Analysis on Drug-Resistance-Associated Mutations among Multidrug-Resistant Mycobacterium tuberculosis Isolates in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Drug Susceptibility Test
2.3. DNA Extraction
2.4. Drug-Resistance-Related Genes and DNA Sequencing
2.5. Statistical Analysis
3. Results
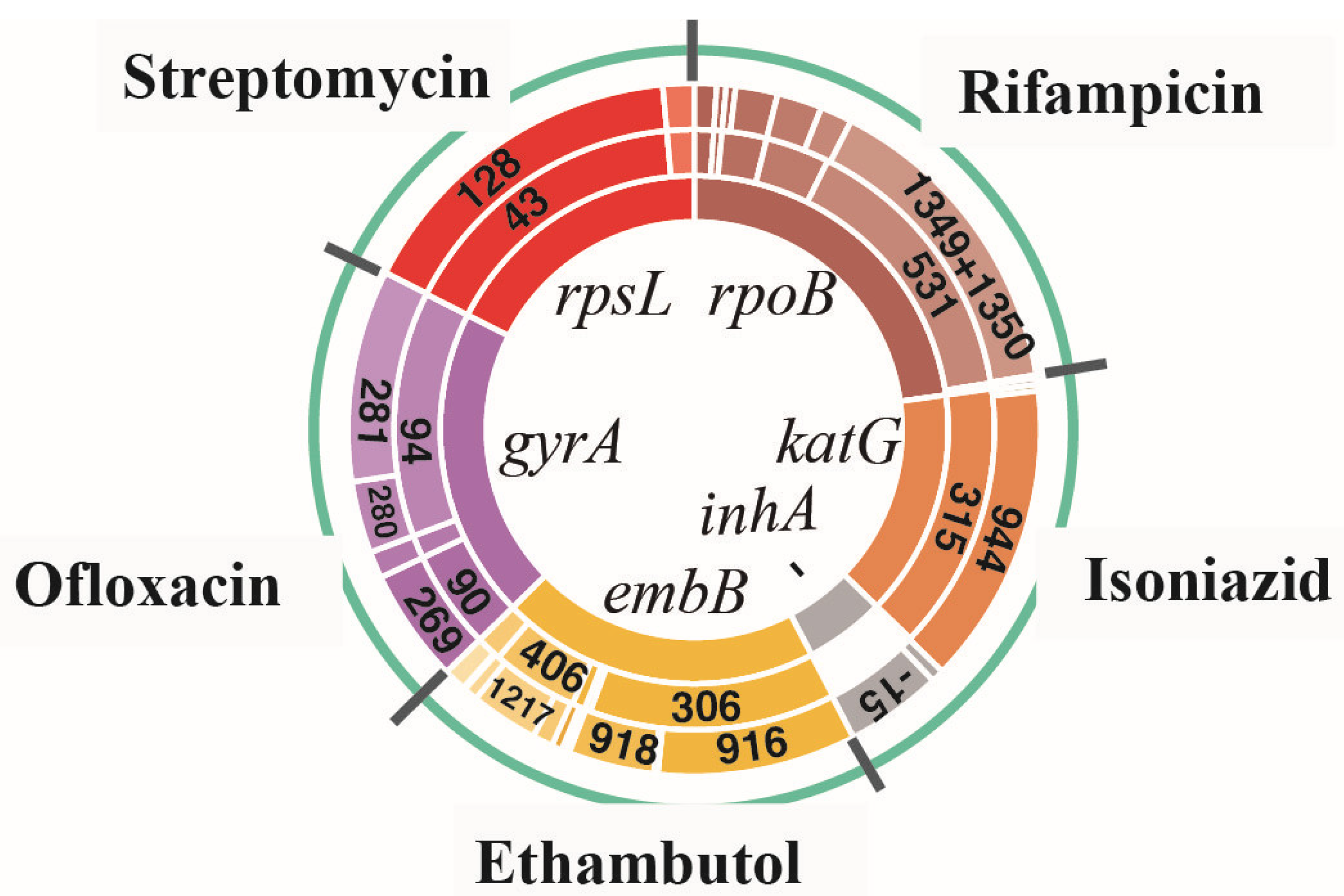
3.1. Amino Acid Missense Mutation Types and Their Corresponding SNP Mutation Types for All Drug-Resistant Isolates
3.2. Analysis of Linked Missense Mutations Found in All Drug-Resistant Isolates
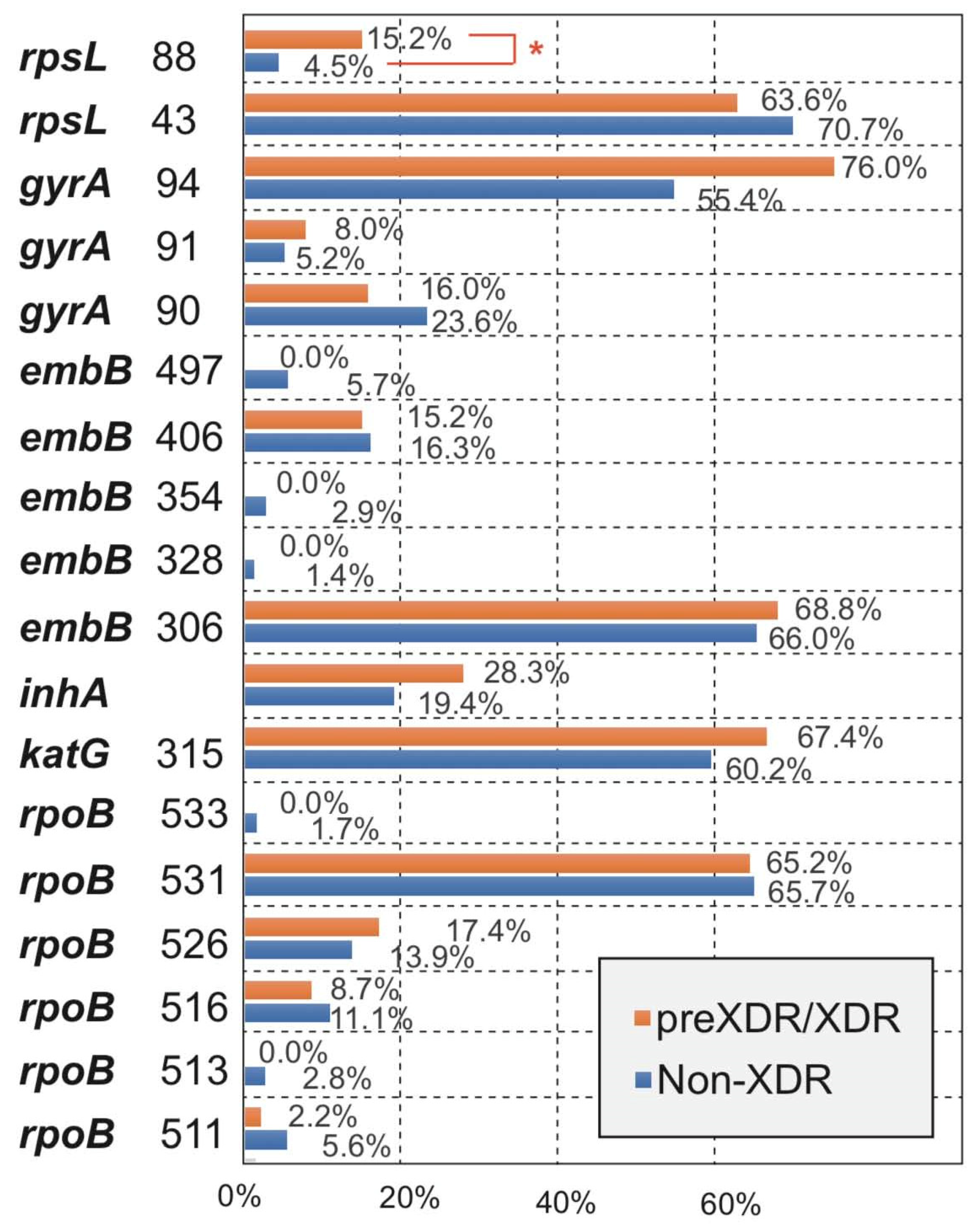
3.3. Comparison for the Occurrence of Amino Acid Missense Mutation Types between Non-XDR and preXDR/XDR Groups
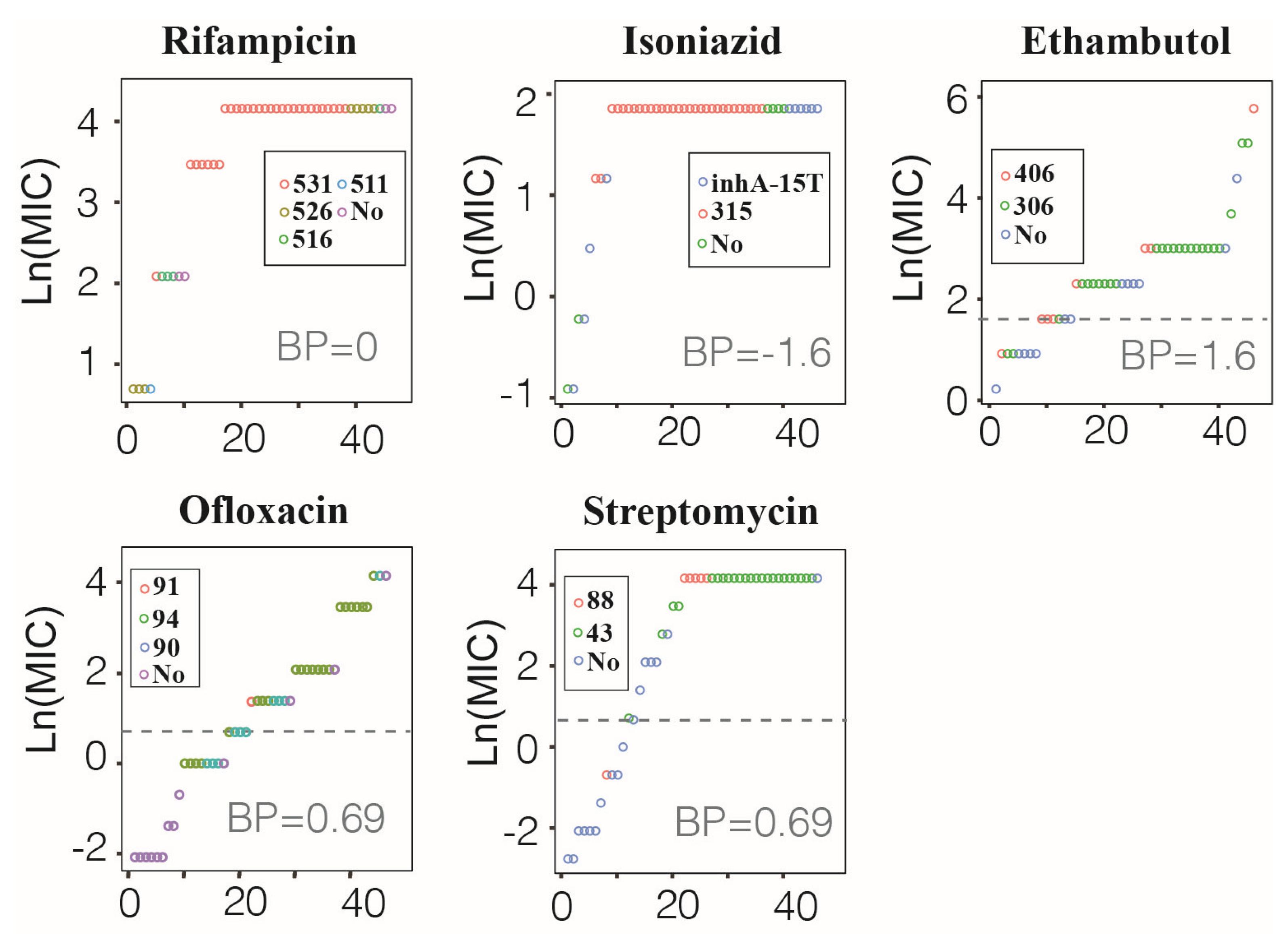
3.4. Correspondence between MM Type and MIC Value in preXDR/XDR Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Marisa, K.; Robin, M.W.; Cindy, H.; Nicolaas, C.G.P.; Elizabeth, M.S.; Borna, M.; Frederick, A.S.; Mamisa, C.; Ebrahim, H.; Gerrit, C.; et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 2013, 19, 449–455. [Google Scholar]
- Xie, Y.L.; Chakravorty, S.; Armstrong, D.T.; Hall, S.L.; Via, L.E.; Song, T.; Yuan, X.; Mo, X.; Zhu, H.; Xu, P.; et al. Evaluation of a rapid molecular drug-susceptibility test for tuberculosis. N. Engl. J. Med. 2017, 377, 1043–1054. [Google Scholar] [CrossRef]
