antimicrobial agents is primarily based on the electrostatic appeal to the cell membrane’s negative-loaded bilayer; the suggestion has also been supported by the discovery that anionic particles are not toxic while cationic particles are [243]. Near-infrared (NIR) light can absorb gold nanocages, nanoparticles, nanorods, and nanoshells often used to cure a bacterial infection under intense laser beams of sufficient wavelength [252]. AuNPs, coupled with antibiotics or antimicrobials, were shown to have elevated effectiveness, and modifications have been studied of the specific antimicrobial effects [255]. In addition, bacterial killing was attributed to the intense laser-induced hyperthermic effect couped with the bubbles forming around cluster AuNPs [256]. The gold nanoparticles have been found to cause strong antimicrobial effects on Gram-negative and Gram-positive bacteria with an antibiotic (i.e., streptomycin-coated Au NPs, gentamicin, and neomycin). In contrast with conventional free ampicillin, nanotechnology advances show that AuNPs coated with chitosan and ampicillin demonstrate at least a two-fold rise in their antimicrobial activity [243]. Gold nanoparticles encourage adjuvants to replace antibiotic therapy to treat severe bacterial infections, including multi-drug-resistant bacteria, with low doses and minimal adverse effects.
Zinc Oxide Nanoparticles
Zinc, as a metal oxide, an antibacterial agent ZnO NPs, and its antibacterial activity can conserve agricultural goods and food against specific foodborne bacterial pathogens such as E. coli [242,257]. ZnO NPs’ less expensive production, the ability to obstruct UV, and its white appearance make it more useful than Ag NPs. It showed the peerless capability of the multilayer avowal of Nano-ZnO on cotton fabric with antibacterial activity against S. aureus on its analysis part [258]. ZnO NPs’ potential to destroy the bacteria by destroying the cell membrane (proteins and lipids) and removing the intracellular material makes them suitable antibacterial particles. The ZnO-based nanoparticles are additionally broad-spectrum bactericidal NM [259] and have shown a wide variety of antibiotic activity against different microbes that mainly depend on the particle size and concentration chosen [260]. In addition, it also generates H2O2 and Zn+2 ions vital to bacterial cells [260]. Polyvinyl alcohol (PVA)-veiled ZnO shows enhanced permeability of the membrane, cell idealization, and intracellular structural adjustments [261].
Titanium Dioxide Nanoparticles
Titanium Dioxide (TiO2) is a nontoxic antimicrobial with a potential bactericidal application. It is most frequently employed as a photocatalyst disinfecting material. Titanium dioxide nanoparticles (TiO2 NPs) have been widely studied and compared with other preferred nanoparticles to assess their antimicrobial photocatalyst activity [262]. The inhibition of microbial growth is found to be higher after irradiation with near-UV light, and the bactericidal behavior of TiO2 NP improves noticeably when combined with UV-A [263]. TiO2 behavior largely depends on the scale, intensity, and wavelength of light (100–1000 ppm levels are required to destroy bacteria). The UV-A irradiated TiO2 antibacterial agent has shown reduction efficacy in microbial growth (in the declining order) of P. aeruginosa, E. coli, S. aureus, C. albicans, and E. faecium based Cell membrane [264]. It was also suggested that the antibacterial photocatalytic efficacy of TiO2 conditional density of the microbial surface morphology starts to fall in the sequence of viruses > bacterial wall > bacterial spores [264]. Growth inhibition characteristics of TiO2 NPs with UV-A irradiation show inhibition against Enterobacter cloacae, E. coli, and P. aeruginosa were surprisingly less successful against Enterobacter cloacae than P. aeruginosa and E. coli [265].
Reactive oxygen species (ROS) production, free hydroxyl radicals, and peroxides render TiO2 a useful antibacterial photocatalytic agent [266]. TiO2 NP oxidative strike has been attained with hydroxyl radicals (potent oxidants) created from TiO2 photocatalyst, which have broad (nonselective) reactivity and mainly attack microbial surfaces. The composition and the integrity of the cell wall are essential to sustain semi-permeability, respiration, and other phosphorylation reaction. TiO2 NPs photocatalytic action can interrupt the normal cell function compromising the bacterial cell membrane structure. The TiO2 photocatalyst action leads to lipid peroxidation reaction (oxidative lipid degradation), which subsequently leads to cell death [267]. Thus, while the usage of radiation improves the photocatalytic antibacterial activity of TiO2 NPs, it also causes individual bacterial mortality from irradiation [268]. The potential for visible light activation (e.g., sunlight) makes TiO2 far more remarkable antibacterial agent possible.
Metal doping improves the TiO2 antibacterial properties, leading to enhanced bacterial and viral photocatalytic inactivation [269]. Composite Ag/(C, S)-TiO2 NPs have shown strong light-independent antimicrobial activity when tested against B. subtilis and E. coli spores [270]. TiO2 can also be used as a low-cost, high-efficiency, stable, nontoxic alternative to traditional chemical disinfectants, which may produce toxic byproducts, and as such, it would be of particular value for water treatment systems in developing countries. [227]. TiO2 reactivity toward microorganisms can be applied to improve food safety, hygiene, and cosmetics using TiO2 photocatalytic disinfection of nanocomposite antimicrobial surface coating capable of destroying UV radiation-tolerant microbes [269]. TiO2 nanocomposite surface coating for orthodontic products, toothbrushes, dental implants, and screws has shown effective antibacterial activity against Lactobacillus acidophilus [266].
Copper Nanoparticles
Copper is a beneficial antibacterial agent since it is a structural part of many microorganism enzymes. When Cu2+ ions are high, ROS can be formed, disrupting amino acid formation and a more toxic DNA [271,272]. The basic principle behind copper microbial inactivation is the so-called contact-killing. Other antimicrobial mechanisms such as inoculation methods and incubation period, which are important, also rely on the copper contact method. The efficiency of this touch extinction has been improved by factors such as elevated temperatures [273], high metal content in copper [274], and decreased relative moisture [275]. Cu NPs with great affinity to the amines and carboxyl group on the surface of the organism kill Bacillus subtilis and have demonstrated better action than Ag NPs [276]. The chemical and physical quality of copper oxide (CuO) compared with silver is less expansive [277].
Aluminum Oxide Nanoparticles
The bacterial cell wall was damaged at a higher concentration of aluminum oxide (Al2O3) nanoparticles [278]. Alumina NPs are high-temperature thermodynamically stable particles. The chemical arrangement includes oxygen atoms that fill a matrix to two-thirds of the octahedral sites [278], which conform to hexagonally wrapped alumina ions. The aluminum oxide NPs had an effect on the surface charge, shape, and particle size [279]. In contact with organic matter, nanoparticles appear to mix with hard water and seawater in soil. Such aggregations rely on the pH and salt content of the particles. Most toxicity studies focus on the detailed configuration and characterization of the solution parameters, including thickness, distribution, structure, morphologies, surface areas, surface chemistry, and particle reactivity. They are believed to have moderate inhibitory properties and may be used in combination. Ag/Meso-Al2O3 nanoparticles displayed extensive S. aureus and P. aeruginosa inhibitory activity.
Nitric Oxide (NO)—Releasing Nanoparticles
Nitric oxide (NO) has an immune function as a diatomic free radical and is an effective antimicrobial agent acting against infection in two ways; at a low concentration at which it promotes the growth of immune cells, and at a high concentration where it inhibits or kills pathogens (via binding with DAN, proteins, and lipids). For combination therapy, NO is commonly used, but the performance of individual NO donors, based on sparse evaluation, shows limited results. The difficulty of processing or administering NO as an antibacterial agent has been a restricting factor. Lately, a strong trend of vulnerability has been found in Gram-negative and Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus, by utilizing a gaseous NO (gNO) donor delivery platform [280].
NP-based scaffolding could hold large NO loads, which can be released at appropriate sites, physiological pH, or temperature under aquatic conditions [281]. The NO and NO-releasing Silica Nanoparticle antibacterial experiments have been conducted to destroy Gram-positive and Gram-negative fungi and bacteria. In vitro MRSA study has shown pathogen behavior in silane with nanoparticles releasing NO [280]. These nanoparticles can associate with the pathogen surface causing pathogen inhibition and leading to bactericide. The NO-releasing NPs are versatile with unique properties dependent on nanoparticles’ size and adjustable NO donor design. NO-releasing NPs have been used for treating contaminated wounds and subsequently for successful healing of wounds in diabetic mice [282]. Human studies have been limited thus far, but given the versatility and potential of NPs that make them highly suitable for NO delivery, there is a need for intensified research into the human application of this technology.
Magnesium Oxide Nanoparticles
Magnesium oxide (MgO-NPs) is an essential mechanism behind antimicrobial activity; it produces ROS and other nanoparticles [283,284]. MgO-NPs are associated physically with the cell surface as some other nanoparticles and cause a dysfunction of the cell membrane integrity which contributes to membrane leakage [8]. Therefore, MgO-NPs destroy the cells through intracellular biomolecules irreversible oxidation. Furthermore, some studies have shown that with the lack of lipid peroxidation and ROS, MgO-NPs generate a high antibacterial effect. Research suggests that the relationship between MgO-NPs antibacterial behavior and nanoparticle association to the microbial cellular membrane changes pH and releases Mg+2 [285]. Moreover, the antibiotic behavior of MgO-NPs was found to be caused by the adsorption of halogen molecules onto the exterior of MgO [8].
Iron Oxide Nanoparticles
Iron oxide nanoparticles are highly ferromagnetic and have reduced oxidation sensitivity. There has been considerable attention towards iron oxide nanoparticles since they are known as nontoxic and biologically compatible materials because of the presence of Fe (II/III) ions [286]. Release and termination of toxic ions, oxidation damage caused by catalysis, the ion cell membrane transport activity variation, and lipid peroxidation or surfactant products are chemical pathways associated with reactive oxygen species (ROS). In nanotoxicology, ROS is considered a key rudimentary chemical mechanism, which can result in secondary processes that can eventually destroy cells and even cause cell death. In addition, ROS are a major inflammatory factor. The inflammatory action is believed to occur by up-regulation, stimulated by the activation of specific transcription factors in genes involved in a proinflammatory response (NF-ŢB, AP-2). Cell stability can also be directly affected by free radical formation [287,288]. Nano–bio-interface physical processes are primarily governed by particle size and surface properties. They involve membrane function, membrane transport mechanism destruction, protein conformation or folding, and the accumulation of proteins [289]. The iron oxide nanoparticles can impede the production of B. subtilis as well as E. coli. The maximum inhibition (29 mm) in S. aureus compared to E. coli and P. aeruginosa was observed with 015 mg/mL of iron oxide nanoparticles [290]. In another analysis, Fe2O3 and Ag/Fe2O3 were found to have antibacterial properties, and nanoparticles against S. dysenteriae had antibacterial activity much more significant than that of Fe2O3 nanoparticles individually [291].
Super-Paramagnetic Iron Oxide (SPION)
SPION is a modern approach to using magnetic particles; it induces local hyperthermia in the presence of a magnetic field [292] or may otherwise be protected in other nano-materials. Biofilms can be penetrated and degraded by Ag and Au and their magnetic effect [293,294,295].
4.3.3. Organic-Based Nanomaterials
Organic-Based Nanomaterials involve nanomaterial primarily composed of organic material, with the exception of inorganic or carbon nanomaterial. The use of noncovalent (weak) molecular interactions tends to convert organic nanomaterial into desired structures, including liposomes, micelles, dendrimers, and polymeric NPs. Compared with inorganic materials, organic antibacterial materials are known to be less stable in nature, particularly at higher temperatures (Table 4).
Poly-ε-Lysine
A cationic homopeptide of L-lysine known as Poly-ε-lysine is effective against Gram-positive and Gram-negative bacteria. Additionally, it acts against spores of B. subtilis, B. stearothermophilus, and B. coagulans [296,297]. Some scientists have developed a technique to fight deadly antibiotic-resistant bacteria using nanocargos of gold nanoparticles (AuNPs) combined with ε-polylysine. These nanocargos were 15–20-fold more antibacterial compared with free poly-e-lysine when measured against carbapenem-resistant Acinetobacter baumannii (CRAB) referral strains and Staphylococcus aureus (MRSA) [298].
Quaternary Ammonium Compounds
The proven disinfectants are the quaternary ammonium compounds, including cetrimonium chloride, benzalkonium, and stearalkonium chloride. The antimicrobial action of quaternary ammonium compound relies on bash of the N-alkyl chain length and lipophilicity [299].
The mechanism of electrostatic interaction between a positively charged compound moieties and a negative charge bacterial membrane, followed by the integration of the hydrophobic tail compound into a hydrophobic membrane core where structural enzymes and proteins are denatured, culminated in initial encounters with the bacterial wall [300].
N–Halamine Compounds
N–Halamine complexes comprise one or multiple covalent bonds between nitrogen and halogen, which are common imide, amide, or amine group halogenations that provide stabilization and gradual emissions into the atmosphere of freely activated halogen species. These oxidizing halogenic agents facilitate the direct transfer of the active substance to the required biological locations or isolation from the aqueous medium to the halogen-free medium. These free, reactive halogens cause a microbial cell to inhibit or inactivate [301].
Polysiloxanes
Polysiloxanes, linear silicone oxide polymers, are yet another major type of polymer. Sauvet et al. synthesized a block of copolymers and statistical siloxanes having ammonium quaternary salt as a lateral substitution. These polymers show high antimicrobial action against Escherichia coli and Staphylococcus aureus. In block polymers or statistical copolymers, there was no significant difference in the activity [302].
Benzoic Acid, Phenol, and p-Hydroxy Benzoate Esters
Benzoic acid, phenol, and p-hydroxybenzoate esters are the most widely used disinfectants and preservatives. As monomers, the antibiotic activity of these compounds has already been identified. Attempts were made to combine them into a polymer backbone to synthesize the new, improved activity of antimicrobial polymers. Phydroxyphenyl acrylate has been more beneficial for both bacteria and fungi in a descriptive analysis of the antimicrobial activity of p-hydroxyphenyl acrylate, p-2-propane oxyphenol [303].
Quaternary Phosphonium or Sulfonium Groups
A broad spectrum of antimicrobial activity was found in quaternary ammonium compounds against both the Gram-positive and Gram-negative bacteria. Quaternary ammonium polyethylenimines (QPEI) provide a wide range of bacterial targets. When embedded in the different polymeric matrices, the polyamines have been shown to be an effective antimicrobial nanoparticles [304]. Polymers that have phosphonium quaternary or sulfonium groups have structures that are close to those of the compound ammonium quaternary group. Phosphonium-based polycationic biocides are more effective than quaternary ammonium salt polymers with respect to antimicrobial action. The NIPAAm and methacryloyloxyethyl trialkylphosphine chlorides experiments on water-soluble copolymer thermal sensitivity demonstrate that antimicrobial activity rises incrementally in polymer alkyl chain length and units of phosphonium in the polymer [305].
Triclosan
Triclosan is among the most common antimicrobial agents. A Triclosan solution was combined with water-based styrene–acrylate emulsion, and Enterococcus faecalis was tested. Depending on an agar diffusion test, triclosan liberation relies mainly on the solvent, either nonexistent or very sluggish in water and relatively fast with n-heptanes [306]. In addition to organic/aqua solutions, triclosan has been integrated, and PVA nanoparticles, which are water-dispersible, display higher antibacterial activity in relation to Corynebacterium [307].
Chitosan
The broad spectrum of antibacterial action has been identified in deacetylated chitin known as chitosan [308]. Chitosan nano-scale and its compounds have only recently been shown to possess antimicrobial action against microbes, viruses, and molds [309]. It is much more efficacious than bacteria against viral and fungal infections, and it has often been found to become more selective for Gram-positive than Gram-negative bacteria [306]. The molecular weight of Chitosan has a significant function in its antibacterial action, which also relies on variation in targeted bacteria’s cell wall: low molecular weight chitosan exhibits strong antimicrobial effects towards Gram (−) bacteria and high molecular weight towards Gram (+) [310]. Mechanisms through which chitosan works as an antimicrobial agent are described in multiple hypotheses:
- a.
- Adding it to the negatively loaded cell surface, inducing agglutination and even microbial cell permeation, allowing leaks of intracellular substances [309].
- b.
- Chitosan chelation characteristics used for the chelation of trace metals blocks the action of certain enzymes, causing cell death.
- c.
- Fungal chitosan produced through host hydrolytic enzymes from the fungal wall prevents RNA and protein synthesis [310].
Chitosan antimicrobial activities and water-soluble chitosan derivatives are observed to have significantly greater efficacy in combating bacterial membranes over others. Chitosan is a suitable, low-cost, and effective disinfectant for developing nations with a wide range of action and far less toxicity to mammalian cells [311].
4.3.4. Composite-Based Nanomaterial
Composite nanomaterial consists of multiphase nanoparticles with a single nano-dimensional layer that can either mix nanoparticles with other nanoparticles or combine nanoparticles with smaller or larger materials (e.g., hybrid nanofibers) or more complex frameworks, such as metalorganic framework systems (Table 4).
Ceramic Matrix Nano-Composites (CMNC)
Nano-composites in the ceramic matrix are primarily Al2O3 or silicon carbide (SiC). Most of the research so far has shown that, after adding low-volume (approximately 10%) SiC particles of appropriate size and the heat pressure of the final mixture, the Al2O3 matrix has shown significant strengthening.
In a study, the coating system with a nano-TiO2 antimicrobial agent was used to prepare the antibacterial corrugating medium, and the antimicrobial efficacy was tested using the zone of inhibition approach. Furthermore, various concentrations of TiO2 antimicrobial agents have been observed in the mechanical properties of corrugating media, such as thickness, rigidity, bursting strength, tensile strength, and folding tolerance [312].
Metal Matrix Nanocomposites (MMNC)
Nanocomposites in a metal matrix (MMNC) refer to materials that comprise a matrix of a ductile metal or alloy that are implanted in a nano-sized reinforcing material. Metallic and ceramic materials combine these composites.
Metal nanocomposite active antimicrobial packages are developed by the incorporation of polymeric films of metal NPs. In nanocomposite antimicrobial mechanisms, the performance of NEs is effective primarily because of the high surface to volume proportion and large surface reactivity of the antimicrobial/metal oxide nano-sized particles, which enable them to more easily inactivate microbes [313]. Silver (Ag), gold (Au), zinc oxide (ZnO), silicon (SiO2), titanium dioxide (TiO2), alumina (Al2O3), and iron oxides (Fe3O4, Fe2O3) are some of metal and metal oxides nanomaterial widely used as antimicrobial agent.
Polymer Matrix Nano-Composites (PMNC)
The nanocomposites of the polymeric matrix are used extensively in heavy industry to make manufacturing simpler, lightweight, and flexible. They do, however, exhibit several drawbacks compared with metals and ceramics, such as low modulus and power.
Bio-nano-composite films built on poly (lactic acid) (PLA) and strengthened with nanoclay C30B (5.0 percent w/w) are infused with thymol and cinnamaldehyde active compounds at a high concentration of 11 to 17 percent w/w. The addition of active substances and nanoclay produces structural, thermal, mechanical, and antimicrobial properties to alter against specified Gram-negative and Gram-positive bacteria [314].


Table 4.
Types of Nanomaterials with their efficacy against bacteria.
Table 4.
Types of Nanomaterials with their efficacy against bacteria.
| Nanoparticles | Particle Size (nm) | Targeted Bacteria and Antibiotic Resistance | Antibacterial Mechanisms | Factors Affecting Antimicrobial Activity | References |
|---|---|---|---|---|---|
| Inorganic Nanomaterials | |||||
| Fe2O3 NP | 1–100 |
|
|
|
|
| Ag NP | 1–100 |
|
|
|
|
| ZnO NP | 10–100 |
|
|
|
|
| Cu NP | 2–350 |
|
|
|
|
| Au NP | 1–100 |
|
|
|
|
| TiO2 NP | 30–45 |
|
|
|
|
| Si NP | 20–400 |
|
|
|
|
| MgO NP | 15–100 |
|
|
|
|
| Al NP | 10–100 |
|
|
|
|
| SPIONS | 15–25 |
|
|
|
|
| Organic Nanomaterials | |||||
| Poly-ε- lysine | 1–100 |
|
|
|
|
| Chitosan | 200 |
|
|
|
|
| Quaternary ammonium compounds | 1–100 |
|
|
|
|
| N-halamine compounds | 1–10 |
|
|
|
|
| Quaternary bis-phosphonium and ammonium | 1–100 |
|
|
|
|
| Carbon-Based Nanomaterials | |||||
| Fullerenes | 200 |
|
|
|
|
| CNTs | 1–100 |
|
|
|
|
| Graphene Oxide NPs | 12 |
|
|
|
|
| Composite-Based Nanomaterials | |||||
| Ceramic Matrix Nano-composites | 1–100 |
|
|
|
|
| Metal Matrix Nano-composites | 1–100 |
|
|
|
|
| Polymer Matrix Nano-composites | 1–100 |
|
|
|
|
4.4. Mechanism of Action of Nanoparticles
The use of nanoparticles to fight bacteria is a fairly modern approach that may provide a viable solution to the crisis of global antibiotic resistance. Theoretically, it would be difficult for microbes to establish a resistance to several destroying pathways utilizing nanoparticles because of their multiple-target and action capability.