- McGrath, M.; van Pittius, G.N.C.; Van Helden, P.D.; Warren, R.M.; Warner, D.F. Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2014, 69, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Hu, Q.; Zhang, H.; Xu, B. Overview of the epidemic of drug-resistant tuberculosis. Chin. J. Antituberc. 2010, 32, 351–353. [Google Scholar]
- Boehme, C.C.; Nicol, M.P.; Nabeta, P.; Michael, J.S.; Gotuzzo, E.; Tahirli, R.; Gle, R.M.T.; Blakemore, R.; Worodria, W.; Gray, C.; et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: A multicentre implementation study. Lancet 2011, 377, 1495–1505. [Google Scholar] [CrossRef]
- Hameed, H.M.A.; Islam, M.M.; Chhotaray, C.; Wang, C.; Liu, Y.; Tan, Y.; Li, X.; Tan, S.; Delorme, V.; Yew, W.W.; et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis strains. Front. Cell. Infect. Microbiol. 2018, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.V.; Reich, R.; Dou, S.J.; Jasperse, L.; Pan, X.; Wanger, A.; Quitugua, T.; Graviss, E.A. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 1241–1250. [Google Scholar] [CrossRef]
- Hazbón, M.H.; Brimacombe, M.; Bobadilla, d.V.M.; Cavatore, M.; Guerrero, M.I.; Varma-Basil, M.; Billman-Jacobe, H.; Lavender, C.; Fyfe, J.; García-García, L.; et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2006, 50, 2640–2649. [Google Scholar] [CrossRef]
- Zhao, L.L.; Sun, Q.; Liu, H.C.; Wu, X.C.; Xiao, T.Y.; Zhao, X.; Li, G.L.; Jiang, Y.; Zeng, C.Y.; Wan, K.L. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob. Agents Chemother. 2015, 59, 2045–2050. [Google Scholar] [CrossRef]
- Mohammadi, B.; Ramazanzadeh, R.; Nouri, B.; Rouhi, S. Frequency of codon 306 mutations in embB gene of Mycobacterium tuberculosis resistant to ethambutol: A systematic review and meta-analysis. Int. J. Prev. Med. 2020, 11, 112. [Google Scholar]
- Jagielski, T.; Ignatowska, H.; Bakuła, Z.; Dziewit, Ł.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Zwolska, Z.; Bielecki, J. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE 2014, 9, e100078. [Google Scholar] [CrossRef]
- Wan, L.; Liu, H.; Li, M.; Jiang, Y.; Zhao, X.; Liu, Z.; Wan, K.; Li, G.; Guan, C.X. Genomic analysis identifies mutations concerning drug-resistance and Beijing genotype in multidrug-resistant Mycobacterium tuberculosis isolated from China. Front. Microbiol. 2020, 11, 1444. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, J.; Lu, J.; Huang, X.; Hu, Z. Association of mutation patterns in gyrA/B genes and ofloxacin resistance levels in Mycobacterium tuberculosis isolates from East China in 2009. BMC Infect. Dis. 2011, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines used in the Treatment of Drug-Resistant Tuberculosis; World Health Organization: Geneva, Switzerland, 2018; Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- John, J.; Richard, E.; Alexander, P.; Tim, G.; Michael, F.; Olaf, R.; Kathryn, T.; Russ, B.; Augustin, Ž.; Anna, P.