Many experiments have been conducted to test the metal-based nanoparticles’ mechanism of action. The nanoparticles are known for their strong antibacterial activity against a wide array of pharmaco-resistant species in silver and silver oxide. They use several mechanisms to work against bacteria, rendering them most efficient. The silver nanoparticles work by generating vast amounts of silver ions, which influence the permeability of the cell membrane and help to inhibit energy transfer through the transport chain of electrons. In addition, microbial cell damage to DNA [336,337,338] has also been identified. Zinc oxide nanoparticles are another type of nanoparticle which grows in the cells and emit Zn+2 ions there. These involve hydrogen peroxide production and destruction of the cell membranes [339,340]. Titanium dioxide nanoparticles participate in reactive oxygen production and subsequently influence the integrity of the cell membrane [341,342,343]. In contrast to metal nanoparticles, numerous nanomaterials dependent on polymers, liposomes, or carbons, each of which has its own mechanisms of action, are often used for fighting pathogens. Nanoparticles with chitosan work by boost permeability and breakout of the membrane. They as well engage within enzyme inactivation of the microbial machinery [227,309]. Carbon nanotubes act primarily in producing and oxidative degradation of the cell membrane, lipid, and proteins through reactive oxygen species [344]. A relatively recent family of nanoparticles that involves fullerenes works by rising neutrophil infiltration and is active in cell membrane disruption [345,346]. These various action pathways are ways in which nanoparticles can successfully attack the microbial machinery (Figure 5).
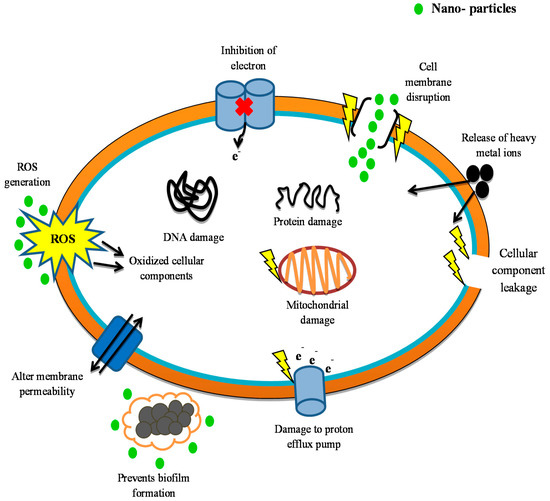
Figure 5.
Different pathways for nanoparticle antimicrobial behavior (organic, inorganic, carbon-based, composite-based nanomaterials). ROS: Reactive Oxygen Species.
4.5. Drug Release Kinetics of Nanoantibiotics
The effectiveness and efficiency of a nano-formulation filled with medicines depend mainly upon the kinetic release. For maximum effectiveness, it is essential to have medicines released for nano-carrier slow before reaching the site of action to prevent medication loss [347,348,349]. This procedure must be tested and continuously monitored to guarantee optimal and safe activity at the site and to avoid off-site behavior and possible subsequent adverse reactions. The drug release kinetics is the key factor in the effect nanoparticles, designed as antibiotic nano-carriers, will have on medication delivery [350,351]. Popular techniques for evaluating the release of NPs include dynamic ultracentrifugation, ultrafiltration, dialysis, and others. Dynamic dialysis is often preferred since additional phases in the isolation and release of medication from NPs are omitted at different kinetic periods. Furthermore, the exterior stress applied in several other approaches during the separation often reduces the efficiency of those methods [352]. Commonly, a broad range of technics is used to include antibiotics, such as liposomes, nano-emulsion, polymeric NPs, micelle systems, dendrimers, and strong lipid NPs, in targeted, regulated, and appropriate manner [353]. The drug release kinetics for polymeric NPs depends largely on the following:
- i.
- Surface-bound or adsorbed product desorption
- ii.
- Drugs diffusion from polymeric NPs
- iii.
- Polymeric NP erosion and a cumulative erosion/diffusion effect
Unless the rate of membrane degradation becomes more significant than the product diffusion from the polymer layer, diffusion happens as a consequence of drug exposure. The initial “burst-free release” effect is, typically, a drug release from polymeric NPs, which implies the surface release of a compound bound or adsorbed in contrast with that used within the matrix [354]. Compared with polymeric NPs, the release of the drug from liposome depends largely on the following:
- i.
- Lipid membrane composition
- ii.
- Nature of drug involved
- iii.
- The percentage of drug’s permeability
- iv.
- Environmental issues to consider such as temperature, pH, or exterior stimulations like degradation of the enzymes, ultrasound, or interaction of the proteins [91,92,93,94]
A significant focus has been placed on designing nanoparticles with improved drug delivery mechanisms. Several techniques for regulation of product release kinetics have been implemented, including the use of anionic gemini surfactant and covalent bonds, which can disrupt the pH of polymer NPs within an acidic environment [355]. In order to produce new nanoparticles with improved product supply and antibacterial action, the filling of the drug with polyelectrolytes is now an emergent trend [356]. Numerous experiments have examined nanoparticle drug loading and release kinetics for directed and controlled release. The chitosan-magnetic iron oxide NPs (Strep-CS-MNP) packed with streptomycin exhibited a significant burst discharge preceded by controlled drug release kinetics based on the physical mixture of chitosan–streptomycin, as well as magnetic iron oxide NPs, implicit the need for monitored drug release pathways. The analysis found that particles observed kinetics with a correlation (R) valuation of 0.9863 in the quasi-2nd order. The analysis found that Strep-CS-MNP had greater antibacterial activity than Streptomycin only toward MRSA. Other studies have found an improved production of the pH-responsive vancomycin-loaded chitosan NP at pH 6.5 relative to pH 7.4. The treatment further revealed that the risk of MRSA in a design of skin infection of the mouse was 8-fold reduced compared with those handled with pure vancomycin [353].
Newly published research indicates that the 90% loaded release of the drug around 2.5 h after first-order release kinetics has been generated by single-walled carbon nanotubes (SWCNTs). The drug demonstrated 16 times greater efficacy in contrast to ciprofloxacin against P. Aeruginosa and S. aureus and eight times against E. coli [357]. Different structures were used together to incorporate metal and antibiotic, antibacterial results. In yet another study, 120 h of vancomycin release profile with high antibacterial impact toward MRSA were shown in vancomycin-loaded aragonite NP. Such NPs, with a broad release profile, will serve as a strong local antibiotic supply mechanism for osteomyelitis treatment [355]. Research further showed the design of well-ordered, biocompatible Cu-doped MSNs with an extensive specific surface area, subsequently encapsulated with tetracycline (TET) species and coated with ultrasmall silver nanoparticles-stabilized polyethyleneimine (PEI-SNP) complex layer for combating antibiotic bacterial resistance. Outcomes from the bioassay showed that SNPs increased the antibacterial potential of the strain of MDR E. coli substantially by cell membrane sensitization and enhanced intracellular access to nanoscale containers for pH-caused drug cargo transport. In addition, the degradation of MDR bacteria was aided by massive ROS levels owing to Cu species [358]. An effective approach has been proposed for the delivery of antimicrobial agents, in particular at acidic pH. Nanoparticles were immobilized with silver–indole-3 acetic acid hydrazide (IAAH–Ag) complexes via a pH-sensitive hydrazone bond, which acts as a model drug. The synthesized IAAH-Ag pH-sensitive complex was shown to have a robust antimicrobial ability in the planktonic and biofilm states towards multi-drug-resistant bacterial isolates. Nanoconjugates have been shown to be of strong effectiveness in the treatment of bacterial mice infection [359].
4.6. Synthesis of Nanoparticles
The special features of biologically synthesized NPs are more significant than nanomaterial developed by chemical and physical methods. Nanoparticles may be synthesized physically/chemically [360]. These processes may pose a variety of issues, such as the use of dangerous chemicals, the generation of hazardous by-products, and geometrical errors [361]. Chemical processes typically consist of using one or more organic compounds or molecules, which aid particle toxicity and sensitivity and could create a risk to the environment and human health [361].
Green synthesis particulates are clearly distinguished from physical–chemical ones. The bottom-up method of green synthesis is similar to a chemical reduction in which expensive chemical-reducing compound is substituted with extracts of natural substances, such as fruits or tree leaves for metal or metal oxides NP synthesis, or by using different biological organisms. The nanoparticles produced through green synthesis are known as ‘biogenic nanoparticles’. Biological species have enormous production potential for NPs. Biogenic reductions of metal precursors to link NPs are environment friendly [362], safe [363], chemical-free [364,365], and can be used in large amounts. In addition, expensive metals like gold or silver can be recycled through the production of biological nanoparticles. These metals have limited sources, and their costs fluctuate [366]. NPs’ can easily communicate with other biological molecules, a capacity that improves antimicrobial behavior by enhancing the interaction mechanism of microorganisms, primarily with sugars enzymes, proteins, and even the entire cells [367]. The biological composition of the biogenic NPs allows for smooth drug isolation or up-concentration through centrifugation of biogenic NPs from the reaction media [368].
For biogenic NP synthesis, biological removal of metal precursors generally occurs in vitro or in vivo. Carbohydrates, enzymes, and plant chemicals such as flavonoids, phenolics, terpenoids, cofactors, and others, are thus mostly reduced and stabilized [369,370]. It has been suggested that fungi, bacteria, algae, plants, and yeast produce biogenic NPs in-vivo [371,372,373]. Biological extracts mostly used in in vitro synthesis include the purification of organic agents and mixing them in a controlled manner for the related metal precursor into an aqueous solution. This spontaneous reaction takes place at around room temperature [374], but it is often vital to involve stirring and heating [375]. These items are environmentally safe for the processing of biogenic NPs and their waste materials [376].
4.6.1. Green Synthesis of Nanoparticles
The green synthesis of NPs will typically adopt either the bottom-up or top-down method (Figure 6). NPs are generated by reducing the size and via different chemical and physical techniques in the top-down approach [377]. NPs are formed by smaller structures, such as molecules and atoms, with the key effect on oxidation/reduction in the bottom-up synthesis. NPs with homogenous chemical characteristics and limited errors are extracted in this sustainable and green pathway. Plant extracts and microorganisms are commonly used in the biological method of NPs production (Figure 7) [378,379,380]. The choice of the suitable species or extracts will consider the specific features, such as phytochemical substances, biochemical processes, enzyme functions, circumstances of cell development, and optimum reaction [381].

Figure 6.
General flow chart of different procedures of nanoparticles synthesis.

Figure 7.
Biological synthesis of Nanoparticles.
Fungi
Fungi are secondary metabolite contributors and active biomolecules, which are very important for NP synthesis. Fungal species such as Foxysporum produce proteins, polymers, and enzymes that willingly support the development of metal NPs [382]. These components enhance NP outcome and stability. In studies, some fungal organisms were reported to generate NPs with traces of extracellular amino acid. For example, the surface of the yeast includes glutamic acid and aspartic acid, which, in the presence of sufficient light, transforms silver into silver ions [383]. Ahmad et al. (2003) have found that fungal species such as Foxysporum have cytosol reductase enzymes that, in the presence of NADH+ reduction element, reduce silver from silver ions [381]. The phytochelatin class is strongly able to suppress silver ions in silver metal [384], which are present primarily in fungus. The population of fungal Coriolusversicolor was used by Sanghi and Verma (2009) for the silver NP synthesis (Ag NPs). In this study, FTIR data revealed the existence of the hydroxyl group in fungal mycelium, which contributes electrons to the silver ion and decreases them to transform Ag NPs into bare metal. In addition, it is reported that aromatic and aliphatic amines and other proteins in the fungal extract used to maintain Ag-NPs developed as a capping agent. It was further shown how silver powder became stabilized by the amide coupling of protein [385]. The role in the stabilization and capping of Ag NPs by SH group comprising fungal extract protein has also been documented by Tan et al. (2002) [386]. Das et al. (2009) utilized Roryzae mycelia for synthesizing gold (Au) nanoparticles by reducing the in situ of acidic chloroauric acid (HAuCl4) (pH 3) [387]. Verticillium fungus is indeed an excellent mediator for silver NPs synthesis. Biomass of fungus has generally been discovered to produce intracellular NPs on AgNO3 treatment in an acidic medium (pH 5.5–6) [388] (Table 5).

Table 5.
Fungal and Yeast species in the green synthesis of nanoparticles.
Bacteria
NP synthesis aided by bacteria is produced by two methods: intracellular and extracellular. Less time-consuming, extracellular synthesis of NPs is more advantageous than the intracellular system, provided that no downstream process is needed in order to gather NPs from organisms [396,397]. On the inside of the cell, the bacteria contain reductase enzyme, which catalyzes metal ion decrease into metal NPs. Dradioduran species have high antioxidant activity and are highly resistant to oxidative stress and radiation. This makes it suitable to use Au NPs in green synthesis from their ionic form (Table 6).

Table 6.
Bacterial species in the green synthesis of nanoparticles.
Leptothrix bacteria were used to synthesize AU-NPs by reducing gold salt on an aqueous medium in a new analysis by Kunoh Tatsuki et al. (2017). It has been documented that guanine RNA molecules and 2-deoxy guanosine residues minimize gold salt (Figure 6).
Plant
Plant-aided NP synthesis is far more effective than microbial synthesis. Plants possess numerous biochemicals (e.g., polyphenols) and many metabolites, which can act as stabilizers and reduce the synthesis factor of biogenic NPs. The synthesis of NPs in plants is environmentally sustainable and cost-effective. Plant-based NPs have been shown to be somewhat more stable than microbial and fungal NPs [396]. The controlled plant synthesis of the NPs is divided into three groups; phytochemical, intracellular, and extracellular materials. When plant extract is used as the raw material for the synthesis of NPs, the extracellular method is used. Intracellular synthesis of NPs occurs within plant tissue cells utilizing cellular enzymes. After synthesis, by destroying the plant cell wall, the NPs are restored. Synthesis of plant extract NP is relatively inexpensive and results in higher yields because more plant extract phytochemicals can stabilize or transform ions of metal into metal NPs [404]. Phytochemical controlled NP synthesis is not a commonly used method, as it involves understanding the specific phytochemical required for balanced NP synthesis [405]. Shakeel Ahmed et al. (2015) produced Ag NPs in a spherical form via A. indica leaves extract. In the FTIR study, the flavonoids and phytochemical compounds of the plant extract were found to act as reducers and stabilizers during the synthesis of NP. There was a high antibiotic response in these NPs [406]. Suman et al. (2013) isolated Au NPs through M. citrifolia root extract that had also been reported to be antimicrobial in nature [407] (Table 7).

Table 7.
Plant species in the green synthesis of nanoparticles.
4.6.2. Purification of Nanoparticles Extracted Biologically
Purification of nanoparticles after the synthesis is quite important before using them in any sort of application (Figure 6). Scientists have increasingly used the centrifugation process to purify nanoparticles since the procedure is simple and time-effective. Therefore, for the isolation and purification of metallic nanoparticles produced biologically, in order to isolate unreacted bioactive molecules, frequent washing and high-speed centrifugation are performed [405]. However, the process has several drawbacks, such as centrifugation which may induce NP agglomeration. Because of the disunion of the passivating agent from NPs and a transition in the NP’s underlying properties, NP destabilization occurs. The process of dialysis, using the exact cutting of the membrane, is another common form of purification. The dialysis membrane can quickly transfer tiny organic molecules contained in plant extracts, although organic molecules found as surface-passivizing agents stay in place and are connected with NPs in the dialysis membrane. This filtration process requires time which normally takes more than 24 h. However, for bio-fabricated nanoparticles, the diafiltration method is not used as it is insoluble in soil. The application of external magnetic power effectively splits up the magnetic nanoparticles such as Fe2O3 and Fe3O4. Moreover, it is always challenging to extract closely bound biomolecules from the nanoparticle layer (Figure 6).
4.6.3. Nanoparticles Coating for Antibacterial Activity
Some researchers have obtained four types of antibacterial materials and coatings during the last decade. The first types of antibacterial coatings are those to which bacteria do not show a natural tendency to attach. Frequently, this form of coating is focused on hydrophilic polymers like polyethylene glycol (PEG), oxazolines, radicals in nitroxide, and chlorinated plasma polymers [419,420,421]. The second type of coatings or compounds that are modified can destroy bacteria while in contact. Studies have shown that an RNH4+ bonded nitrogen concentration of 4.18% and a surface potential of +120.4 mV is needed for the killing of Escherichia coli efficiently [422,423], with a carefully planned amount density gradient of QAC. Figure 8 presents strategies that provide excellent approaches to avoid the colonization of bacteria on the device surface while the bacteria that have penetrated the implantation site are not removed. An exposed wound is therefore infected by opportunistic pathogens. Coatings or materials that release antibacterial agents have been produced for pathogen neutralization. A significant range of antibacterial substances, including classical antibiotics [424,425], nitrous oxide [426,427], antibacterial polymers, and peptides, may be issued under this strategy [428,429,430]. The last category consists of coatings and materials that trigger antibacterial material only when the product has been infected or engulfed by photogenic bacteria (Figure 8) [431,432,433].

Figure 8.
Types of antibacterial coatings: (a) the repelling of bacterial contact or formation of biofilms; (b) Contact killing; (c) release of antibacterial agents; (d) triggers the secretion of receptive compounds in the presence of bacteria.
4.7. Factors Influencing the Synthesis of Various NPs
Three main factors influence the synthesis of various NPs. They are temperature, pH, and reaction time (Figure 9).

Figure 9.
Factors influencing the synthesis of the Nanoparticles.
4.7.1. Temperature
Significant research is being currently being carried out worldwide on the question of temperature regulation in NPs. Temperature is one of the most critical parameters that affect the morphology and synthesis of the NPs. The different forms, dimensions, and synthetic structure of NPs are temperature-dependent (triangles, platelets of octahedral, spherical, and rod-like). The rate of the reaction, as well as the formation of nuclear centers, tends to increase with the increase in temperature [434,435,436]. Sneha et al. (2010) analyzed the temperature effects on the morphology of Piper leaf extract synthesized Au NPs [437]. They found triangular NP types at 20 °C, while octahedral NPs of 5–500 nm were produced at temperatures from 30 to 40 °C. Therefore, the size distribution of the NPs was much more appropriate and spherical in the form at increased temperature (50–60 °C). Iravani and Zolfaghari (2013) [438] documented the biological development of Ag NPs via P. eldarica bark extracts under different temperatures. The synthesis was conducted at 25, 50, 100, and 150 °C. Triangular NP types were found at 20 °C, while octahedral NPs of 5–500 nm were produced at temperatures from 30 to 40 °C. The NPs were produced at 20 °C. Electron micrographs scan of NPs confirmed this result. In the synthesis of the AgNPs using biocompatible polymer PEG [439], the effect and temperature of atmospheric oxidation have been investigated by Fleitas-Salazar et al. (2017). The analysis showed that PEG had a tendency towards decreasing silver salt at 100 °C. It was also suggested that the functional PEG groups strongly interacting with Ag molecules at 100 °C resulted in a more balanced and stabilized Ag-NP structure. At 60 °C, Ag ions were reduced by hydroxyl group oxidations found in PEG. The researchers have thus confirmed that there are many methods for Ag NP synthesis operating at different temperatures. The synthesis procedure for AU NPs in aqueous poly (ethylene oxide)-poly (propylene oxide) solution at various temperature levels was developed by Islam et al. (2011). Their findings revealed that the NPs are regulated by changes in polymer morphology occurring at relatively low temperatures, distribution, and size. However, modification in polymer chemistry is regulated at higher temperatures [440]. The impact of temperatures upon the encapsulation and development of Au NPs was studied by Tans et al. (2015) using the standard PNIPAm/PEI. The TEM analysis showed that the optimum encapsulation of Au NPs, and that of stable Au-PNIPAm/PEI composite particle production, is between 25–30 °C, with homogeneous particle distribution across the template. The encapsulation at a lower temperature (15 °C) was relatively poor [441]. In another review of the synthesis of cobalt ferrite NPs doped by Manganese [442], the number of NPs was shown to rise with the increase in temperature.
4.7.2. pH
The pH of the reaction plays a crucial role in forming NPs. The formation of nuclear centers is also regulated by the pH-like temperature. With the increase in pH, there is a parallel rise in the number of nuclear sites, hence the growing number of metal NPs. Several studies on the effectiveness of pH in developing the morphology and scale of NPs have been conducted. Armendariz et al. (2004) have observed the synthesis of A. sativa Au NPs at various pH levels. The lower pH (pH 2) revealed fewer NPs but a much greater NPs size (25–85 nm). They indicated that Au NPs do not generate any new nuclear centers at a lower pH but instead had a tendency to aggregate into larger NPs [443]. In contrast, small NPs were developed with a slightly higher pH (pH 3–4). Fan et al. performed an additional analysis of the synthesis of pH-dependent NPs. They monitored the release of NIPAm poly-(N-isopropylacrylamide)/chitosan NPs filled with Camptothecin into the tumor and noted that when the NIPAm and chitosan ratio is 4:1 (w/w), the loaded NPs are released to the target. At pH 6.8 the rate of release of Camptothecin was considered to be ideal. However, they noted that the release rate decreased as the pH decreased or increased to the 37 °C temperature. Okitsu et al. (2009) [444] performed further experimental analysis to examine the impact of pH on dimensions of the average size of the gold nanorods and concluded that the ratio and the size decreased with an increase in pH.