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucl. Acids Res. 2021, 49, 458–460. [Google Scholar] [CrossRef]
- Pang, Y.; Lu, J.; Wang, Y.; Song, Y.; Wang, S.; Zhao, Y. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2013, 57, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Koga, H.; Kohno, S.; Tashiro, T.; Hara, K. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob. Agents. Chemother. 1996, 40, 1053–1056. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Musser, J.M. Molecular genetic basis of anti-microbial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 1998, 79, 3–29. [Google Scholar] [CrossRef]
- Jagielski, T.; Bakuła, Z.; Brzostek, A.; Minias, A.; Stachowiak, R.; Kalita, J.; Napiórkowska, A.; Augustynowicz-Kopeć, E.; Żaczek, A.; Vasiliauskiene, E.; et al. Characterization of mutations conferring resistance to rifampin in Mycobacterium tuberculosis clinical strains. Antimicrob. Agents Chemother. 2018, 62, e01093-18. [Google Scholar] [CrossRef] [PubMed]
- Kozhamkulov, U.; Akhmetova, A.; Rakhimova, S.; Belova, E.; Alenova, A.; Bismilda, V.; Chingissova, L.; Ismailov, S.; Ramanculov, E.; Momynaliev, K. Molecular characterization of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Kazakhstan. Jpn. J. Infect. Dis. 2011, 64, 253–255. [Google Scholar]