4.7.3. Reaction Time
The reaction time, along with the temperature and pH, is considered an essential factor affecting the NP morphology. Karade et al. (2018) [445] conducted an analysis of the impact of the reaction time on magnetic NPs. Fe NPs have been synthesized with green tea extract using Ferric nitrate solution. It was reported that the reaction time affected both the structural and magnetic properties of magnetic NPs. The size of the particles enlarges from 7.5–12 nm as the reaction time rises. Increasing reaction time has also been observed to increase the magnetic saturation (Ms) of NPs. Additionally, the impact of the duration of reaction on cadmium selenide NP particle size has been studied [446]. The estimated particle sizes at 4, 8, 12, and 16 h reaction times were determined to be 15.8, 10.5, 6.7, and 111.7 nm, respectively. The size of cadmium selenide NPs decreased with an increase in reaction time. It was reported that the unusual size increase at 16 h resulted from particle accumulation. Furthermore, Flor et al. (2004) studied the impact of response time on the particle sizes of ZnO and Cerium doped ZnO [447]. The study indicated the linear increase in particle size with reaction time rise. It was found that at constant periods of reaction, cerium doped particle sizes are greater than standard ZnO. Finally, the impact of the reaction time on stability, size, and reduction of AuNPs synthesized from oil palm oil extracted (E. guineensis) was examined, and it was observed that with the rise in reaction time, the reduction rate of AgNPs was also increased.
4.8. Factors Influencing the Activity of Nanoparticles
Nanoparticle behavior toward the microbes (bacteria and fungi) may be caused and influenced by various factors. Here, we identified several that could, theoretically, affect nanoparticle antimicrobial behavior. Chemical structure or size, composition, the concentration of nanoparticles, along with their form, target microorganism nanoparticles are acting against, and the effect of photoactivation are addressed below (Figure 10).

Figure 10.
Factors influencing the activity of Nanoparticles.
4.8.1. Chemical Composition of Nanoparticle
Chemical Composition is the crucial feature of the antibacterial mechanism in nanostructures. Many studies have focused on the choice of a substance suitable for nanoparticle synthesis and the potential effects such choices would entail for the antibacterial mechanisms [281,448]. Analysis revealed the significance of the chemical compositions of nanoparticles and the way nanoparticles are sprayed against bacterial cells. There were substantial effects in copper nanoparticles compared with iron, size 30–40 nm and 30–70 nm, respectively, of ten drug-resistant S. aureus. Recent research interest has turned to the combined production of nanoparticles with the objective of achieving chemical and antibacterial activities.
4.8.2. Shape of Nanoparticles
Nanoparticle shape has been shown to affect the effectiveness of several antibacterial nanoparticles. The shape-dependent behavior of silver nanoparticles was defined by three distinct shapes (sphere, elongated, and truncated triangular silver nano-plates). Analysis of E. coli activity was based on the percentage of the active facets. Increased antimicrobial activity with more facets [111] with high atomic density facets was observed [224].
4.8.3. The Target Organisms
The micro-organisms targeted by the nanoparticle have significant effects on its activities. A higher percentage of AgNP activity towards Gram-negative rods than Gram-positive cocci and higher tolerance to E. coli than to S. aureus for silver nanoparticles are good examples. The results were linked to peptidoglycan that is absent in mammalian cells in the S. aureus cell wall. The cell walls of both Gram-negative and Gram-positive bacteria, in contrast, were disordered by ZnO NPs [449,450]. In ZnO NPs, the operation against S. aureus was higher than that against E. coli [263,451]. These findings demonstrate the target-dependent nature of the relationship between antibacterial activity and nanoparticles.
4.8.4. The Photoactivation
Photoactivation is considered the most relevant parameter of the antibacterial behavior of the nanoparticle. When exposed to UV radiation, the TiO2 NPs were considerably more active against E. coli [452]. TiO2 nanoparticles with exposure to standard laboratory lightning exhibited a 20% increase in growth control properties, and ZnO nanoparticles, when exposed to visible light and UV radiation resulted, also showed increased activity [453].
5. Characterization of Nanoparticles
After the synthesis of the NPs, a range of methodologies are used to evaluate their conformational specifications for size, form, homogeneity dispersity, and morphology. Dynamic Light Scattering (DLS), UV-Vis Absorption Spectroscopy, X-Ray Diffraction (XRD), Electron Microscopy Transmission (TEM), Energy Dispersive X-ray Analysis (EDAX), and Scanning Electron Microscopy (SEM) are methods most frequently used to characterize NPs (Figure 11).
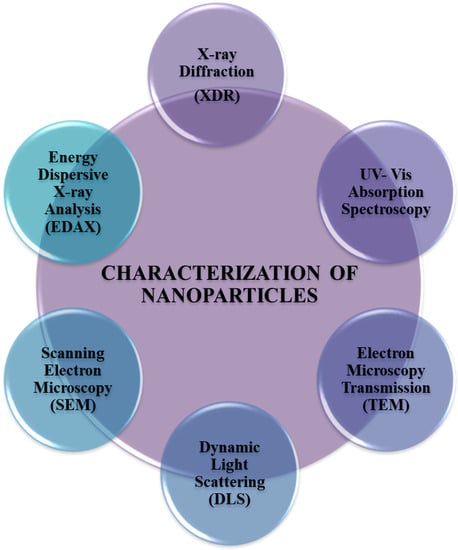
Figure 11.
Methods for the characterization of Nanoparticles.
The UV—Vis absorption spectra are used for the aqueous suspension of NPs in size and form [454]. To identify the NPs of approximately 2 to 100 nm [455], wavelengths from 300 to 800 nm are usually used. For instance, UV—Vis emission spectra of Aloe Vera extract synthesized ZnO particles display high UV spectrum absorption with a higher wavelength of 358 nm to 375 nm because of their membrane Plasmon resonance [456].
The SEM and TEM describe the morphology and scale of NPs [457]. ZnO-NP (25 to 55 nm) seen in the electron microscopy study is compatible with the XRD [458]. The green synthesized carbon nanotubes were analyzed by SEM and TEM, which were completely coated with polyaniline layers [459]. TiO2 particles were commonly spherically agglomerated within the range of 10 to 30 nm in the TEM examination. In addition, a crystalline structure was shown by Selected Area Electron Diffraction (SAED) [460].
XRD contains data regarding symmetry, sizes, and the state of metallic NP detection [461]. X-rays penetrate nanomaterials. The division sequence collected is correlated with structural knowledge requirements. XRD peaks (2 h) at 28.51, 33.06, and 47.42 angles of 111, 200, and 220 planes, respectively, and normal separation peaks at the front-and-center cubic step of the CeO2 NPs [462]. The XRD analysis confirmed the existence in the Scherer equation (Elango and Roopan 2015) of the crystalline patterns of PbNPs and the average particle size of 47 nm.
FTIR Spectroscopy is designed to identify different types of functional groups or metabolites that may contribute to the reduction and stabilization of NPs at the surface of NPs [378]. The functional group bands observed at 3450, 3266, and 2932 cm−1 have been allocated to stretching the amines frequencies, O–H to stretch alcohols, and C–H to extend the alkanes for NPs using Aloe-Vera extracts. ZnO is allocated for the peaks in the region of 600–400 cm−1 [463]. In 1648, 1535, 1450, and 1019 cm−1 and 1450 cm−1 peaks of carboxyl ions, the FTIR range of AgNP synthesized using the Solanumtorvum leaf extract was reportedly responsible for stabilization of the Ag NPs [464].
DLS and EDAX are used to study the flow of size of liquids and essential components of NPs accordingly [465,466,467].
6. Comparison of Antibiotics with Nanoparticles
Although antibiotics have been important historically, nanoparticles are increasingly being used in emerging medical research and application. If the efficacy and potential of antibiotics and nanoparticles are to be compared, their combined action will prove to be more significant than that of either one alone. Some comparisons are offered below in Table 8.

Table 8.
Comparison of Nanoparticles with antibiotics and their combination.
7. Antimicrobials and Nanoparticles in Combination
As already stated, NPs do not only defend themselves against bacterial and microbial resistance but can also be the “medium and carrier” for antibiotics. The methodologies of NP-based drugs vary based on the pathways discussed earlier.
The essential features of NPs as a carrier for the delivery of antibiotics, in contrast with conventional delivery systems, are:
- Size: the ultra-small and customizable size of NPs is ideal for performing antimicrobial activities and combating intracellular bacteria [467,468].
- Protection: NP carriers can help protect the drugs from resistance by target bacteria by increasing the serum levels of antibiotics [280,469]
- Precision and Safety: NP carriers can help find a contaminated region with antibiotics and eliminate systemic adverse reactions [470,471].
- Controllability: safe and controllable antibiotic release can be flexibly achieved [472,473,474].
- Combination: the same NP may be used to combine several antibiotics or drugs, and NPs can be combined with others to enhance the antimicrobial properties of the antibacterial agents [282,472,476]
Recently, researchers have combined the gold and silver nanoparticles with Ampicillin. They reported that silver nanoparticles have an inherent ability to combat the microorganism, although gold nanoparticles have an antimicrobial effect only when in surface communication with ampicillin. Broad-spectrum action against the Gram-negative bacteria and Gram-positive bacteria is accomplished by the ampicillin-functioning of silver and gold nanoparticles. Silver and gold combined with ampicillin are very effective in treating bacteria resistant to antibiotics [477].
Any effective antibiotic, including certain penicillins, can be recovered through the use of antibiotics with metallic nanoparticles [245,252,261,267]. In addition, the combined use of nanoparticles with antibiotics or other antimicrobials allows these agents to decrease the toxicity in human cells [269]. The majority of research is dedicated to the study of the interactions of nanoparticles and antibiotics and their various combinations, especially with b-lactams (ampicillin, amoxicillin) and glycopeptides (vancomycin), which have shown promising enhancing effects in vitro. These effects are much more likely due to the increased penetration of these nanoparticles in the cell wall. While the interaction of other metallic nanoparticles with antibiotics has still not been thoroughly investigated, ZnO and TiO2 nanoparticles have shown, with limited evidence that they can respond to efflux pumps that cause the resistance of many clinically important antibiotics, such as fluoroquinolones and many more [478].
8. Antimicrobial Applications of Nanoparticles on Animal Model
Although there are currently several applications of the nanoparticles, only a few studies have been conducted in the context of antimicrobial activity. Some of the antimicrobial applications of nanoparticles on different animal models are shown in Table 9.

Table 9.
Nano-particles and their applications on animal models.
9. Challenges for Nanoparticle
The benefits of nanotechnology in numerous fields, including medicine, are well-established, but the potential implications or harmful effects of these nano-sized particles are not well-explained. In the medical context, as an alternative antibiotic, a whole range of functions and relationships of the nanoparticles can be found [483]. A number of groundbreaking studies are underway in nanotechnology, while the critics of these new technologies find it incredibly challenging to reconcile the side of nanoparticles with their reported strengths. As nanotechnology development is gaining momentum, concerns have been raised about its safety, particularly in medicine [484]. Nano-formulation plays an essential role in the development of nano-based drugs and thus faces regulatory issues relating to medicines. To design a new nanodrug based on the form of drugs already existing, specific regulatory manufacturing criteria must be provided and rigorously adhered to throughout drug production. A shorter approval process is pursued for nanodrugs developed from previously licensed micro-formulations; however, when a novel product is formulated, the paths of assessment and approval are more stringent [485]. Manufacturers must follow the guidance of the FDA (Current Appropriate Manu—Invoicing Practices) and the Quality Control Regulations for the production of new Nano drugs.
Nanoparticles have significant benefits and advancements in the management of infectious diseases over conventional antibiotics; however, their delivery technique is still difficult in clinical applications. For proper therapeutic results and effective clinical application, an assessment of the potential nanoparticle interactions with tissues, organs, or cells, their doses, and potential mode of administration, is needed [486]. Nanoparticle toxicity is necessary for effective clinical translation to take place. [487]. However, intravenous (I.V.) NP administration can lead to NPs accumulating in bone marrow, spleen, liver, and lung [488]. Due to their small size and effective cell absorption, inhaled NPs can enter the liver, brain, spleen, heart, and lung [489]. Moreover, the toxicity of several nanoparticles is not well known [483]. NP therapeutic administration can produce nanotoxicity of multiorganisms. All toxic cases show NPs association with oxidative stress-inducing cells contributing to hepatotoxicity and pulmonary toxicity [490,491]. Suggested metabolic modifications such as mitochondrial dysfunction, decreased ketogenesis, beta-oxidation of fatty acids, and glycolysis contribute to hepatotoxicity and nephrotoxicity [492]. While preexisting in vitro methods have certain advantages, there are no popular practices for universal NP dosing [484,488], as nanoparticles demonstrate size-specific action. To solve these problems, additional characterization methods are required [490]. More recent work mainly focuses on delivering targeted bacteria [493,494].
10. Conclusions
Antimicrobial resistance has posed an unprecedented global threat to human life. Conventional antibiotics are losing their potency against the ever-evolving multiple drug-resistant pathogens. In search for a viable alternative strategy, a nanotechnology-offers promise and nanotechnology-based drug delivery device for the production of future nano-antibiotics (nAbts) is seen as a weapon in the 21st century technological revolution. To evolve novel pharmaceutical drugs over time, the newly developed field of nanotechnology is still in its infancy and requires significant effort and investment. Compared with traditional antibiotics, nanoparticles can have numerous benefits, such as longevity, absorption, controlled release, distribution, and delivery.
Furthermore, for the resistant, antimicrobial environment, nanoparticles can be cost-effective and ecological, as well as flexible. Nanoparticles have distinct and well-defined physical and chemical characteristics that can be customized for desirable purposes. Moreover, owing to the excessive volumetrically surface area, they have a strong antimicrobial performance that gives them an advantage over their chemical counterparts facing drug resistance challenges. The advances in nanotechnology and nanoparticle synthesis have opened the floodgates for groundbreaking strategies in the production of new antimicrobial agents. A multitude of processes that vary in traits, including size, morphology, electrical charge, and surface coatings, allow researchers to develop novel composite antimicrobial substances for different applications used to perform antimicrobial activities. The antimicrobial activity of inorganic nanoparticles and carbon-based nanoparticles can be applied to various research, medical, and industrial uses in the future and offer a solution to the crisis of antimicrobial resistance to traditional approaches. In addition, nanomaterials provide a wide range of opportunities for infection prevention, diagnosis, treatment, and biofilm control. However, a detailed evaluation of these nanomaterials is required to identify their effects on natural organic tissues and to assess their impact on humans and the environment before large-scale industry implementations are carried out.
Author Contributions
Conceptualization, M.S.N., and I.K.; Writing—original draft preparation, B.M., A.N.A., R.R., and I.U., Writing—review and editing, S.S.I., S.A., M.M.G., and S.I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
Authors are thankful to our supporting fellows.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Etebu, E.; Arikekpar, I. Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 2016, 4, 90–101. [Google Scholar]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Genet. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Padiyara, P.; Inoue, H.; Sprenger, M. Global Governance Mechanisms to Address Antimicrobial Resistance. Infect. Dis. Res. Treat. 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. J. Manag. Care Hosp. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- Baluja, Z.; Nabi, N.; Ray, A. Challenges in Antimicrobial Resistance: An Update, EC Pharmacol. Toxicology 2018, 6, 865–877. [Google Scholar]
- Davis, M.D.M.; Whittaker, A.; Lindgren, M.; Djerf-Pierre, M.; Manderson, L.; Flowers, P. Understanding media publics and the antimicrobial resistance crisis. Glob. Public Health 2017, 13, 1158–1168. [Google Scholar] [CrossRef]
- Blecher, K.; Nasir, A.; Friedman, A. The growing role of nanotechnology in combating infectious disease. Virulence 2011, 2, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef]
- Teixeira, M.; Sanchez-Lopez, E.; Espina, M.; Calpena, A.; Silva, A.; Veiga, F.; Garcia, M.L.; Souto, E. Chapter 9—Advances in antibiotic nanotherapy: Overcoming antimicrobial resistance. In Emerging Nanotechnologies in Immunology; Elsevier: Boston, MA, USA, 2018; pp. 233–259. [Google Scholar]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven Ways to Preserve the Miracle of Antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef]
- Adeniji, F. Global analysis of strategies to tackle antimicrobial resistance. Int. J. Pharm. Pract. 2018, 26, 85–89. [Google Scholar] [CrossRef]
- Wong, I.Y.; Bhatia, S.N.; Toner, M. Nanotechnology: Emerging tools for biology and medicine. Genes Dev. 2013, 27, 2397–2408. [Google Scholar] [CrossRef]
- Jena, M.; Mishra, S.; Jena, S.; Mishra, S. Nanotechnology-future prospect in recent medicine: A review. Int. J. Basic Clin. Pharmacol. 2013, 2, 353–359. [Google Scholar] [CrossRef]
- Allen, H.; Moe, L.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse Beta-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef]
- Donato, J.J.; Moe, L.A.; Converse, B.J.; Smart, K.D.; Berklein, F.C.; McManus, P.S.; Handelsman, J. Metagenomic Analysis of Apple Orchard Soil Reveals Antibiotic Resistance Genes Encoding Predicted Bifunctional Proteins. Appl. Environ. Microbiol. 2010, 76, 4396–4401. [Google Scholar] [CrossRef]
- Georgiev, V.S. National Institute of Allergy and infectious diseases. In NIH: Volume 2: Impact on Global Health; Springer Science & Business Media: North Bethesda, MD, USA, 2009. [Google Scholar]
- Morgun, A.; Dzutsev, A.; Dong, X.; Greer, R.; Sexton, D.J.; Ravel, J.; Schuster, M.; Hsiao, W.; Matzinger, P.; Shulzhenko, N. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 2015, 64, 1732–1743. [Google Scholar] [CrossRef] [PubMed]
- Scully, J.L. What is a disease? Disease, disability and their definitions. Eur. Mol. Biol. Organ. 2004, 7, 650–653. [Google Scholar]
- Friswell, M.; Campbell, B.; Rhodes, J. The Role of Bacteria in the Pathogenesis of Inflammatory Bowel Disease. Gut Liver 2010, 4, 295–306. [Google Scholar] [CrossRef]
- Baker, S.J.; Hui, X.; Maibach, H.I. Progress on new therapeutics for fungal nail infections. Ann. Rep. Med. Chem. 2005, 40, 323. [Google Scholar]
- Peiris, J.S.; Guan, Y.; Yuen, K.Y. Severe acute respiratory syndrome. Nat. Med. 2004, 10, S88–S97. [Google Scholar] [CrossRef]
- Mosqueda, J.; Olvera-Ramirez, A.; Aguilar-Tipacamu, G.; Canto, G.J. Current Advances in Detection and Treatment of Babesiosis. Curr. Med. Chem. 2012, 19, 1504–1518. [Google Scholar] [CrossRef]
- Seok, J.Y.; Lee, Y.; Lee, H.; Yi, S.Y.; Oh, H.E.; Song, J.-S. Human Cutaneous Protothecosis: Report of a Case and Literature Review. Korean J. Pathol. 2013, 47, 575–578. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Lepp, P.W.; Relman, D.A. Archaea and Their Potential Role in Human Disease. Infect. Immun. 2003, 71, 591–596. [Google Scholar] [CrossRef]
- Ishaq, S.L.; Moses, P.L.; Wright, A.D. The pathology of methanogenic archaea in human gastrointestinal tract disease. Gut Microbiome Implic. Hum. Dis. 2016. [Google Scholar] [CrossRef]
- De Jesus, C.; Whiting, R. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 2003, 66, 1611–1617. [Google Scholar] [CrossRef]
- Mohr, K.I. History of Antibiotics Research. In Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2016; Volume 398, pp. 237–272. [Google Scholar]
- Bankova, V. Chemical diversity of propolis and the problem of standardization. J. Ethnopharmacol. 2005, 100, 114–117. [Google Scholar] [CrossRef]
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharm. 2017, 133, 4–19. [Google Scholar] [CrossRef]
- Oliver, S.P.; Murinda, S.E.; Jayarao, B.M. Impact of Antibiotic Use in Adult Dairy Cows on Antimicrobial Resistance of Veterinary and Human Pathogens: A Comprehensive Review. Foodborne Pathog. Dis. 2011, 8, 337–355. [Google Scholar] [CrossRef]
- Armelagos, G.J.; Kolbacher, K.; Collins, K.; Cook, J.; Krafeld-Daugherty, M. Tetracycline consumption in prehistory. In Tetracyclines in Biology, Chemistry and Medicine; Springer: Cham, Switzerland, 2001; pp. 219–236. [Google Scholar]
- Ryu, C.; Lee, K.; Yoo, C.; Seong, W.K.; Oh, H.-B. Sensitive and Rapid Quantitative Detection of Anthrax Spores Isolated from Soil Samples by Real-Time PCR. Microbiol. Immunol. 2003, 47, 693–699. [Google Scholar] [CrossRef]
- Lipp, E.K.; Huq, A.; Colwell, R.R. Effects of Global Climate on Infectious Disease: The Cholera Model. Clin. Microbiol. Rev. 2002, 15, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Ostensvik, O.; Skulberg, O.M.; Underdal, B.; Hormazabal, V. Antibacterial properties of extracts from selected planktonic freshwater cyanobacteria—A comparative study of bacterial bioassays. J. Appl. Microbiol. 1998, 84, 1117–1124. [Google Scholar] [CrossRef]
- Arai, T.; Mikami, Y.; Fukushima, K.; Utsumi, T.; Yazawa, K. A new antibiotic, leucinostatin, derived from penicillium lilacinum. J. Antibiot. 1973, 26, 157–161. [Google Scholar] [CrossRef]
- Nadar, V.S.; Chen, J.; Dheeman, D.S.; Galván, A.E.; Yoshinaga-Sakurai, K.; Kandavelu, P.; Sankaran, B.; Kuramata, M.; Ishikawa, S.; Rosen, B.P.; et al. Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic. Commun. Biol. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Cole, H.N. The use of antisyphilitic remedies. J. Am. Med. Assoc. 1936, 107, 2123. [Google Scholar] [CrossRef]
- Xiong, G.M.; Venkatraman, K.; Venkatraman, S. The magic bullet as cancer therapeutic—Has nanotechnology failed to find its mark? Prog. Biomed. Eng. 2020, 2, 042004. [Google Scholar] [CrossRef]
- Giesbrecht, P.; Kersten, T.; Maidhof, H.; Wecke, J. Staphylococcal cell wall: Morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 1998, 62, 1371–1414. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, D. The effects of biocidal treatments on metabolism in soil—IV. The decomposition of fumigated organisms in soil. Soil Biol. Biochem. 1976, 8, 203–208. [Google Scholar] [CrossRef]
- Bentley, R. Different roads to discovery; Prontosil (hence sulfa drugs) and penicillin (hence β-lactams). J. Ind. Microbiol. Biotechnol. 2009, 36, 775–786. [Google Scholar] [CrossRef]
- Yousef, F.; Mansour, O.; Herbali, J. Sulfonamides: Historical Discovery Development (Structure-Activity Relationship Notes). In-Vitro In-Vivo In-Silico J. 2018, 1, 1–15. [Google Scholar]
- Genç, Y.; Özkanca, R.; Bekdemir, Y. Antimicrobial activity of some sulfonamide derivatives on clinical isolates of Staphylococus aureus. Ann. Clin. Microbiol. Antimicrob. 2008, 7, 17. [Google Scholar] [CrossRef]
- Wood, W.B.; Long, P.H. Observations upon the experimental and clinical use of sulfapyridine. III. The mechanism of recovery from pneumococcal pneumonia in patients treated with sulfapyridine. Ann. Intern. Med. 1939, 13, 612. [Google Scholar] [CrossRef]
- Stauss-Grabo, M.; Atiye, S.; Le, T.; Kretschmar, M. Decade-long use of the antimicrobial peptide combination tyrothricin does not pose a major risk of acquired resistance with gram-positive bacteria and Candida spp. Die Pharm. 2014, 69, 838–841. [Google Scholar]
- Takada, Y.; Itoh, H.; Paudel, A.; Panthee, S.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Discovery of gramicidin A analogues with altered activities by multidimensional screening of a one-bead-one-compound library. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Stark, B.J.; Earl, H.S.; Gross, G.N.; Lumry, W.R.; Goodman, E.L.; Sullivan, T.J. Acute and chronic desensitization of penicillin-allergic patients using oral penicillin. J. Allergy Clin. Immunol. 1987, 79, 523–532. [Google Scholar] [CrossRef]
- Genvert, I.G.; Cohen, E.J.; Donnenfeld, E.D.; Blecher, M.H. Erythema multiforme after use of topical sulfacetamide. Am. J. Ophthalmol. 1985, 99, 465–468. [Google Scholar] [CrossRef]
- Singha, P.; Locklin, J.; Handa, H. A review of the recent advances in antimicrobial coatings for urinary catheters. Acta Biomater. 2017, 50, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Abeylath, S.C.; Turos, E. Drug delivery approaches to overcome bacterial resistance to β-lactam antibiotics. Expert Opin. Drug Deliv. 2008, 5, 931–949. [Google Scholar] [CrossRef]
- Forrest, D.M.; Schellenberg, R.R.; Thien, V.V.S.; King, S.; Anis, A.H.; Dodek, P.M. Introduction of a Practice Guideline for Penicillin Skin Testing Improves the Appropriateness of Antibiotic Therapy. Clin. Infect. Dis. 2001, 32, 1685–1690. [Google Scholar] [CrossRef]
- Brooks, D.; Garrett, G.; Hollihead, R. Sulphadimidine, co-trimoxazole, and a placebo in the management of symptomatic urinary tract infection in general practice. J. R. Coll. Gen. Pract. 1972, 22, 695–703. [Google Scholar]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2017, 36, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Chapple, D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Hossain, G.G.; Amoroso, A.; Banu, A.; Malik, K. Syntheses and characterisation of mercury complexes of sulfadiazine, sulfamerazine and sulfamethazine. Polyhedron 2007, 26, 967–974. [Google Scholar] [CrossRef]
- Shama, G.; Shama, G. Zones of inhibition? The transfer of information relating to penicillin in Europe during World War II. Adv. Appl. Microbiol. 2009, 69, 133–158. [Google Scholar]
- Lipsky, B.A.; Hoey, C. Topical Antimicrobial Therapy for Treating Chronic Wounds. Clin. Infect. Dis. 2009, 49, 1541–1549. [Google Scholar] [CrossRef]
- Harir, M.; Bendif, H.; Bellahcene, M.; Pogni, Z.F.A.R. Streptomyces Secondary Metabolites. Basic Biol. Appl. Actinobacteria 2018, 6, 99–122. [Google Scholar] [CrossRef]
- Strohl, W.R. Antimicrobials. In Microbial Diversity and Bioprospecting; ASM Press: Washington, DC, USA, 2014; pp. 336–355. [Google Scholar] [CrossRef]
- Figueroa, R.A.; Leonard, A.; Mackay, A.A. Modeling Tetracycline Antibiotic Sorption to Clays. Environ. Sci. Technol. 2004, 38, 476–483. [Google Scholar] [CrossRef]
- He, Z.; Kisla, D.; Zhang, L.; Yuan, C.; Green-Church, K.B.; Yousef, A.E. Isolation and Identification of a Paenibacillus polymyxa Strain That Coproduces a Novel Lantibiotic and Polymyxin. Appl. Environ. Microbiol. 2006, 73, 168–178. [Google Scholar] [CrossRef]
- Gadebusch, H.H.; Basch, H.I. New Antimicrobial Nitrofuran, trans-5-Amino-3-[2-(5-Nitro-2-Furyl) Vinyl]-Δ2-1,2,4-Oxadiazole: Antibacterial, Antifungal, and Antiprotozoal Activities In Vitro. Antimicrob. Agents Chemother. 1974, 6, 263–267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wulf, N.R.; Matuszewski, K.A. Sulfonamide cross-reactivity: Is there evidence to support broad cross-allergenicity? Am. J. Health Pharm. 2013, 70, 1483–1494. [Google Scholar] [CrossRef]
- Stypulkowska, K.; Blazewicz, A.; Fijalek, Z.; Warowna-Grześkiewicz, M.; Srebrzynska, K. Determination of neomycin and related substances in pharmaceutical preparations by reversed-phase high performance liquid chromatography with mass spectrometry and charged aerosol detection. J. Pharm. Biomed. Anal. 2013, 76, 207–214. [Google Scholar] [CrossRef]
- Das, S.; Al Faysal, M.N.; Ferdous, J.; Sachi, S.; Islam, M.S.; Sikder, M.H. Detection of oxytetracycline and doxycycline residue in different growth stages of commercial broiler. Bangladesh J. Vet. Med. 2009, 17, 7–14. [Google Scholar]
- George, A.M.; Levy, S.B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: Involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 1983, 155, 531–540. [Google Scholar] [CrossRef]
- Yoneyama, H.; Katsumata, R. Antibiotic Resistance in Bacteria and Its Future for Novel Antibiotic Development. Biosci. Biotechnol. Biochem. 2006, 70, 1060–1075. [Google Scholar] [CrossRef]
- Cannon, M.; Harford, S.; Davies, J. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J. Antimicrob. Chemother. 1990, 26, 307–317. [Google Scholar] [CrossRef]
- Amin, M.M.; Zilles, J.L.; Greiner, J.; Charbonneau, S.; Raskin, L.; Morgenroth, E. Influence of the Antibiotic Erythromycin on Anaerobic Treatment of a Pharmaceutical Wastewater. Environ. Sci. Technol. 2006, 40, 3971–3977. [Google Scholar] [CrossRef] [PubMed]
- Crank, C.W.; O’Driscoll, T. Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015, 8, 217–230. [Google Scholar] [CrossRef]
- Abraham, E.P. Cephalosporins 1945–1986. Drugs 1987, 34, 1–14. [Google Scholar] [CrossRef]
- Zhong, P.; Shortridge, V.D. The role of efflux in macrolide resistance. Drug Resist. Updat. 2000, 3, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.E.; Okeke, N.L.; Hicks, C.B. Treatment of syphilis: A systematic review. JAMA 2014, 312, 1905–1917. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Wu, L.; Zhu, M.; He, G.; Chen, X.; Sun, F.; Liu, Q.; Wang, X.; Zhang, W. Cycloserine for treatment of multidrug-resistant tuberculosis: A retrospective cohort study in China. Infect. Drug Resist. 2019, 12, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Valentini, P.; Annunziata, M.L.; Angelone, D.F.; Masini, L. Role of spiramycin/cotrimoxazole association in the mother-to-child transmission of toxoplasmosis infection in pregnancy. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Flynt, L.K.; Kenney, R.M.; Zervos, M.J.; Davis, S.L. The Safety and Economic Impact of Cefazolin versus Nafcillin for the Treatment of Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections. Infect. Dis. Ther. 2017, 6, 225–231. [Google Scholar] [CrossRef]
- Weisblum, B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 1995, 39, 797–805. [Google Scholar] [CrossRef]
- Gikalo, M.B.; Nosova, E.Y.; Krylova, L.Y.; Moroz, A.M. The role of eis mutations in the development of kanamycin resistance in Mycobacterium tuberculosis isolates from the Moscow region. J. Antimicrob. Chemother. 2012, 67, 2107–2109. [Google Scholar] [CrossRef]
- Schiewe, H.J.; Zeeck, A. Cineromycins, γ-butyrolactones and ansamycins by analysis of the secondary metabolite pattern created by a single strain of Streptomyces. J. Antibiot. 1999, 52, 635–642. [Google Scholar] [CrossRef]
- Cheng, I.-L.; Chen, Y.-H.; Lai, C.-C.; Tang, H.-J. Intravenous Colistin Monotherapy versus Combination Therapy against Carbapenem-Resistant Gram-Negative Bacteria Infections: Meta-Analysis of Randomized Controlled Trials. J. Clin. Med. 2018, 7, 208. [Google Scholar] [CrossRef]
- Müller, M. Reductive activation of nitroimidazoles in anaerobic microorganisms. Biochem. Pharm. 1986, 35, 37–41. [Google Scholar] [CrossRef]
- Enwemeka, C.S.; Williams, D.; Enwemeka, S.K.; Hollosi, S.; Yens, D. Blue 470-nm Light Kills Methicillin-Resistant Staphylococcus aureus (MRSA) in Vitro. Photomed. Laser Surg. 2009, 27, 221–226. [Google Scholar] [CrossRef]
- Mitchell, D.A. Metronidazole: Its use in clinical dentistry. J. Clin. Periodontol. 1984, 11, 145–158. [Google Scholar] [CrossRef]
- Harkins, C.P.; Pichon, B.; Doumith, M.; Parkhill, J.; Westh, H.; Tomasz, A.; De Lencastre, H.; Bentley, S.D.; Kearns, A.M.; Holden, M.T.G. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 18, 130. [Google Scholar] [CrossRef]
- Klein, J.O. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 1997, 16, S5–S8. [Google Scholar] [CrossRef]
- Riedel, S.; Vijayakumar, D.; Berg, G.; Kang, A.D.; Smith, K.P.; Kirby, E.J. Evaluation of apramycin against spectinomycin-resistant and -susceptible strains of Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2019, 74, 1311–1316. [Google Scholar] [CrossRef]
- Jönsson, S.; Davidse, A.; Wilkins, J.; Van Der Walt, J.-S.; Simonsson, U.S.H.; Karlsson, M.O.; Smith, P.; McIlleron, H. Population Pharmacokinetics of Ethambutol in South African Tuberculosis Patients. Antimicrob. Agents Chemother. 2011, 55, 4230–4237. [Google Scholar] [CrossRef][Green Version]
- Werner, A.; Russell, A. Mupirocin, fusidic acid and bacitracin: Activity, action and clinical uses of three topical antibiotics. Veter Dermatol. 1999, 10, 225–240. [Google Scholar] [CrossRef]
- Andriole, V.T. The Quinolones: Past, Present, and Future. Clin. Infect. Dis. 2005, 41, S113–S119. [Google Scholar] [CrossRef]
- Imbuluzqueta, E.; Elizondo, E.; Gamazo, C.; Moreno-Calvo, E.; Veciana, J.; Ventosa, N.; Blanco-Prieto, M.; María, J. Novel bioactive hydrophobic gentamicin carriers for the treatment of intracellular bacterial infections. Acta Biomater. 2011, 7, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Bonomo, A.R. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: β-lactams in peril! Curr. Opin. Microbiol. 2005, 8, 518–524. [Google Scholar] [CrossRef]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.D.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015, 22, 90–101. [Google Scholar] [CrossRef]
- Crumplin, G.C.; Smith, J.T. Nalidixic Acid: An Antibacterial Paradox. Antimicrob. Agents Chemother. 1975, 8, 251–261. [Google Scholar] [CrossRef]
- Holmes, N.E.; Charles, P.G. Safety and Efficacy Review of Doxycycline. Clin. Med. 2009, 1, CMT-S2035. [Google Scholar] [CrossRef]
- Momtaz, H.; Khamesipour, F.; Tavakol, M.; Awosile, B. Determination of Antimicrobial Resistance and Resistant Genes in Acinetobacter Baumannii from Human Clinical Samples. West Indian Med. J. 2015, 66, 56–64. [Google Scholar] [CrossRef][Green Version]
- Kasten, M.J. Clindamycin, Metronidazole, and Chloramphenicol. Mayo Clin. Proc. 1999, 74, 825–833. [Google Scholar] [CrossRef]
- Korenromp, E.L.; Scano, F.; Williams, B.G.; Dye, C.; Nunn, P. Effects of Human Immunodeficiency Virus Infection on Recurrence of Tuberculosis after Rifampin-Based Treatment: An Analytical Review. Clin. Infect. Dis. 2003, 37, 101–112. [Google Scholar] [CrossRef]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- Brogden, R.N.; Carmine, A.A.; Heel, R.C.; Speight, T.M.; Avery, G.S. Trimethoprim: A review of its antibacterial activity, pharmacokinetics and therapeutic use in urinary tract infections. Drugs 1982, 23, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Reffert, J.L.; Smith, W.J. Fosfomycin for the treatment of resistant gram-negative bacterial infections: Insights from the society of infectious diseases pharmacists. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2014, 34, 845–857. [Google Scholar] [CrossRef]
- Ball, A.P.; Gray, J.A.; Murdoch, J.M. Antibacterial Drugs Today; Springer: New York, NY, USA, 2012. [Google Scholar]
- Lyu, Y.; Yang, X.; Goswami, S.; Gorityala, B.K.; Idowu, T.; Domalaon, R.; Zhanel, G.G.; Shan, A.; Schweizer, F. Amphiphilic Tobramycin–Lysine Conjugates Sensitize Multidrug Resistant Gram-Negative Bacteria to Rifampicin and Minocycline. J. Med. Chem. 2017, 60, 3684–3702. [Google Scholar] [CrossRef]
- Ward, A.; Campoli-Richards, D.M. Mupirocin. Drugs 1986, 32, 425–444. [Google Scholar] [CrossRef]
- Birnbaum, J.; Kahan, F.M.; Kropp, H.; Macdonald, J.S. Carbapenems, a new class of beta-lactam antibiotics: Discovery and development of imipenem/cilastatin. Am. J. Med. 1985, 78, 3–21. [Google Scholar] [CrossRef]
- Garrido-Mesa, N.; Camuesco, D.; Arribas, B.; Comalada, M.; Bailón, E.; Cueto-Sola, M.; Utrilla, P.; Nieto, A.; Zarzuelo, A.; Rodríguez-Cabezas, M.E.; et al. The intestinal anti-inflammatory effect of minocycline in experimental colitis involves both its immunomodulatory and antimicrobial properties. Pharmacol. Res. 2011, 63, 308–319. [Google Scholar] [CrossRef]
- Cañete, R.; Escobedo, A.A.; González, M.E.; Almirall, P.; Cantelar, N. A randomized, controlled, open-label trial of a single day of mebendazole versus a single dose of tinidazole in the treatment of giardiasis in children. Curr. Med. Res. Opin. 2006, 22, 2131–2136. [Google Scholar] [CrossRef]
- Klastersky, J.; Vamecq, G.; Cappel, R.; Swings, G.; Vandenborre, L. Effects of the Combination of Gentamicin and Carbenicillin on the Bactericidal Activity of Serum. J. Infect. Dis. 1972, 125, 183–186. [Google Scholar] [CrossRef]
- Liu, Q.; Li, M.; Zhang, F.; Yu, H.; Zhang, Q.; Liu, X. The removal of trimethoprim and sulfamethoxazole by a high infiltration rate artificial composite soil treatment system. Front. Environ. Sci. Eng. 2017, 11, 12. [Google Scholar] [CrossRef]
- Goldberg, E.; Bishara, J. Contemporary unconventional clinical use of co-trimoxazole. Clin. Microbiol. Infect. 2012, 18, 8–17. [Google Scholar] [CrossRef]
- Kawaguchi, H. Discovery, Chemistry, and Activity of Amikacin. J. Infect. Dis. 1976, 134, S242–S248. [Google Scholar] [CrossRef]
- Stapley, E.O.; Birnbaum, J.; Miller, A.K.; Wallick, H.; Hendlin, D.; Woodruff, H.B. Cefoxitin and Cephamycins: Microbiological Studies. Clin. Infect. Dis. 1979, 1, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; A Hansford, K.; Blaskovich, M.A.T.; Halai, R.; Cooper, A.M. Glycopeptide antibiotics: Back to the future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef]
- Bozkurt, A.; Deniz, M.; Yegen, B.C. Cefaclor, a cephalosporin antibiotic, delays gastric emptying rate by a CCK-A receptor-mediated mechanism in the rat. Br. J. Pharm. 2000, 131, 399–404. [Google Scholar] [CrossRef]
- Grover, N.; Sahni, A.; Bhattacharya, S. Therapeutic challenges of ESBLS and AmpC beta-lactamase producers in a tertiary care center. Med. J. Armed. Forces India 2013, 69, 4–10. [Google Scholar] [CrossRef]
- Owens, R.C., Jr.; Ambrose, P.G. Clinical use of the fluoroquinolones. Med. Clin. N. Am. 2000, 84, 1447–1469. [Google Scholar] [CrossRef]
- Sinha, M.; Srinivasa, H.; Macaden, R. Antibiotic resistance profile & extended spectrum beta-lactamase (ESBL) production in Acinetobacter species. Indian J. Med. Res. 2007, 126, 63. [Google Scholar] [PubMed]
- Chakravarty, I.; Kundu, K.; Kundu, S. Daptomycin: Discovery, development and perspectives. The battle against microbial pathogens: Basic science, technological advances and educational programs. Microbiology 2015, 2, 895–903. [Google Scholar]
- Bradley, J.; Garau, J.; Lode, H.; Rolston, K.; Wilson, S.; Quinn, J. Carbapenems in clinical practice: A guide to their use in serious infection. Int. J. Antimicrob. Agents 1999, 11, 93–100. [Google Scholar] [CrossRef]
- Bonten, M.J.; Willems, R.; A Weinstein, R. Vancomycin-resistant enterococci: Why are they here, and where do they come from? Lancet Infect. Dis. 2001, 1, 314–325. [Google Scholar] [CrossRef]
- Solomkin, J.S.; Mazuski, J. Intra-abdominal Sepsis: Newer Interventional and Antimicrobial Therapies. Infect. Dis. Clin. N. Am. 2009, 23, 593–608. [Google Scholar] [CrossRef]
- Ezelarab, H.A.A.; Abbas, S.H.; Hassan, H.A.; Abuo-Rahma, G.E.-D.A. Recent updates of fluoroquinolones as antibacterial agents. Arch. Pharm. 2018, 351, e1800141. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef]
- El Amin, N.; Giske, C.G.; Jalal, S.; Keijser, B.; Kronvall, G.; Wretlind, B. Carbapenem resistance mechanisms in Pseudomonas aeruginosa: Alterations of porin OprD and efflux proteins do not fully explain resistance patterns observed in clinical isolates. APMIS 2005, 113, 187–196. [Google Scholar] [CrossRef]
- Hooper, D.C. Mechanisms of fluoroquinolone resistance. Drug Resist. Updat. 1999, 2, 38–55. [Google Scholar] [CrossRef]
- Harris, M.D. Infectious Disease in Athletes. Curr. Sports Med. Rep. 2011, 10, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Marin, A.C.; McNicholl, A.G.; Gisbert, J.P. A review of rescue regimens after clarithromycin-containing triple therapy failure (for Helicobacter pylori eradication). Expert Opin. Pharmacother. 2013, 14, 843–861. [Google Scholar] [CrossRef]
- Huwyler, T.; Lenggenhager, L.; Abbas, M.; Lorenzini, K.I.; Hughes, S.; Huttner, B.; Karmime, A.; Uçkay, I.; Von Dach, E.; Lescuyer, P.; et al. Cefepime plasma concentrations and clinical toxicity: A retrospective cohort study. Clin. Microbiol. Infect. 2017, 23, 454–459. [Google Scholar] [CrossRef]
- Van Hal, S.J.; Paterson, D.L. Systematic Review and Meta-Analysis of the Significance of Heterogeneous Vancomycin-IntermediateStaphylococcus aureusIsolates. Antimicrob. Agents Chemother. 2010, 55, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Lamb, H.M.; Figgitt, D.P.; Faulds, D. Quinupristin/Dalfopristin. Drugs 1999, 58, 1061–1097. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.W.; Kucia, M.L.; Salata, R.A. Use of Linezolid, an Oxazolidinone, in the Treatment of Multidrug-Resistant Gram-Positive Bacterial Infections. Clin. Infect. Dis. 2000, 30, 146–151. [Google Scholar] [CrossRef]
- Krueger, W.A.; Unertl, K.E. New treatment option for gram-positive infections in critically ill patients-overview over linezolid. Anasthesiol. Intensivmed. Notf. Schmerzther. AINS 2002, 37, 199. [Google Scholar] [CrossRef] [PubMed]
- Shain, C.S.; Amsden, G.W. Telithromycin: The First of the Ketolides. Ann. Pharm. 2002, 36, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Pennefather, P.M.; Kaye, S.B.; Hart, C.A. Fluoroquinolones. Drugs 2001, 61, 747–761. [Google Scholar] [CrossRef]
- Mendes, R.E.; Deshpande, L.M.; Jones, R.N. Linezolid update: Stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 2014, 17, 1–12. [Google Scholar] [CrossRef]
- Duval, R.E.; Grare, M.; Demoré, B. Fight against Antimicrobial Resistance: We Always Need New Antibacterials but for Right Bacteria. Molecular 2019, 24, 3152. [Google Scholar] [CrossRef]
- Chang, S.; Sievert, D.M.; Hageman, J.C.; Boulton, M.L. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 2003, 348, 1342. [Google Scholar] [CrossRef]
- Pirri, G.; Giuliani, A.; Nicoletto, S.; Pizzuto, L.; Rinaldi, A. Lipopeptides as anti-infectives: A practical perspective. Open Life Sci. 2009, 4, 258–273. [Google Scholar] [CrossRef]
- Tally, F.P.; Zeckel, M.; Wasilewski, M.M.; Carini, C.; Berman, C.L.; Drusano, G.L.; Oleson, F.B., Jr. Daptomycin: A novel agent for Gram-positive infections. Expert Opin. Investig. Drugs 1999, 8, 1223–1238. [Google Scholar] [CrossRef]
- Ackermann, G.; Rodloff, A.C. Drugs of the 21st century: Telithromycin (HMR 3647)—The first ketolide. J. Antimicrob. Chemother. 2003, 51, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Rose, W.E.; Rybak, M.J. Tigecycline: First of a new class of antimicrobial agents. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2006, 26, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Crook, D.W.; Walker, A.S.; Kean, Y.; Weiss, K.; Cornely, O.A.; Miller, M.A.; Esposito, R.; Louie, T.J.; Stoesser, N.E.; Young, B.C.; et al. Fidaxomicin Versus Vancomycin for Clostridium difficile Infection: Meta-analysis of Pivotal Randomized Controlled Trials. Clin. Infect. Dis. 2012, 55, S93–S103. [Google Scholar] [CrossRef] [PubMed]
- Deoghare, S. Bedaquiline: A new drug approved for treatment of multidrug-resistant tuberculosis. Indian J. Pharm. 2013, 45, 536–537. [Google Scholar] [CrossRef] [PubMed]
- Sandrock, C.E.; Shorr, A.F. The Role of Telavancin in Hospital-Acquired Pneumonia and Ventilator-Associated Pneumonia. Clin. Infect. Dis. 2015, 61, 79–86. [Google Scholar] [CrossRef]
- Sharma, C.; Singh, C.; Sharma, L.N.; Purvia, R.; Adlakha, M. Antibiotic resistant organism: An emerging public health problem and role of ayurveda (an overview). Int. J. Ayurveda Pharm. Res. 2014, 2, 17–29. [Google Scholar]
- O’Brien, J.J.; Campbell, N.; Conaghan, T. Effect of cooking and cold storage on biologically active antibiotic residues in meat. Epidemiol. Infect. 1981, 87, 511–523. [Google Scholar] [CrossRef]
- Wu, G.; Abraham, T.; Lee, S. Ceftazidime-Avibactam for Treatment of Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2016, 63, 1147–1148. [Google Scholar] [CrossRef] [PubMed]
- Giltrap, A. Total Synthesis of Teixobactin. In Total Synthesis of Natural Products with Antimicrobial Activity; Springer: Singapore, 2018; Volume 2, pp. 33–69. [Google Scholar] [CrossRef]
- Tucker, A.T.; Leonard, S.P.; Dubois, C.D.; Knauf, G.A.; Cunningham, A.L.; Wilke, C.O.; Trent, M.S.; Davies, B.W. Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface-Displayed Peptide Libraries. Cell 2018, 172, 618–628.e13. [Google Scholar] [CrossRef]
- Racine, E.; Gualtieri, M. From worms to drug candidate: The story of odilorhabdins, a new class of antimicrobial agents. Front. Microbiol. 2019, 10, 2893. [Google Scholar] [CrossRef] [PubMed]
- Skovbakke, S.L.; Holdfeldt, A.; Forsman, H.; Bylund, J.; Franzyk, H. The Role of Formyl Peptide Receptors for Immunomodulatory Activities of Antimicrobial Peptides and Peptidomimetics. Curr. Pharm. Des. 2018, 24, 1100–1120. [Google Scholar] [CrossRef]
- Coates, A.R.M.; Hu, Y. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharm. 2007, 152, 1147–1154. [Google Scholar] [CrossRef]
- Sodhi, V.; Kronsberg, K.A.; Clark, M.; Cho, J.C. Tebipenem pivoxil hydrobromide-No PICC, no problem! Pharmacotherapy 2021, 41, 748–761. [Google Scholar] [CrossRef]
- Sharon, O.; Czech, C.; Robilotti, E.; Holubar, M. Cefiderocol: A new cephalosporin stratagem against multidrug resistant gram-negative bacteria. Clin. Infec. Dis. 2021, ciab757. [Google Scholar] [CrossRef]
- Adzitey, F. Antibiotic classes and antibiotic susceptibility of bacterial isolates from selected poultry; a mini review. World Veter. J. 2015, 6, 36. [Google Scholar] [CrossRef]
- Livermore, D.M.; Warner, M.; Mushtaq, S.; Doumith, M.; Zhang, J.; Woodford, N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents 2011, 37, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Pandit, N.; Singla, R.K.; Shrivastava, B. Current Updates on Oxazolidinone and Its Significance. Int. J. Med. Chem. 2012, 2012, 159285. [Google Scholar] [CrossRef] [PubMed]
- Dowling, A. Antibiotics: Mode of Action and Mechanisms of Resistance; Formatex Research Center: Badajoz, Spain, 2017; p. 2017. [Google Scholar]
- Bush, L.M. Overview of Bacteria; Schmidt College of Medicine, Florida Atlantic University: Boca Raton, FL, USA, 2017. [Google Scholar]
- Levy, S.; Chadwick, J.G. Antibiotic Resistance: An Ecological Imbalance, in Ciba Foundation Symposium. Antibiot. Resist. Orig. Evol. Sel. Spread 2007, 1, 1–14. [Google Scholar]
- Levy, S.B. From Tragedy the Antibiotic Age is Born. In The Antibiotic Paradox; Springer International Publishing: Cham, Switzerland, 1992; pp. 1–12. [Google Scholar]
- Hoge, C.W. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin. Infect. Dis. 1998, 26, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.E. Managing neonatal and early childhood syndromic sepsis in sub-district hospitals in resource poor settings: Improvement in quality of care through introduction of a package of interventions in rural Bangladesh. PLoS ONE 2017, 12, e0170267. [Google Scholar] [CrossRef]
- Abraham, C.E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- McEwen, P.J. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002, 34 (Suppl. S3), S93–S106. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Loeffler, M.; Terjung, N. The antimicrobial paradox: Why preservatives lose activity in foods. Curr. Opin. Food Sci. 2015, 4, 69–75. [Google Scholar] [CrossRef]
- Garau, J. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 1999, 43, 2736–2741. [Google Scholar] [CrossRef]
- Huang, S.S.; Johnson, K.M.; Ray, G.T.; Wroe, P.; Lieu, T.A.; Moore, M.R.; Zell, E.R.; Linder, J.A.; Grijalva, C.G.; Metlay, J.P.; et al. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 2011, 29, 3398–3412. [Google Scholar] [CrossRef]
- Hutton, D.; Jeanette, S.; Vetter, K. Studies on streptococcus pyogenes I. J. Bacteriol. 1956, 71, 236–243. [Google Scholar]
- Kwonjune, J.; Seung, S.K.; Rich, M.L. Multidrug-resistant tuberculosis and extensively drug- resistant tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a017863. [Google Scholar]
- Rasheed, M.U.; Thajuddin, N.; Parveez, A.; Zelalem, T.; Kaiser, J. Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Rev. Inst. Med. Trop. 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Yang, C.; Li, H.; Zhang, T.; Chu, Y.; Zuo, J.; Chen, D. Study on antibiotic susceptibility of Salmonella typhimurium L. forms to the third and forth generation cephalosporins. Sci. Rep. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Chesson, H.W.; Kirkcaldy, R.D.; Gift, T.L.; Owusu-Edusei, K.; Weinstock, H.S. An Illustration of the Potential Health and Economic Benefits of Combating Antibiotic-Resistant Gonorrhea. Sex. Transm. Dis. 2018, 45, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, N.; Fujimoto, Y.; Uno, M. Annual changes in drug sensitivity of gonococci derived from urethritis in men. Jpn. J. Chemother. 2004, 52, 31–34. [Google Scholar]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharm. Outcomes Res. 2010, 10, 441–451. [Google Scholar]
- Raza, T.; Ullah, S.R.; Mehmood, K.; Andleeb, S. Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar] [PubMed]
- Appelbaum, P.C.; Appelbaum, P.C. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12 (Suppl. S1), 16–23. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Brown, G.M. Economics of Antibiotic Resistance: A Theory of Optimal Use. J. Environ. Econ. Manag. 2001, 42, 183–206. [Google Scholar] [CrossRef]
- Levy, S.B. The antimicrobial paradox. How miracle drugs are destroying the miracle. N. Engl. J. Med. 1993, 328, 1792. [Google Scholar]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Alanis, A.J. Resistance to Antibiotics: Are We in the Post-Antibiotic Era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Goossens, H.; Ferech, M.; Vander, S.; Elseviers, M. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Avorn, J.; Barrett, J.F.; Davey, P.G. Antibiotic Resistance: Synthesis of Recommendations by Expert Policy; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- Walsh, C. Molecular mechanisms that confer antibacterial drug resistance. Nature 2000, 406, 775–781. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Genet. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. About NIOSH. Available online: https://www.cdc.gov/niosh/about/default.html (accessed on 21 September 2021).
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef]
- Piddock, L.J.V. The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef]
- Gold, K.; Slay, B.; Knackstedt, M.; Gaharwar, A.K. Antimicrobial Activity of Metal and Metal-Oxide Based Nanoparticles. Adv. Ther. 2018, 1, 1700033. [Google Scholar] [CrossRef]
- Heiligtag, F.J.; Niederberger, M. The fascinating world of nanoparticle research. Mater. Today 2013, 16, 262–271. [Google Scholar] [CrossRef]
- Walter, P.; Welcomme, E.; Hallegot, P.; Zaluzec, N.J.; Deeb, C.; Castaing, J.; Veyssière, P.; Bréniaux, R.; Lévêque, J.-L.; Tsoucaris, G. Early Use of PbS Nanotechnology for an Ancient Hair Dyeing Formula. Nano Lett. 2006, 6, 2215–2219. [Google Scholar] [CrossRef]
- Johnson-McDaniel, D.; Barrett, C.A.; Sharafi, A.; Salguero, T.T. Nanoscience of an Ancient Pigment. J. Am. Chem. Soc. 2013, 135, 1677–1679. [Google Scholar] [CrossRef]
- Schaming, D.; Remita, H. Nanotechnology: From the ancient time to nowadays. Found. Chem. 2015, 17, 187–205. [Google Scholar] [CrossRef]
- Artioli, G.; Angelini, I.; Polla, A. Crystals and phase transitions in protohistoric glass materials. Phase Transit. 2008, 81, 233–252. [Google Scholar] [CrossRef]
- Brun, N.; Mazerolles, L.; Pernot, M. Microstructure of opaque red glass containing copper. J. Mater. Sci. Lett. 1991, 10, 1418–1420. [Google Scholar] [CrossRef]
- Leonhardt, U. Invisibility cup. Nat. Photon. 2007, 1, 207–208. [Google Scholar] [CrossRef]
- Rytwo, G. Clay Minerals as an Ancient Nanotechnology: Historical Uses of Clay Organic Interactions, and Future Possible Perspectives. Macla 2008, 9, 15–17. [Google Scholar]
- Mie, G. Beiträge zur Optik trüber Medien, speziell kolloidaler Metallösungen. Ann. Phys. 1908, 330, 377–445. [Google Scholar] [CrossRef]
- Rittner, M.N.; Abraham, T. Nanostructured materials: An overview and commercial analysis. JOM 1998, 50, 37–38. [Google Scholar] [CrossRef]
- Samsung and its attractions—Asia’s new model company. Economist, 1 October 2011.
- Malsch, I.; Gleiche, M.; Hoffschulz, H.; Bøgedal, M. Benefits, Risks, Ethical, Legal and Social Aspects of Nanotechnology. 2004. Available online: https://www.yumpu.com/en/document/read/29766978/benefits-risks-ethical-legal-and-social-aspects-of-nanotechnology (accessed on 21 September 2021).
- Déry, J.-P.; Borra, E.F.; Ritcey, A.M. Ethylene Glycol Based Ferrofluid for the Fabrication of Magnetically Deformable Liquid Mirrors. Chem. Mater. 2008, 20, 6420–6426. [Google Scholar] [CrossRef]
- O’Regan, B.; Grätzel, M. A low-cost, high-efficiency solar cell based on dye-sensitized colloidal TiO2 films. Nature 1991, 353, 737–740. [Google Scholar] [CrossRef]
- Kreuter, J. Nanoparticles—A historical perspective. Int. J. Pharm. 2007, 331, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.; Santos-Martinez, M.J.; Radomski, A.; I Corrigan, O.; Radomski, M.W. Nanoparticles: Pharmacological and toxicological significance. Br. J. Pharm. 2007, 150, 552–558. [Google Scholar] [CrossRef]
- Surendiran, A.; Sandhiya, S.; Pradhan, S.C.; Adithan, C. Novel applications of nanotechnology in medicine. Indian J. Med. Res. 2009, 130, 689–701. [Google Scholar] [PubMed]
- Wagner, V.; Dullaart, A.; Bock, A.-K.; Zweck, A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006, 24, 1211–1217. [Google Scholar] [CrossRef]
- Freitas, R.A. What is nanomedicine? Nanomed. Nanotechnol. Biol. Med. 2005, 1, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.L. Changing patterns of infectious disease. Nat. Cell Biol. 2000, 406, 762–767. [Google Scholar] [CrossRef]
- Gold, H.S.; Moellering, R.C. Antimicrobial-Drug Resistance. N. Engl. J. Med. 1996, 335, 1445–1453. [Google Scholar] [CrossRef]
- Ding, H.H.; Chigan, J.Z.; Zhen, J.B.; Liu, L.; Xu, Y.S.; Chen, C.; Yang, K.W. Cholesteroled polymer (Chol-b-Lys)-based nanoparticles (CL-NPs) confer antibacterial efficacy without resistance. New J. Chem. 2021, 45, 20743–20750. [Google Scholar] [CrossRef]
- Pitt, T.L.; Sparrow, M.; Warner, M.; Stefanidou, M. Survey of resistance of Pseudomonas aeruginosa from UK patients with cystic fibrosis to six commonly prescribed antimicrobial agents. Thorax 2003, 58, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, N.; Sumathi, C.S.; Balasubramanian, V.; Palaniappan, K.R.; Kannan, V.R. Urinary tract infection and antimicrobial susceptibility pattern of extended spectrum beta lactamase producing clinical isolates. Adv. Biol. Res. 2008, 2, 78–82. [Google Scholar]
- Ramesh, N.; Drishya Nair, V.; Karthiayani, H.; Prasant, M.; Shanthini, T.; Gothandam, K.M. Prevalence of blaNDM-1 among Gram negative bacteria from clinical samples of Tamin Nadu. Int. J. Medicobiol. Res. 2004, 1, 389–393. [Google Scholar]
- Sheng, Z.-K.; Hu, F.; Wang, W.; Guo, Q.; Chen, Z.; Xu, X.; Zhu, D.; Wang, M. Mechanisms of Tigecycline Resistance among Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 2014, 58, 6982–6985. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 2009, 12, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Taylor, P.W.; Stapleton, P.D.; Luzio, J.P. New ways to treat bacterial infections. Drug Discov. Today 2002, 7, 1086–1091. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef]
- Goodman, C.M.; McCusker, C.D.; Yilmaz, T.; Rotelo, V.M. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug. Chem. 2004, 15, 897–900. [Google Scholar] [CrossRef]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef]
- Kumar, N.; Kumbhat, S. Essentials in Nanoscience and Nanotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 189–236. [Google Scholar] [CrossRef]
- Hyung, H.; Fortner, J.D.; Hughes, J.B.; Kim, J.-H. Natural Organic Matter Stabilizes Carbon Nanotubes in the Aqueous Phase. Environ. Sci. Technol. 2007, 41, 179–184. [Google Scholar] [CrossRef]
- Li, Q.; Mahendra, S.; Lyon, D.Y.; Brunet, L.; Liga, M.V.; Li, D.; Alvarez, P.J. Antimicrobial nanomaterials for water disinfection and microbial control: Potential applications and implications. Water Res. 2008, 42, 4591–4602. [Google Scholar] [CrossRef] [PubMed]
- Wick, P.; Manser, P.; Limbach, L.K.; Dettlaff-Weglikowska, U.; Krumeich, F.; Roth, S.; Stark, W.J.; Bruinink, A. The degree and kind of agglomeration affect carbon nanotube cytotoxicity. Toxicol. Lett. 2007, 168, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Wang, H.; Yan, L.; Wang, X.; Pei, R.; Yan, T. Cytotoxicity of carbon nanomaterials: Single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol. 2005, 39, 1378–1383. [Google Scholar] [CrossRef]
- Vecitis, C.D.; Zodrow, K.R.; Kang, S.; Elimelech, M. Electronic-Structure-Dependent Bacterial Cytotoxicity of Single-Walled Carbon Nanotubes. ACS Nano 2010, 4, 5471–5479. [Google Scholar] [CrossRef] [PubMed]
- Brady-Estévez, A.S.; Nguyen, T.H.; Gutierrez, L.; Elimelech, M. Impact of solution chemistry on viral removal by a single-walled carbon nanotube filter. Water Res. 2010, 44, 3773–3780. [Google Scholar] [CrossRef]
- Tegos, G.P.; Demidova, T.N.; Arcila-Lopez, D.; Lee, H.; Wharton, T.; Gali, H.; Hamblin, M.R. Cationic fullerenes are effective and selective antimicrobial photosensitizers. Chem. Biol. 2005, 12, 1127–1135. [Google Scholar] [CrossRef]
- Shvedova, A.A.; Pietroiusti, A.; Fadeel, B.; Kagan, V.E. Mechanisms of carbon nanotube-induced toxicity: Focus on oxidative stress. Toxicol. Appl. Pharm. 2012, 261, 121–133. [Google Scholar] [CrossRef]
- Bellucci, S. Nanoparticles and Nanodevices in Biological Applications; Springer: Cham, Switzerland, 2008. [Google Scholar]
- Deryabin, D.G.; Davydova, O.K.; Yankina, Z.Z.; Vasilchenko, A.S.; Miroshnikov, S.A.; Kornev, A.B.; Ivanchikhina, A.V.; Troshin, P.A. The Activity of [60]Fullerene Derivatives Bearing Amine and Carboxylic Solubilizing Groups against Escherichia coli: A Comparative Study. J. Nanomater. 2014, 2014, 907435. [Google Scholar] [CrossRef]
- Cataldo, F.; Da Ros, T. Medicinal Chemistry and Pharmacological Potential of Fullerenes and Carbon Nanotubes; Springer: Trieste, Italy, 2008. [Google Scholar]
- Lu, Z.; Dai, T.; Huang, L.; Kurup, D.B.; Tegos, G.P.; Jahnke, A.; Wharton, T.; Hamblin, M.R. Photodynamic therapy with a cationic functionalized fullerene rescues mice from fatal wound infections. Nano Med. 2010, 5, 1525–1533. [Google Scholar] [CrossRef]
- Azimi, S.; Behin, J.; Abiri, R.; Rajabi, L.; Derakhshan, A.A.; Karimnezhad, H. Synthesis, Characterization and Antibacterial Activity of Chlorophyllin Functionalized Graphene Oxide Nanostructures. Sci. Adv. Mater. 2014, 6, 771–781. [Google Scholar] [CrossRef]
- Weir, E.; Lawlor, A.; Whelan, A.; Regan, F. The use of nanoparticles in anti-microbial materials and their characterization. Analytical 2008, 133, 835–845. [Google Scholar] [CrossRef]
- Dibrov, P.; Dzioba, J.; Gosink, K.K.; Ha, C.C. Chemiosmotic Mechanism of Antimicrobial Activity of Ag+ in Vibrio cholerae. Antimicrob. Agents Chemother. 2002, 46, 2668–2670. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I. The increasing use of silver-based products as antimicrobial agents: A useful development or a cause for concern? J. Antimicrob. Chemother. 2007, 59, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, L.; Mustapha, A.; Li, H.; Hu, Z.Q.; Lin, M. Antibacterial activities of zinc oxide nanoparticles against Escherichia coli O157:H7. J. Appl. Microbiol. 2009, 107, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Chamundeeswari, M.; Sobhana, S.S.L.; Jacob, J.P.; Kumar, M.G.; Devi, M.P.; Sastry, T.P.; Mandal, A.B. Preparation, characterization and evaluation of a biopolymeric gold nanocomposite with antimicrobial activity. Biotechnol. Appl. Biochem. 2010, 55, 29–35. [Google Scholar] [CrossRef]
- Raimondi, F.; Scherer, G.G.; Kötz, R.; Wokaun, A. Nanoparticles in Energy Technology: Examples from Electrochemistry and Catalysis. Angew. Chem. Int. Ed. 2005, 44, 2190–2209. [Google Scholar] [CrossRef]
- Klasen, H. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 2000, 26, 131–138. [Google Scholar] [CrossRef]
- Fayaz, A.M.; Balaji, K.; Girilal, M.; Yadav, R.; Kalaichelvan, P.T.; Venketesan, R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: A study against gram-positive and gram-negative bacteria. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Schacht, V.J.; Neumann, L.V.; Sandhi, S.K.; Chen, L.; Henning, T.; Klar, P.J.; Theophel, K.; Schnell, S.; Bunge, M. Effects of silver nanoparticles on microbial growth dynamics. J. Appl. Microbiol. 2013, 114, 25–35. [Google Scholar] [CrossRef]
- Ashok Kumar, D.; Palanichamy, V.; Roopan, S.M. Photocatalytic action of AgCl nanoparticles and its antibacterial activity. J. Photochem. Photobiol. B Biol. 2014, 138, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Ninganagouda, S.; Rathod, V.; Singh, D. Growth kinetics and mechanistic action of reactive oxygen species released by silver nanoparticles from Aspergillus niger on Escherichia coli. Biomed. Res. Int. 2014, 2014, 753419. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.; Hussain, S.M.; Schrand, A.M.; Braydich-Stolle, L.K.; Hess, K.L.; Jones, R.L.; Schlager, J.J. Unique Cellular Interaction of Silver Nanoparticles: Size-Dependent Generation of Reactive Oxygen Species. J. Phys. Chem. B 2008, 112, 13608–13619. [Google Scholar] [CrossRef]
- Piao, M.J.; Kang, K.A.; Lee, I.K.; Kim, H.S.; Kim, S.; Choi, J.Y.; Choi, J.; Hyun, J.W. Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis. Toxicol. Lett. 2011, 201, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.J.; Hutchison, G.; Christensen, F.M.; Peters, S.; Hankin, S.; Stone, V. A review of the in vivo and in vitro toxicity of silver and gold particulates: Particle attributes and biological mechanisms responsible for the observed toxicity. Crit. Rev. Toxicol. 2010, 40, 328–346. [Google Scholar] [CrossRef]
- Drake, P.L.; Hazelwood, K.J. Exposure-Related Health Effects of Silver and Silver Compounds: A Review. Ann. Occup. Hyg. 2005, 49, 575–585. [Google Scholar] [CrossRef]
- Braydich-Stolle, L.; Hussain, S.; Schlager, J.J.; Hofmann, M.-C. In Vitro Cytotoxicity of Nanoparticles in Mammalian Germline Stem Cells. Toxicol. Sci. 2005, 88, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Pissuwan, D.; Cortie, C.H.; Valenzuela, S.M.; Cortie, M.B. Functionalised gold nanoparticles for controlling pathogenic bacteria. Trends Biotechnol. 2010, 28, 207–213. [Google Scholar] [CrossRef]
- Mühling, M.; Bradford, A.; Readman, J.W.; Somerfield, P.J.; Handy, R.D. An investigation into the effects of silver nanoparticles on antibiotic resistance of naturally occurring bacteria in an estuarine sediment. Mar. Environ. Res. 2009, 68, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Roselli, M.; Finamore, A.; Garaguso, I.; Britti, M.S.; Mengheri, E. Zinc Oxide Protects Cultured Enterocytes from the Damage Induced by Escherichia coli. J. Nutr. 2003, 133, 4077–4082. [Google Scholar] [CrossRef]
- Ugur, S.S.; Sariisik, M.; Aktas, A.G.; Ucar, M.C.; Erden, E. Modifying of Cotton Fabric Surface with Nano-ZnO Multilayer Films by Layer-by-Layer Deposition Method. Nanoscale Res. Lett. 2010, 5, 1204–1210. [Google Scholar] [CrossRef]
- Palanikumar, L.; Ramasamy, S.N.; Balachandran, C. Size-dependent antimicrobial response of zinc oxide nanoparticles. IET Nanobiotechnol. 2014, 8, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Sawai, J. Quantitative evaluation of antibacterial activities of metallic oxide powders (ZnO, MgO and CaO) by conductimetric assay. J. Microbiol. Methods 2003, 54, 177–182. [Google Scholar] [CrossRef]
- Huang, Z.; Zheng, X.; Yan, D.; Yin, G.; Liao, X.; Kang, Y.; Yao, Y.; Huang, D.; Hao, B. Toxicological Effect of ZnO Nanoparticles Based on Bacteria. Langmuir 2008, 24, 4140–4144. [Google Scholar] [CrossRef] [PubMed]
- Gelover, S.; Gómez, L.A.; Reyes, K.; Leal, M.T. A practical demonstration of water disinfection using TiO2 films and sunlight. Water Res. 2006, 40, 3274–3280. [Google Scholar] [CrossRef]
- Reddy, K.M.; Feris, K.; Bell, J.; Wingett, D.G.; Hanley, C.; Punnoose, A. Selective toxicity of zinc oxide nanoparticles to prokaryotic and eukaryotic systems. Appl. Phys. Lett. 2007, 90, 2139021–2139023. [Google Scholar] [CrossRef]
- Salih, F.M. Enhancement of solar inactivation of Escherichia coli by titanium dioxide photocatalytic oxidation. J. Appl. Microbiol. 2002, 92, 920–926. [Google Scholar] [CrossRef]
- Oppezzo, O.J.; A Pizarro, R. Sublethal effects of ultraviolet A radiation on Enterobacter cloacae. J. Photochem. Photobiol. B Biol. 2001, 62, 158–165. [Google Scholar] [CrossRef]
- Choi, J.-Y.; Kim, K.-H.; Choy, K.-C.; Oh, K.-T.; Kim, K.-N. Photocatalytic antibacterial effect of TiO2 film formed on Ti and TiAg exposed to Lactobacillus acidophilus. J. Biomed. Mater. Res. Part B Appl. Biomater. 2007, 80, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Maness, P.C.; Smolinski, S.; Blake, D.M.; Huang, Z.; Wolfrum, E.J.; Jacoby, W.A. Bactericidal activity of photocatalytic TiO(2) reaction: Toward an understanding of its killing mechanism. Appl. Environ. Microbiol. 1999, 65, 4094–4098. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.K.; Lyon, D.Y.; Alvarez, P.J. Comparative eco-toxicity of nanoscale TiO2, SiO2, and ZnO water suspensions. Water Res. 2006, 40, 3527–3532. [Google Scholar] [CrossRef] [PubMed]
- Muranyi, P.; Schraml, C.; Wunderlich, J. Antimicrobial efficiency of titanium dioxide-coated surfaces. J. Appl. Microbiol. 2009, 108, 1966–1973. [Google Scholar] [CrossRef]
- Hamal, D.B.; Häggström, J.A.; Marchin, G.L.; Ikenberry, M.A.; Hohn, K.; Klabunde, K.J. A Multifunctional Biocide/Sporocide and Photocatalyst Based on Titanium Dioxide (TiO2) Codoped with Silver, Carbon, and Sulfur. Langmuir 2010, 26, 2805–2810. [Google Scholar] [CrossRef]
- Esteban-Tejeda, L.; Malpartida, F.; Esteban-Cubillo, A.; Pecharromán, C.; Moya, J.S. Antibacterial and antifungal activity of a soda-lime glass containing copper nanoparticles. Nanotechnology 2009, 20, 505701. [Google Scholar] [CrossRef]
- Hejazy, M.; Koohi, M.; Bassiri Mohamad Pour, A.; Najafi, D. Toxicity of manufactured copper nanoparticles—A review. Nanomed. Res. J. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- Ruparelia, J.P.; Chatterjee, A.K.; Duttagupta, S.P.; Mukherji, S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008, 4, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Elguindi, J.; Wagner, J.; Rensing, C. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 2009, 106, 1448–1455. [Google Scholar] [CrossRef]
- Wilks, S.; Michels, H.; Keevil, C. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 2005, 105, 445–454. [Google Scholar] [CrossRef]
- Michels, H.T.; Noyce, J.O.; Keevil, C.W. Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett. Appl. Microbiol. 2009, 49, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.-Y.; Byeon, J.H.; Park, J.-H.; Hwang, J. Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci. Total Environ. 2007, 373, 572–575. [Google Scholar] [CrossRef]
- Ren, G.; Hu, D.; Cheng, E.W.; Vargas-Reus, M.A.; Reip, P.; Allaker, R.P. Characterisation of copper oxide nanoparticles for antimicrobial applications. Int. J. Antimicrob. Agents 2009, 33, 587–590. [Google Scholar] [CrossRef]
- Sadiq, I.M.; Chowdhury, B.; Chandrasekaran, N.; Mukherjee, A. Antimicrobial sensitivity of Escherichia coli to alumina nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 282–286. [Google Scholar] [CrossRef]
- Murdock, R.C.; Braydich-Stolle, L.; Schrand, A.M.; Schlager, J.J.; Hussain, S.M. Characterization of Nanomaterial Dispersion in Solution Prior to In Vitro Exposure Using Dynamic Light Scattering Technique. Toxicol. Sci. 2007, 101, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Hetrick, E.M.; Shin, J.H.; Stasko, N.A.; Johnson, C.B.; Wespe, D.A.; Holmuhamedov, E.; Schoenfisch, M.H. Bactericidal Efficacy of Nitric Oxide-Releasing Silica Nanoparticles. ACS Nano 2008, 2, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Weller, R.; Finnen, M.J. The effects of topical treatment with acidified nitrite on wound healing in normal and diabetic mice. Nitric Oxide 2006, 15, 395–399. [Google Scholar] [CrossRef]
- Kirtane, A.R.; Verma, M.; Karandikar, P.; Furin, J.; Langer, R.; Traverso, G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021, 16, 369–384. [Google Scholar] [CrossRef]
- Kostarelos, K. The emergence of nanomedicine: A field in the making. Nano Med. 2006, 1, 1–3. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Health Mater. 2018, 7, e1701503. [Google Scholar] [CrossRef]
- Leung, Y.H.; Ng, A.M.C.; Xu, X.; Shen, Z.; Gethings, L.A.; Wong, M.T.; Chan, C.M.N.; Guo, M.Y.; Ng, Y.H.; Djurišić, A.B.; et al. Mechanisms of Antibacterial Activity of MgO: Non-ROS Mediated Toxicity of MgO Nanoparticles Towards Escherichia coli. Small 2014, 10, 1171–1183. [Google Scholar] [CrossRef]
- Kandpal, N.D. Co-precipitation method of synthesis and characterization of iron oxide nanoparticles. J. Sci. Ind. Res. 2014, 73, 87–90. [Google Scholar]
- Singh, N.; Jenkins, G.J.; Asadi, R.; Doak, S.H. Potential toxicity of superparamagnetic iron oxide nanoparticles (SPION). Nano Rev. 2010, 1, 5358. [Google Scholar] [CrossRef]
- Li, G.-Y.; Jiang, Y.-R.; Huang, K.-L.; Ding, P.; Chen, J. Preparation and properties of magnetic Fe3O4–chitosan nanoparticles. J. Alloys Compd. 2008, 466, 451–456. [Google Scholar] [CrossRef]
- Kim, E.H.; Ahn, Y.; Lee, H.S. Biomedical applications of superparamagnetic iron oxide nanoparticles encapsulated within chitosan. J. Alloys Compd. 2007, 434, 633–636. [Google Scholar] [CrossRef]
- Thukkaram, M.; Sitaram, S.; Kannaiyan, S.K.; Subbiahdoss, G. Antibacterial Efficacy of Iron-Oxide Nanoparticles against Biofilms on Different Biomaterial Surfaces. Int. J. Biomater. 2014, 2014, 716080. [Google Scholar] [CrossRef]
- Kareem, Z.H.; Shareef, H.K.; Alkaim, A.F. Evaluation of antibacterial activity of Fe2O3 nanoparticles against Shigella dysenteriae. J. Pharm. Sci. Res. 2018, 10, 1980–1982. [Google Scholar]
- Park, H.; Park, H.-J.; Kim, J.A. Inactivation of Pseudomonas aeruginosa PA01 biofilms by hyperthermia using superparamagnetic nanoparticles. J. Microbiol. 2011, 84, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, M.J.; Fromm, K.M.; Ashkarran, A.A.; De Aberasturi, D.J.; De Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef]
- Durmus, N.G.; Taylor, E.N.; Kummer, K.M.; Webster, T.J. Enhanced Efficacy of Superparamagnetic Iron Oxide Nanoparticles Against Antibiotic-Resistant Biofilms in the Presence of Metabolites. Adv. Mater. 2013, 25, 5706–5713. [Google Scholar] [CrossRef]
- Bankar, S.B.; Singhal, R.S. Panorama of poly-ε-lysine. RSC Adv. 2013, 3, 8586–8603. [Google Scholar] [CrossRef]
- Taylor, E.N.; Kummer, K.M.; Durmus, N.G.; Leuba, K.; Tarquinio, K.M.; Webster, T.J. Superparamagnetic iron oxide nanoparticles (SPION) for the treatment of antibiotic-resistant bio-films. Small 2012, 8, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.N.; Webster, T.J. The use of superparamagnetic nanoparticles for prosthetic biofilm prevention. Int. J. Nanomed. 2009, 4, 145–152. [Google Scholar]
- Jamil, B.; Habib, H.; Abbasi, S.; Nasir, H.; Rahman, A.; Rehman, A.; Bokhari, H.; Imran, M. Cefazolin loadedchitosan nanoparticles to cure multi drug resistant Gram-negative pathogens. Carbohydr. Polym. 2016, 136, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.H.; Cha, S.-H.; Kim, J.-H.; Yoon, M.; Cho, S.; Park, Y. Silver Nanoparticles Synthesized Using Caesalpinia sappan Extract as Potential Novel Nanoantibiotics Against Methicillin-Resistant Staphylococcus aureus. J. Nanosci. Nanotechnol. 2015, 15, 5543–5552. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, J. Basic and applied studies on ε-polylysine. J. Antibact. Antifung. Agents 1995, 23, 349–354. [Google Scholar]
- Muñoz-Bonilla, A.; Fernández-García, M. Polymeric materials with antimicrobial activity. Prog. Polym. Sci. 2012, 37, 281–339. [Google Scholar] [CrossRef]
- Ansari, M.; Khan, H.M.; Khan, A.; Cameotra, S.S.; Saquib, Q.; Musarrat, J. Interaction of Al2O3 nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Appl. Microbiol. 2014, 116, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Denyer, S.; Stewart, G. Mechanisms of action of disinfectants. Int. Biodeterior. Biodegrad. 1998, 41, 261–268. [Google Scholar] [CrossRef]
- Sauvet, G.; Fortuniak, W.; Kazmierski, K.; Chojnowski, J. Amphiphilic block and statistical siloxane copolymers with antimicrobial activity. J. Polym. Sci. Part A Polym. Chem. 2003, 41, 2939–2948. [Google Scholar] [CrossRef]
- Park, E.-S.; Moon, W.-S.; Song, M.-J.; Kim, M.-N.; Chung, K.-H.; Yoon, J.-S. Antimicrobial activity of phenol and benzoic acid derivatives. Int. Biodeterior. Biodegrad. 2001, 47, 209–214. [Google Scholar] [CrossRef]
- Nonaka, T.; Hua, L.; Ogata, T.; Kurihara, S. Synthesis of water-soluble thermosensitive polymers having phosphonium groups frommethacryloyloxyethyl trialkyl phosphonium chlorides-N-isopropylacrylamide copolymers and their functions. J. Appl. Polym. Sci. 2003, 87, 386–393. [Google Scholar] [CrossRef]
- Chung, D.; Papadakis, S.E.; Yam, K.L. Evaluation of a polymer coating containing triclosan as the antimicrobial layer for packaging materials. Int. J. Food Sci. Technol. 2003, 38, 165–169. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, D.; Butler, R.; Campbell, N.L.; Long, J.; Tan, B.; Duncalf, D.J.; Foster, A.J.; Hopkinson, A.; Taylor, D. Formation and enhanced biocidal activity of water-dispersable organic nanoparticles. Nat. Nanotechnol. 2008, 3, 506–511. [Google Scholar] [CrossRef]
- Bankova, M.; Manolova, N.; Markova, N. Hydrolysis and antibacterial activity of polymers containing 8-quinolinyl acrylate. J. Bioact. Compat. Polym. 1997, 12, 294–307. [Google Scholar] [CrossRef]
- Chung, Y.C.; Wang, H.L.; Chen, Y.M.; Li, S.L. Effect of abiotic factors on the antibacterial activity of chitosan against waterborne pathogens. Bioresour. Technol. 2003, 88, 179–184. [Google Scholar] [CrossRef]
- Qi, L.; Xu, Z.; Jiang, X.; Hu, C.; Zou, X. Preparation and antibacterial activity of chitosan nanoparticles. Carbohydr. Res. 2004, 339, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.; Tavaria, F.K.; Fonseca, S.C.; Ramos, O.S.; E Pintado, M.; Malcata, F.X. In vitro screening for anti-microbial activity of chitosans and chitooligosaccharides, aiming at potential uses in functional textiles. J. Microbiol. Biotechnol. 2010, 20, 311–318. [Google Scholar] [CrossRef]
- Rabea, E.I.; Badawy, M.E.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as antimicrobial agent: Applications and mode of action. Biomacromolecules 2003, 4, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Kenawy, E.R.; Worley, S.D.; Broughton, R. The chemistry and applications of antimicrobial polymers: A state-of-the-art review. Biomacromolecules 2007, 8, 1359–1384. [Google Scholar] [CrossRef]
- Shvero, D.K.; Zatlsman, N.; Hazan, B.; Weiss, E.I.; Beyth, N.I. Characterisation of the antibacterial effect of polyethyleneimine nanoparticles in relation to particle distribution in resin composite. J. Dent. 2015, 43, 287–294. [Google Scholar]
- Beyth, N.; Ira, Y.-F.; Perez-Davidi, N.; Domb, A.J.; Weiss, E.I. Polyethyleneimine nanoparticles incorporated into resin composite cause cell death and trigger biofilm stress in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 22038–22043. [Google Scholar] [CrossRef]
- Feng, Y.; Tan, H.; Li, C.; Wang, Y.; Zhang, Y.; Wen, P.; Xu, L. Preparation and characterization of nanoTiO2 antibacterial corrugating medium. J. Nanosci. Nanotechnol. 2017, 17, 8912–8917. [Google Scholar] [CrossRef]
- Emamifar, A.; Kadivar, M.; Shahedi, M.; Soleimanian-Zad, S. Evaluation of nanocomposite packaging containing Ag and ZnO on shelf life of fresh orange juice. Innov. Food Sci. Emerg. Technol. 2010, 11, 742–748. [Google Scholar] [CrossRef]
- Villegas, C.; Arrieta, M.; Rojas, A.; Torres, A.; Faba, S.; Toledo, M.; Gutierrez, M.; Zavalla, E.; Romero, J.; Galotto, M.; et al. PLA/organoclay bionanocomposites impregnated with thymol and cinnamaldehyde by supercritical impregnation for active and sustainable food packaging. Compos. Part B Eng. 2019, 176, 107336. [Google Scholar] [CrossRef]
- Rudramurthy, G.R.; Swamy, M.K.; Sinniah, U.R.; Ghasemzadeh, A. Nanoparticles: Alternatives against drug-resistant pathogenic microbes. Molecules 2016, 21, 836. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Misba, L.; Khan, A.U. Nano-therapeutics: A revolution in infection control in post antibiotic era. Nanomedicine 2017, 13, 2281–2301. [Google Scholar]
- Dizaj, S.M.; Lotfipour, F.; Barzegar-Jalali, M.; Zarrintan, M.H.; Adibkia, K. Antimicrobial activity of the metals and metal oxide nanoparticles. Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 44, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Hemeg, A.H. Nanomaterials for alternative antibacterial therapy. Int. J. Nanomed. 2017, 12, 8211–8225. [Google Scholar] [CrossRef]
- Vandebriel, R.J.; De Jong, W.H. A review of mammalian toxicity of ZnO nanoparticles. Nanotechnol. Sci. Appl. 2012, 5, 61–71. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Chakraborty, R.; Basu, T. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology 2014, 25, 135101. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.W.; Hsu, C.Y.; Lai, S.M.; Syu, W.J.; Wang, T.Y.; Lai, P.S. Metal nanobullets for multidrug resistant bacteria and biofilms. Adv. Drug Deliv. Rev. 2014, 78, 88–104. [Google Scholar] [CrossRef]
- Dulińska-Litewka, J.; Łazarczyk, A.; Hałubiec, P.; Szafrański, O. Superparamagnetic Iron Oxide Nanoparticles—Current and Prospective Medical Applications. Materials 2019, 12, 617. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Shi, Y.; Xing, B.; Hou, Y.; Cui, J.; Jia, S. The antimicrobial effects and mechanism of ε-poly-lysine against Staphylococcus aureus. Bioresour. Bioprocess. 2019, 6, 11. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceuticals 2017, 9, 53. [Google Scholar] [CrossRef]
- McBain, A.J.; Ledder, R.G.; Moore, L.E.; Catrenich, C.E.; Gilbert, P. Effects of Quaternary-Ammonium-Based Formulations on Bacterial Community Dynamics and Antimicrobial Susceptibility. Appl. Environ. Microbiol. 2004, 70, 3449–3456. [Google Scholar] [CrossRef]
- Demir, B.; Broughton, R.M.; Qiao, M.; Huang, T.-S.; Worley, S.D. N-Halamine Biocidal Materials with Superior Antimicrobial Efficacies for Wound Dressings. Molecules 2017, 22, 1582. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.V.; Zeldi, M.I.; Pugachev, M.V.; Sapozhnikov, S.V.; Shtyrlin, N.V.; Kuznetsova, S.V.; Evtygin, V.E.; Bogachev, M.I.; Kayumov, A.R.; Shtyrlin, Y.G. Antibacterial effects of quaternary bis-phosphonium and ammonium salts of pyridoxine on Staphylococcus aureus cells: A single base hitting two distinct targets? World J. Microbiol. Biotechnol. 2015, 32, 1–7. [Google Scholar] [CrossRef]
- Bakry, R.; Vallant, R.M.; Najam-Ul-Haq, M.; Rainer, M.; Szabo, Z.; Huck, C.W.; Bonn, G.K. Medicinal applications of fullerenes. Int. J. Nanomed. 2007, 2, 639–649. [Google Scholar]
- Venkataraman, A.; Amadi, E.V.; Chen, Y.; Papadopoulos, C. Carbon Nanotube Assembly and Integration for Applications. Nanoscale Res. Lett. 2019, 14, 1–47. [Google Scholar] [CrossRef]
- Donaldson, K.; Aitken, R.; Tran, L.; Stone, V.; Duffin, R.; Forrest, G.; Alexander, A. Carbon Nanotubes: A Review of Their Properties in Relation to Pulmonary Toxicology and Workplace Safety. Toxicol. Sci. 2006, 92, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Aunkor, M.; Toasin Hossain, T.; Topu, R.; Shamsul, H.P.; Metselaar, H.S.C. Antibacterial activity of graphene oxide nanosheet against multidrug resistant superbugs isolated from infected patients. R. Soc. Open Sci. 2020, 7, 200640. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Huo, P.; Zhang, R.; Liu, B. Antibacterial Properties of Graphene-Based Nanomaterials. Nanomaterials 2019, 9, 737. [Google Scholar] [CrossRef]
- Karwowska, E. Antibacterial potential of nanocomposite-based materials–a short review. Nanotechnol. Rev. 2017, 6, 243–254. [Google Scholar] [CrossRef]
- Pinto, R.J.B.; Daina, S.; Sadocco, P.; Neto, C.P.; Trindade, T. Antibacterial Activity of Nanocomposites of Copper and Cellulose. Biomed Res. Int. 2013, 2013, 280512. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Li, Y.; Tjong, S.C. Antibacterial Activities of Aliphatic Polyester Nanocomposites with Silver Nanoparticles and/or Graphene Oxide Sheets. Nanomaterials 2019, 9, 1102. [Google Scholar] [CrossRef] [PubMed]
- Klasen, H.J. Historical review of the use of silver in the treatment of burns. I. Early uses. Burns 2000, 26, 117–130. [Google Scholar] [CrossRef]
- De Aberasturi, D.J.; Serrano-Montes, A.B.; Liz-Marzán, L.M. Modern applications of plasmonic nanoparticles: From energy to health. Adv. Optic. Mater. 2015, 3, 602–617. [Google Scholar] [CrossRef]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Dastjerdi, R.; Montazer, M. A review on the application of inorganic nano-structured materials in the modification of textiles: Focus on anti-microbial properties. Colloids Surf. B Biointerfaces 2010, 79, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Tania, I.S.; Ali, M. Coating of ZnO Nanoparticle on Cotton Fabric to Create a Functional Textile with Enhanced Mechanical Properties. Polymers 2021, 13, 2701. [Google Scholar] [CrossRef]
- Pratap Reddy, M.; Venugopal, A.; Subrahmanyam, M. Hydroxyapatite-supported Ag-TiO2 as Escherichia coli disinfection photocatalyst. Water Res. 2007, 41, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.P.; Chaberny, I.F.; Massholder, K.; Stickler, M.; Benz, V.W.; Sonntag, H.-G.; Erdinger, L. Disinfection of surfaces by photocatalytic oxidation with titanium dioxide and UVA light. Chemosphere 2003, 53, 71–77. [Google Scholar] [CrossRef]
- Shrivastava, R.; Kushwaha, P.; Bhutia, Y.C.; Flora, S.J. Oxidative stress following exposure to silver and gold nanoparticles in mice. Toxicol. Ind. Health 2016, 32, 1391–1404. [Google Scholar] [CrossRef]
- Kang, S.; Pinault, M.; Pfefferle, L.D.; Elimelech, M. Single-Walled Carbon Nanotubes Exhibit Strong Antimicrobial Activity. Langmuir 2007, 23, 8670–8673. [Google Scholar] [CrossRef]
- Sayes, C.M.; Gobin, A.M.; Ausman, K.D.; Mendez, J.; West, J.L.; Colvin, V.L. Nano-C60 cytotoxicity is due to lipid peroxidation. Biomaterials 2005, 26, 7587–7595. [Google Scholar] [CrossRef]
- Marković, Z.; Todorovic-Markovic, B.; Kleut, D.; Nikolic, N.; Vranješ-Djurić, S.; Misirkić, M.; Vučićević, L.; Janjetovic, K.; Isakovic, A.; Harhaji, L.; et al. The mechanism of cell-damaging reactive oxygen generation by colloidal fullerenes. Biomaterials 2007, 28, 5437–5448. [Google Scholar] [CrossRef]
- Loew, S.; Fahr, A.; May, S. Modeling the Release Kinetics of Poorly Water-Soluble Drug Molecules from Liposomal Nanocarriers. J. Drug Deliv. 2011, 2011, 376548. [Google Scholar] [CrossRef]
- Zeng, L.; An, L.; Wu, X. Modeling Drug-Carrier Interaction in the Drug Release from Nanocarriers. J. Drug Deliv. 2011, 2011, 370308. [Google Scholar] [CrossRef] [PubMed]
- Drummond, D.C.; Noble, C.O.; Guo, Z.; Hong, K.; Park, J.W.; Kirpotin, D.B. Development of a Highly Active Nanoliposomal Irinotecan Using a Novel Intraliposomal Stabilization Strategy. Cancer Res. 2006, 66, 3271–3277. [Google Scholar] [CrossRef]
- Johnston, M.J.; Semple, S.C.; Klimuk, S.K.; Edwards, K.; Eisenhardt, M.L.; Leng, E.C.; Karlsson, G.; Yanko, D.; Cullis, P.R. Therapeutically optimized rates of drug release can be achieved by varying the drug-to-lipid ratio in liposomal vincristine formulations. Biochim. Biophys. Acta Biomembr. 2006, 1758, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Joguparthi, V.; Anderson, B.D. Liposomal Delivery of Hydrophobic Weak Acids: Enhancement of Drug Retention Using a High Intraliposomal pH. J. Pharm. Sci. 2008, 97, 433–454. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Anderson, B.D. Determination of Drug Release Kinetics from Nanoparticles: Overcoming Pitfalls of the Dynamic Dialysis Method. Mol. Pharm. 2013, 10, 3076–3089. [Google Scholar] [CrossRef] [PubMed]
- Kalhapure, R.S.; Suleman, N.; Mocktar, C.; Seedat, N.; Govender, T. Nanoengineered Drug Delivery Systems for Enhancing Antibiotic Therapy. J. Pharm. Sci. 2015, 104, 872–905. [Google Scholar] [CrossRef]
- Lu, X.Y.; Wu, D.C.; Li, Z.J.; Chen, G.Q. Polymer nanoparticles. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2011; Volume 104, pp. 299–323. [Google Scholar]
- Saidykhan, L.; Bin Abu Bakar, Z.; Rukayadi, Y.; Kura, A.U.; Latifah, S.Y. Development of nanoantibiotic delivery system using cockle shell-derived aragonite nanoparticles for treatment of osteomyelitis. Int. J. Nanomed. 2016, 11, 661–673. [Google Scholar] [CrossRef]
- Sikwal, D.R.; Kalhapure, R.S.; Rambharose, S.; Vepuri, S.; Soliman, M.; Mocktar, C.; Govender, T. Polyelectrolyte complex of vancomycin as a nanoantibiotic: Preparation, in vitro and in silico studies. Mater. Sci. Eng. C 2016, 63, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Assali, M.; Zaid, A.N.; Abdallah, F.; Almasri, M.; Khayyat, R. Single-walled carbon nanotubes-ciprofloxacin nanoantibiotic: Strategy to improve ciprofloxacin antibacterial activity. Int. J. Nanomed. 2017, 12, 6647–6659. [Google Scholar] [CrossRef] [PubMed]
- Kankala, R.K.; Lin, W.-Z.; Lee, C.-H. Combating Antibiotic Resistance through the Synergistic Effects of Mesoporous Silica-Based Hierarchical Nanocomposites. Nanomaterials 2020, 10, 597. [Google Scholar] [CrossRef] [PubMed]
- Kuthati, Y.; Kankala, R.K.; Lin, S.X.; Weng, C.F.; Lee, C.H. pH-triggered controllable release of silver–indole-3 acetic acid complexes from mesoporous silica nanoparticles (IBN-4) for effectively killing malignant bacteria. Mol. Pharm. 2015, 12, 2289–2304. [Google Scholar] [CrossRef]
- Singh, A.; Singh, N.B.; Hussain, I.; Singh, H.; Singh, S. Plantnanoparticle interaction: An approach to improve agricultural practices and plant productivity. Int. J. Pharm. Sci. Invent. 2015, 4, 25–40. [Google Scholar]
- Li, X.; Xu, H.; Chen, Z.-S.; Chen, G. Biosynthesis of Nanoparticles by Microorganisms and Their Applications. J. Nanomater. 2011, 2011, 270974. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Rahuman, A.A.; Kirthi, A.V.; Marimuthu, S.; Santhoshkumar, T.; Bagavan, A.; Gaurav, K.; Karthik, L.; Rao, K.B. Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 90, 78–84. [Google Scholar] [CrossRef]
- Gopinath, K.; Karthika, V.; Gowri, S.; Senthilkumar, V.; Kumaresan, S.; Arumugam, A. Antibacterial activity of ruthenium nanoparticles synthesized using Gloriosa superba L. leaf extract. J. Nanostruct. Chem. 2014, 4, 83. [Google Scholar] [CrossRef]
- Chandran, S.P.; Chaudhary, M.; Pasricha, R.; Ahmad, A.; Sastry, M. Synthesis of Gold Nanotriangles and Silver Nanoparticles Using Aloe vera Plant Extract. Biotechnol. Prog. 2006, 22, 577–583. [Google Scholar] [CrossRef]
- Huang, J.; Li, Q.; Sun, D.; Lu, Y.; Su, Y.; Yang, X. Biosynthesis of silver and gold nanoparticles by novel sundried Cinnamomum camphora leaf. Nanotechnology 2007, 18, 105104–105115. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Ma, C.; Song, R.; Hou, H.; Li, D. Enrichment and separation of silver from waste solutions by metal ion imprinted membrane. Hydrometallurgy 2009, 100, 82–86. [Google Scholar] [CrossRef]
- Botes, M.; Cloete, T.E. The potential of nanofibers and nanobiocides in water purification. Crit. Rev. Microbiol. 2010, 36, 68–81. [Google Scholar] [CrossRef]
- Sintubin, L.; De Windt, W.; Dick, J.; Mast, J.; Van Der Ha, D.; Verstraete, W.; Boon, N. Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl. Microbiol. Biotechnol. 2009, 84, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Thakkar, M.S.; Snehit, S.; Mhatre, M.S.; Rasesh, Y.; Parikh, M.S. Biological synthesis of metallic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 257–262. [Google Scholar]
- Kharissova, O.V.; Dias, H.R.; Kharisov, B.I.; Pérez, B.O.; Pérez, V.M.J. The greener synthesis of nanoparticles. Trends Biotechnol. 2013, 31, 240–248. [Google Scholar] [CrossRef]
- Torresdy, J.L.G.; Gomez, E.; Videa, J.R.P.; Parsons, J.G.; Troiani, H.; Yacaman, J.M. Alfalfa sprouts: A natural source for the synthesis of silver nanoparticles. Langmuir 2003, 19, 1357–1361. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Byrne, J.M.; Coker, V.S. Biotechnological synthesis of functional nanomaterials. Curr. Opin. Biotechnol. 2011, 22, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Gour, A.; Jain, N.K. Advances in green synthesis of nanoparticles. Artif. Cells Nanomed. Biotechnol. 2019, 47, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Rajakumara, G.; Rahumana, A.A.; Roopan, S.M.; Khannac, V.G.; Elangoa, G.; Kamaraja, C. Fungus-mediated biosynthesis and characterization of TiO2 nanoparticles and their activity against pathogenic bacteria. Spectrochim. Acta Part A 2012, 9, 123–129. [Google Scholar] [CrossRef]
- Sankar, R.; Rizwana, K.; Shivashangari, K.S.; Ravikumar, V. Ultra-rapid photocatalytic activity of Azadirachta indica engineered colloidal titanium dioxide nanoparticles. Appl. Nanosci. 2015, 5, 731–736. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, G.; Jang, N.R.; Yun, J.M.; Song, J.Y.; Kim, B.S. Biological synthesis of copper nanoparticles using plant extract. Nanotechnology 2011, 1, 371–374. [Google Scholar]
- Singh, S.C.; Mishra, S.K.; Srivastava, R.K.; Gopal, R. Optical Properties of Selenium Quantum Dots Produced with Laser Irradiation of Water Suspended Se Nanoparticles. J. Phys. Chem. C 2010, 114, 17374–17384. [Google Scholar] [CrossRef]
- Dameron, C.T.; Reese, R.N.; Mehra, R.K.; Kortan, A.R.; Carroll, P.J.; Steigerwald, M.L. Biosynthesis of cadmium sulfide quantum semiconductor crystallites. Lett. Nat. 1989, 338, 596–597. [Google Scholar] [CrossRef]
- Bharde, A.; Wani, A.; Shouche, Y.; Joy, P.A.; Prasad, B.L.V.; Sastry, M. Bacterial Aerobic Synthesis of Nanocrystalline Magnetite. J. Am. Chem. Soc. 2005, 127, 9326–9327. [Google Scholar] [CrossRef]
- Naika, H.R.; Lingaraju, K.; Manjunath, K.; Kumar, D.; Nagaraju, G.; Suresh, D.; Nagabhushana, H. Green synthesis of CuO nanoparticles using Gloriosa superba L. extract and their antibacterial activity. J. Taibah Univ. Sci. 2015, 9, 7–12. [Google Scholar] [CrossRef]
- Ahmad, A.; Mukherjee, P.; Senapati, S.; Mandal, D.; Khan, M.; Kumar, R.; Sastry, M. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf. B Biointerfaces 2003, 28, 313–318. [Google Scholar] [CrossRef]
- Riddin, T.; Gericke, M.; Whiteley, C. Analysis of the inter-and extracellular formation of platinum nanoparticles by Fusarium oxysporum f. sp. lycopersici using response surface methodology. Nanotechnology 2006, 17, 3482. [Google Scholar] [CrossRef]
- Nam, K.; Lee, Y.; Krauland, E.; Kottmann, S. Belcher, Peptidemediated reduction of silver ions on engineered biological scaffolds. ACS Nano 2008, 2, 1480–1486. [Google Scholar] [CrossRef]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef]
- Sanghi, R.; Verma, P. Biomimetic synthesis and characterisation of protein capped silver nanoparticles. Bioresour. Technol. 2009, 100, 501–504. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, Y.; Jiang, L.; Zhu, D. Thiosalicylic Acid-Functionalized Silver Nanoparticles Synthesized in One-Phase System. J. Colloid Interface Sci. 2002, 249, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Das, A.R.; Guha, A.K. Gold Nanoparticles: Microbial Synthesis and Application in Water Hygiene Management. Langmuir 2009, 25, 8192–8199. [Google Scholar] [CrossRef]
- Mukherjee, P.; Ahmad, A.; Mandal, D.; Senapati, S.; Sainkar, S.R.; Khan, M.I.; Parishcha, R.; Ajaykumar, P.V.; Alam, M.; Kumar, R.; et al. Fungus-Mediated Synthesis of Silver Nanoparticles and Their Immobilization in the Mycelial Matrix: A Novel Biological Approach to Nanoparticle Synthesis. Nano Lett. 2001, 1, 515–519. [Google Scholar] [CrossRef]
- Apte, M.; Sambre, D.; Gaikawad, S.; Joshi, S.; Bankar, A.; Kumar, A.R.; Zinjarde, S. Psychrotrophic yeast Yarrowia lipolytica NCYC 789 mediates the synthesis of antimicrobial silver nanoparticles via cell-associated melanin. AMB Express 2013, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef]
- Schreurs, W.J.; Rosenberg, H. Effect of silver ions on transport and retention of phosphate by Escherichia coli. J. Bacteriol. 1982, 152, 7–13. [Google Scholar] [CrossRef]
- Składanowski, M.; Wypij, M.; Laskowski, D.; Golińska, P.; Dahm, H.; Rai, M. Silver and gold nanoparticles synthesized from Streptomyces sp. isolated from acid forest soil with special reference to its antibacterial activity against pathogens. J. Clust. Sci. 2017, 28, 59–79. [Google Scholar] [CrossRef]
- Singh, T.; Jyoti, K.; Patnaik, A.; Singh, A.; Chauhan, R.; Chandel, S. Biosynthesis, characterization and antibacterial activity of silver nanoparticles using an endophytic fungal supernatant of Raphanus sativus. J. Genet. Eng. Biotechnol. 2017, 15, 31–39. [Google Scholar] [CrossRef]
- Maliszewska, I.; Sadowski, Z. Synthesis and antibacterial activity of of silver nanoparticles. J. Phys. Conf. Ser. 2009, 146, 012024. [Google Scholar] [CrossRef]
- Priyadarshini, E.; Pradhan, N.; Sukla, L.B.; Panda, P.K. Controlled Synthesis of Gold Nanoparticles UsingAspergillus terreusIF0 and Its Antibacterial Potential against Gram Negative Pathogenic Bacteria. J. Nanotechnol. 2014, 2014, 653198. [Google Scholar] [CrossRef]
- Singh, P.; Kim, Y.-J.; Zhang, D.; Yang, D.-C. Biological Synthesis of Nanoparticles from Plants and Microorganisms. Trends Biotechnol. 2016, 34, 588–599. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Ma, X.; Tian, B.; Li, T.; Yu, J.; Dai, S.; Weng, Y.; Hua, Y. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. Int. J. Nanomed. 2016, 11, 5931–5944. [Google Scholar] [CrossRef]
- Mirzajani, F.; Ghassempour, A.; Aliahmadi, A.; Esmaeili, M.A. Antibacterial effect of silver nanoparticles on Staphylococcus aureus. Res. Microbiol. 2011, 162, 542–549. [Google Scholar] [CrossRef]
- Lok, C.-N.; Ho, C.-M.; Chen, R.; He, Q.-Y.; Yu, W.-Y.; Sun, H.; Tam, P.K.-H.; Chiu, J.-F.; Che, C.-M. Proteomic Analysis of the Mode of Antibacterial Action of Silver Nanoparticles. J. Proteome Res. 2006, 5, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.; Liu, H.-L.; Dai, A.S.; Fu, K.-Y. Antibacterial properties of silver nanoparticles in three different sizes and their nanocomposites with a new waterborne polyurethane. Int. J. Nanomed. 2010, 5, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Chaloupka, K.; Malam, Y.; Seifalian, A.M. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Biotechnol. 2010, 28, 580–588. [Google Scholar] [CrossRef]
- Samadi, N.; Golkaran, D.; Eslamifar, A.; Jamalifar, H.; Fazeli, M.R.; Mohseni, F.A. Intra/extracellular biosynthesis of silver nanoparticles by an autochthonous strain of Proteus mirabilis isolated from photographic waste. J. Biomed. Nanotechnol. 2009, 5, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Ashajyothi, C.; Manjunath, N.R.; Chandrakanth, R.K. Antibacterial Activity of Biogenic Zinc Oxide Nanoparticals Synthesised From Enterococcus faecalis. Int. J. Chemtech. Res. USA 2014, 6, 3131–3136. [Google Scholar]
- Mohammadinejad, R.; Shavandi, A.; Raie, D.S.; Sangeetha, J.; Soleimani, M.; Hajibehzad, S.S.; Thangadurai, D.; Hospet, R.; Popoola, J.O.; Arzani, A.; et al. Plant molecular farming: Production of metallic nanoparticles and therapeutic proteins using green factories. Green Chem. 2019, 21, 1845–1865. [Google Scholar] [CrossRef]
- Dauthal, P.; Mukhopadhyay, M. Noble Metal Nanoparticles: Plant-Mediated Synthesis, Mechanistic Aspects of Synthesis, and Applications. Ind. Eng. Chem. Res. 2016, 55, 9557–9577. [Google Scholar] [CrossRef]
- Ahmed, S.; Saifullah, M.; Ahmad, B.L.; Swami, S. Green synthesis of silver nanoparticles using Azadirachta indica aqueous leaf extract. J. Radiat. Res. Appl. Sci. 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Suman, T.; Rajasree, S.R.; Ramkumar, R.; Rajthilak, C.; Perumal, P. The Green synthesis of gold nanoparticles using an aqueous root extract of Morinda citrifolia L. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 118, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Deng, K.K.; Kim, N.-J.; Ross, L.; Surampalli, R.Y.; Hu, Z. The inhibitory effects of silver nanoparticles, silver ions, and silver chloride colloids on microbial growth. Water Res. 2008, 42, 3066–3074. [Google Scholar] [CrossRef]
- Velayutham, K.; Rahuman, A.A.; Rajakumar, G.; Santhoshkumar, T.; Marimuthu, S.; Jayaseelan, C.; Bagavan, A.; Kirthi, A.V.; Kamaraj, C.; Zahir, A.A.; et al. Evaluation of Catharanthus roseus leaf extract-mediated biosynthesis of titanium dioxide nanoparticles against Hippobosca maculata and Bovicola ovis. Parasitol. Res. 2011, 111, 2329–2337. [Google Scholar] [CrossRef]
- Órdenes-Aenishanslins, N.A.; Saona, L.A.; Durán-Toro, V.M.; Monrás, J.P.; Bravo, D.M.; Pérez-Donoso, J.M. Use of titanium dioxide nanoparticles biosynthesized by Bacillus mycoides in quantum dot sensitized solar cells. Microb. Cell Factories 2014, 13, 90. [Google Scholar] [CrossRef]
- Korbekandi, H.; Iravani, S.; Abbasi, S. Production of nanoparticles using organisms. Crit. Rev. Biotechnol. 2009, 29, 279–306. [Google Scholar] [CrossRef]
- Singh, K.; Panghal, M.; Kadyan, S.; Chaudhary, U.; Yadav, J.P. Green silver nanoparticles of Phyllanthus amarus: As an antibacterial agent against multi drug resistant clinical isolates of Pseudomonas aeruginosa. J. Nanobiotechnol. 2014, 12, 1–9. [Google Scholar] [CrossRef]
- Rao, B.; Tang, R.-C. Green synthesis of silver nanoparticles with antibacterial activities using aqueous Eriobotrya japonica leaf extract. Adv. Nat. Sci. Nanosci. Nanotechnol. 2017, 8, 015014. [Google Scholar] [CrossRef]
- Bose, D.; Chatterjee, S. Antibacterial Activity of Green Synthesized Silver Nanoparticles Using Vasaka (Justicia adhatoda L.) Leaf Extract. Indian J. Microbiol. 2015, 55, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Senthil, M.; Ramesh, C. Biogenic synthesis of Fe3O4 nanoparticles using tridax procumbens leaf extract and its antibacterial activity on Pseudomonas aeruginosa. Dig. J. Nanomater. Biostructures 2012, 7, 1655–1661. [Google Scholar]
- Naseem, T.; Farrukh, M.A. Antibacterial activity of green synthesis of iron nanoparticles using Lawsonia inermis and Gardenia jasminoides leaves extract. J. Chem. 2015, 2015, 912342. [Google Scholar] [CrossRef]
- Seetharaman, P.; Chandrasekaran, R.; Gnanasekar, S.; Mani, I.; Sivaperumal, S. Biogenic gold nanoparticles synthesized using Crescentia cujete L. and evaluation of their different biological activities. Biocatal. Agric. Biotechnol. 2017, 11, 75–82. [Google Scholar] [CrossRef]
- Suresh, J.; Pradheesh, G.; Alexramani, V.; Sundrarajan, M.; Hong, S.I. Green synthesis and characterization of zinc oxide nanoparticle using insulin plant ( Costus pictus D. Don ) and investigation of its antimicrobial as well as anticancer activities. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 015008. [Google Scholar] [CrossRef]
- Cavallaro, A.A.; MacGregor-Ramiasa, M.N.; Vasilev, K. Antibiofouling Properties of Plasma-Deposited Oxazoline-Based Thin Films. ACS Appl. Mater. Interfaces 2016, 8, 6354–6362. [Google Scholar] [CrossRef]
- Ramiasa, M.N.; Cavallaro, A.A.; Mierczynska, A.; Christo, S.N.; Gleadle, J.M.; Hayball, J.D.; Vasilev, K. Plasma polymerised polyoxazoline thin films for biomedical applications. Chem. Commun. 2015, 51, 4279–4282. [Google Scholar] [CrossRef]
- Michl, T.D.; Coad, B.R.; Hüsler, A.; Valentin, J.D.P.; Vasilev, K.; Griesser, H.J. Effects of Precursor and Deposition Conditions on Prevention of Bacterial Biofilm Growth on Chlorinated Plasma Polymers. Plasma Process. Polym. 2016, 13, 654–662. [Google Scholar] [CrossRef]
- Cavallaro, A.; Mierczynska, A.; Barton, M.; Majewski, P.; Vasilev, K. Influence of immobilized quaternary ammonium group surface density on antimicrobial efficacy and cytotoxicity. Biofouling 2016, 32, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, A.; Majewski, P.; Barton, M.; Vasilev, K. Substrate Independent Approach for Immobilisation of Quaternary Ammonium Compounds to Surfaces to Reduce Bio-Burden. Mater. Sci. Forum 2014, 783, 1389–1395. [Google Scholar] [CrossRef]
- Vasilev, K.; Poulter, N.; Martinek, P.; Griesser, H.J. Controlled Release of Levofloxacin Sandwiched between Two Plasma Polymerized Layers on a Solid Carrier. ACS Appl. Mater. Interfaces 2011, 3, 4831–4836. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, A.; Vasilev, K. Controlled and sustained release of pharmaceuticals via single step solvent-free encapsulation. Chem. Commun. 2014, 51, 1838–1841. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michl, T.D.; Coad, B.R.; Doran, M.; Osiecki, M.; Kafshgari, M.H.; Voelcker, N.H.; Hüsler, A.; Vasilev, K.; Griesser, H.J. Nitric oxide releasing plasma polymer coating with bacteriostatic properties and no cytotoxic side effects. Chem. Commun. 2015, 51, 7058–7060. [Google Scholar] [CrossRef]
- Kafshgari, M.H.; Delalat, B.; Harding, F.J.; Cavallaro, A.; Mäkilä, E.; Salonen, J.; Vasilev, K.; Voelcker, N.H. Antibacterial properties of nitric oxide-releasing porous silicon nanoparticles. J. Mater. Chem. B 2016, 4, 2051–2058. [Google Scholar] [CrossRef]
- Michl, T.D.; Locock, K.E.S.; Stevens, N.; Hayball, J.D.; Vasilev, K.; Postma, A.; Qu, Y.; Traven, A.; Haeussler, M.; Meagher, L.; et al. RAFT-derived antimicrobial polymethacrylates: Elucidating the impact of end-groups on activity and cytotoxicity. Polym. Chem. 2014, 5, 5813–5822. [Google Scholar] [CrossRef]
- Locock, K.; Michl, T.; Valentin, J.; Vasilev, K.; Hayball, J.; Qu, Y.; Traven, A.; Griesser, H.; Meagher, L.; Haeussler, M. Guanylated polymethacrylates: A class of potent antimicrobial polymers with low hemolytic activity. Biomacromolecules 2013, 14, 4021–4031. [Google Scholar] [CrossRef]
- Locock, K.E.S.; Michl, T.D.; Stevens, N.; Hayball, J.D.; Vasilev, K.; Postma, A.; Griesser, H.J.; Meagher, L.; Haeussler, M. Antimicrobial Polymethacrylates Synthesized as Mimics of Tryptophan-Rich Cationic Peptides. ACS Macro Lett. 2014, 3, 319–323. [Google Scholar] [CrossRef]
- Cavallaro, A.; Taheri, S.; Vasilev, K. Responsive and smart antibacterial surfaces: Common approaches and new developments (Review). Biointerphases 2014, 9, 029005. [Google Scholar] [CrossRef]
- Baier, G.; Cavallaro, A.; Vasilev, K.; Mailänder, V.; Musyanovych, A.; Landfester, K. Enzyme responsive hyaluronic acid nanocapsules containing polyhexanide and their exposure to bacteria to prevent infection. Biomacromolecules 2013, 14, 1103–1112. [Google Scholar] [CrossRef]
- Baier, G.; Cavallaro, A.; Friedemann, K.; Müller, B.; Glasser, G.; Vasilev, K.; Landfester, K. Enzymatic degradation of poly(l-lactide) nanoparticles followed by the release of octenidine and their bactericidal effects. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Devatha, C.P.; Thalla, A.K. Synthesis of Inorganic Nanomaterials. In Advances and Key Technologies; Elsevier: Amsterdam, The Netherlands, 2018; pp. 169–184. [Google Scholar]
- Patra, J.K.; Baek, K.-H. Green Nanobiotechnology: Factors Affecting Synthesis and Characterization Techniques. J. Nanomater. 2014, 2014, 417305. [Google Scholar] [CrossRef]
- Sneha, K.; Sathishkumar, M.; Kim, S.; Yun, Y.-S. Counter ions and temperature incorporated tailoring of biogenic gold nanoparticles. Process. Biochem. 2010, 45, 1450–1458. [Google Scholar] [CrossRef]
- Kumari, M.; Mishra, A.; Pandey, S.; Singh, S.P.; Chaudhry, V.; Mudiam, M.K.; Shukla, S.; Kakkar, P.; Nautiyal, C.S. Physico-chemical condition optimization during biosynthesis lead to development of improved and catalytically efficient gold nano particles. Sci. Rep. 2016, 6, 1–4. [Google Scholar] [CrossRef]
- Iravani, S.; Zolfaghari, B. Green Synthesis of Silver Nanoparticles UsingPinus eldaricaBark Extract. Biomed Res. Int. 2013, 2013, 639725. [Google Scholar] [CrossRef]
- Jagathesan, G.; Rajiv, P. Biosynthesis and characterization of iron oxide nanoparticles using Eichhornia crassipes leaf extract and assessing their antibacterial activity. Biocatal. Agric. Biotechnol. 2018, 13, 90–94. [Google Scholar] [CrossRef]
- Islam, A.M.; Mukherjee, M. Effect of temperature in synthesis of silver nanoparticles in triblock copolymer micellar solution. J. Exp. Nanosci. 2011, 6, 596–611. [Google Scholar] [CrossRef]
- Tan, N.P.B.; Lee, C.H.; Li, P. Influence of temperature on the formation and encapsulation of gold nanoparticles using a temperature-sensitive template. Data Brief 2015, 5, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Kumar, E.R.; Jayaprakash, R.; Arunkumar, T.; Kumar, S. Effect of reaction time on particle size and dielectric properties of manganese substituted CoFe2O4 nanoparticles. J. Phys. Chem. Solids 2013, 74, 110–114. [Google Scholar] [CrossRef]
- Armendariz, V.; Herrera, I.; Peralta-Videa, J.R.; Jose-Yacaman, M.; Troiani, H.; Santiago, P.; Gardea-Torresdey, J.L. Size controlled gold nanoparticle formation by Avena sativa biomass: Use of plants in nanobiotechnology. J. Nanopart. Res. 2004, 6, 377–382. [Google Scholar] [CrossRef]
- Okitsu, K.; Sharyo, K.; Nishimura, R. One-Pot Synthesis of Gold Nanorods by Ultrasonic Irradiation: The Effect of pH on the Shape of the Gold Nanorods and Nanoparticles. Langmuir 2009, 25, 7786–7790. [Google Scholar] [CrossRef]
- Karade, V.; Dongale, T.; Sahoo, S.C.; Kollu, P.; Chougale, A.; Patil, P. Effect of reaction time on structural and magnetic properties of green-synthesized magnetic nanoparticles. J. Phys. Chem. Solids 2018, 120, 161–166. [Google Scholar] [CrossRef]
- Rose, I.; Sathish, R.; Rajendran, A.; Sagayaraj, P. Effect of reaction time on the synthesis of cadmium selenide nanoparticles and the efficiency of solar cell. J. Mater. Environ. Sci. 2016, 7, 1589–1596. [Google Scholar]
- Flor, J.; Lima, S.; Davolos, M.R. Effect of Reaction Time on the Particle Size of ZnO and ZnO:Ce Obtained by a Sol–Gel Method; Springer International Publishing: Cham, Switzerland, 2004; pp. 239–243. [Google Scholar]
- Perelshtein, I.; Applerot, G.; Perkas, N.; Wehrschetz-Sigl, E.; Hasmann, A.; Guebitz, G.M.; Gedanken, A. Antibacterial Properties of an in situ Generated and Simultaneously Deposited Nanocrystalline ZnO on Fabrics. ACS Appl. Mater. Interfaces 2008, 1, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.; Mir, A.; Mallik, D.; Sinha, A.; Nayar, S.; Webster, T.J. Bactericidal effect of iron oxide nanoparticles on Staphylococcus aureus. Int. J. Nanomed. 2010, 5, 277–283. [Google Scholar]
- Guo, B.L.; Han, P.; Guo, L.C.; Cao, Y.Q.; Li, A.D.; Kong, J.Z.; Zhai, H.F.; Wu, D. The antibacterial activity of Ta-doped ZnO nanoparticles. Nanosci. Res. Lett. 2015, 10, 1–10. [Google Scholar] [CrossRef]
- Liu, P.; Duan, W.; Wang, Q.; Li, X. The damage of outer membrane of Escherichia coli in the presence of TiO2 combined with UV light. Colloids Surf. B Biointerfaces 2010, 78, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. [Google Scholar] [CrossRef] [PubMed]
- Rispoli, F.; Angelov, A.; Badia, D.; Kumar, A.; Seal, S.; Shah, V. Understanding the toxicity of aggregated zero valent copper nanoparticles against Escherichia coli. J. Hazard. Mater. 2010, 180, 212–216. [Google Scholar] [CrossRef]
- Rajesh, W.R.; Jaya, R.L.; Niranjan, S.K.; Vijay, D.M.; Sahebrao, B.K. Phytosynthesis of silver nanoparticle using Gliricidia sepium. Curr. Nanosci. 2009, 5, 117–122. [Google Scholar]
- Feldheim, D.L.; Foss, C.A. Metal Nanoparticles: Synthesis, Characterization, and Applications; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Sangeetha, G.; Sivaraj, R.; Venckatesh, R. Green synthesis of zinc oxide nanoparticles by Aloe barbadensis miller leaf extract: Structure and optical properties. Mater. Res. Bull. 2011, 46, 2560–2566. [Google Scholar] [CrossRef]
- Schaffer, B.; Hohenester, U.; Trügler, A.; Hofer, F. High-resolution surface plasmon imaging of gold nanoparticles by energy-filtered transmission electron microscopy. Phys. Rev. B 2009, 79, 0414011. [Google Scholar] [CrossRef]
- Ali, K.; Dwivedi, S.; Azam, A.; Saquib, Q.; Al-Said, M.S.; Alkhedhairy, A.A.; Musarrat, J. Aloe vera extract functionalized zinc oxide nanoparticles as nanoantibiotics against multi-drug resistant clinical bacterial isolates. J. Colloid Interface Sci. 2016, 472, 145–156. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Shim, J.-J. Green Synthesis and Characterization of Carbon Nanotubes/Polyaniline Nanocomposites. J. Spectrosc. 2015, 2015, 297804. [Google Scholar] [CrossRef]
- Dhandapani, P.; Maruthamuthu, S.; Rajagopal, G. Bio-mediated synthesis of TiO2 nanoparticles and its photocatalytic effect on aquatic biofilm. J. Photochem. Photobiol. B Biol. 2012, 110, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Murray, C.B.; Weller, D.; Folks, L.; Moser, A. Monodisperse FePt Nanoparticles and Ferromagnetic FePt Nanocrystal Superlattices. Science 2000, 287, 1989–1992. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, A.; Karthikeyan, C.; Hameed, A.S.H.; Gopinath, K.; Gowri, S.; Karthika, V. Synthesis of cerium oxide nanoparticles using Gloriosa superba L. leaf extract and their structural, optical and antibacterial properties. Mater. Sci. Eng. C 2015, 49, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Kumar, N.; Kumar, R.; Salar, R.K. Antimicrobial activity of zinc oxide nanoparticles synthesized from Aloe vera peel extract. SN Appl. Sci. 2019, 1, 1–9. [Google Scholar] [CrossRef]
- Govindaraju, K.; Tamilselvan, S.; Kiruthiga, V.; Singaravelu, G. Biogenic silver nanoparticles by Solanum torvum and their promising antimicrobial activity. J. Biopest. 2010, 3, 349–399. [Google Scholar]
- Jiang, J.; Oberdörster, G.; Biswas, P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 2009, 11, 77–89. [Google Scholar] [CrossRef]
- Strasser, P.; Koh, S.; Anniyev, T.; Greeley, J.; More, K.; Yu, C. Lattice-strain control of the activity in de alloyed core-fuel cell catalysts. Nat. Chem. 2010, 2, 454–460. [Google Scholar] [CrossRef]
- Andrade, F.; Rafael, D.; Videira, M.; Ferreira, D.; Sosnik, A.; Sarmento, B. Nanotechnology and pulmonary delivery to overcome resistance in infectious diseases. Adv. Drug Deliv. Rev. 2013, 65, 1816–1827. [Google Scholar] [CrossRef]
- Ranghar, S.; Sirohi, P.; Verma, P.; Agarwal, V. Nanoparticle-based drug delivery systems: Promising approaches against infections. Braz. Arch. Biol. Technol. 2013, 57, 209–222. [Google Scholar] [CrossRef]
- Liu, Y.; Tee, J.K.; Chiu, G.N.C. Dendrimers in oral drug delivery application: Current explorations, toxicity issues and strategies for improvement. Curr. Pharm. Des. 2015, 21, 2629–2642. [Google Scholar] [CrossRef]
- Qi, G.; Li, L.; Yu, F.; Wang, H. Vancomycin-Modified Mesoporous Silica Nanoparticles for Selective Recognition and Killing of Pathogenic Gram-Positive Bacteria Over Macrophage-Like Cells. ACS Appl. Mater. Interfaces 2013, 5, 10874–10881. [Google Scholar] [CrossRef]
- Xiong, M.-H.; Li, Y.-J.; Bao, Y.; Yang, X.-Z.; Hu, B.; Wang, J. Bacteria-Responsive Multifunctional Nanogel for Targeted Antibiotic Delivery. Adv. Mater. 2012, 24, 6175–6180. [Google Scholar] [CrossRef]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar] [CrossRef]
- Lv, Y.; Hao, L.; Hu, W.; Ran, Y.; Bai, Y.; Zhang, L. Novel multifunctional pH-sensitive nanoparticles loaded into microbubbles as drug delivery vehicles for enhanced tumor targeting. Sci. Rep. 2016, 6, 29321. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shen, Y.; Jiang, W.; Jiang, W.; Shen, Y. Magnetic targeted drug delivery carriers encapsulated with pH-sensitive polymer: Synthesis, characterization and in vitro doxorubicin release studies. J. Biomater. Sci. Polym. Ed. 2016, 27, 1303–1316. [Google Scholar] [CrossRef]
- Jijie, R.; Barras, A.; Teodorescu, F.; Boukherroub, R.; Szunerits, S. Advancements on the molecular design of nanoantibiotics: Current level of development and future challenges. Mol. Syst. Des. Eng. 2017, 2, 349–369. [Google Scholar] [CrossRef]
- Brooks, B.D.; Brooks, A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Cisneros, J.M.; Rodríguez-Baño, J. Nosocomial bacteremia due to Acinetobacter baumannii: Epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 2002, 8, 687–693. [Google Scholar] [CrossRef]
- Aslan, S.; Loebick, C.Z.; Kang, S.; Elimelech, M.; Pfefferle, L.D.; Van Tassel, P.R. Antimicrobial biomaterials based on carbon nanotubes dispersed in poly(lactic-co-glycolic acid). Nanoscale 2010, 2, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Hoet, P.H.; Brüske-Hohlfeld, I.; Salata, O.V. Nanoparticles—Known and unknown health risks. J. Nanobiotechnol. 2004, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Grelling, N.; Wetteland, C.L.; Rosario, R.; Liu, H. Antimicrobial Activities and Mechanisms of Magnesium Oxide Nanoparticles (nMgO) against Pathogenic Bacteria, Yeasts, and Biofilms. Sci. Rep. 2018, 8, 16260. [Google Scholar] [CrossRef] [PubMed]
- Nijhara, R.; Balakrishnan, K. Bringing nanomedicines to market: Regulatory challenges, opportunities, and uncertainties. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Sandhiya, S.; Dkhar, S.A.; Surendiran, A. Emerging trends of nanomedicine—An overview. Fundam. Clin. Pharm. 2009, 23, 263–269. [Google Scholar] [CrossRef] [PubMed]
- El-Ansary, A.; Al-Daihan, S. On the Toxicity of Therapeutically Used Nanoparticles: An Overview. J. Toxicol. 2009, 2009, 754810. [Google Scholar] [CrossRef]
- Hagens, W.I.; Oomen, A.G.; De Jong, W.H.; Cassee, F.R.; Sips, A.J. What do we (need to) know about the kinetic properties of nanoparticles in the body? Regul. Toxicol. Pharm. 2007, 49, 217–229. [Google Scholar] [CrossRef]
- Poma, A.; Di Giorgio, M.L. Toxicogenomics to Improve Comprehension of the Mechanisms Underlying Responses of In Vitro and In Vivo Systems to Nanomaterials: A Review. Curr. Genom. 2008, 9, 571–585. [Google Scholar] [CrossRef]
- De Jong, W.H.; Borm, P.J. Drug delivery and nanoparticles: Applications and hazards. Int. J. Nanomed. 2008, 3, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Wu, C.; Yang, B.; Ma, H.; Shi, C.; Wang, Q.; Wang, Q.; Yuan, Y.; Liao, M. Integrated metabolomic analysis of the nano-sized copper particle-induced hepatotoxicity and nephrotoxicity in rats: A rapid in vivo screening method for nanotoxicity. Toxicol. Appl. Pharm. 2008, 232, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Current in vitro methods in nanoparticle risk assessment: Limitations and challenges. Eur. J. Pharm. Biopharm. 2009, 72, 370–377. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).