- Valim, A.R.; Rossetti, M.L.; Ribeiro, M.O.; Zaha, A. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J. Clin. Microbiol. 2000, 38, 3119–3122. [Google Scholar] [CrossRef] [PubMed]
- Yuen, L.K.; Leslie, D.; Coloe, P.J. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J. Clin. Microbiol. 1999, 37, 3844–3850. [Google Scholar] [CrossRef] [PubMed]
- Afanas’ev, M.V.; Ikryannikova, L.N.; Il’ina, E.N.; Sidorenko, S.V.; Kuz’min, A.V.; Larionova, E.E.; Smirnova, T.G.; Chernousova, L.N.; Kamaev, E.Y.; Skorniakov, S.N.; et al. Molecular characteristics of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis isolates from the Russian Federation. J. Antimicrob. Chemother. 2007, 59, 1057–1064. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bártfai, Z.; Somoskövi, A.; Ködmön, C.; Szabó, N.; Puskás, E.; Kosztolányi, L.; Faragó, E.; Mester, J.; Parsons, L.M.; Salfinger, M. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 2001, 39, 3736–3739. [Google Scholar] [CrossRef]

| Isolate Name | INH | RIF | EMB | OFX | STR | |
|---|---|---|---|---|---|---|
| Non-XDR | DR | 108 | 108 | 209 | 271 | 157 |
| DR and MM(+) | 102 | 82 | 161 | 180 | 117 | |
| preXDR/XDR | DR | 46 | 46 | 32 | 25 | 33 |
| DR and MM(+) | 42 | 40 | 26 | 22 | 27 | |
| Linked Missense Mutations | Incidence Rate % (No./Total AM No.*) | p Value | |
|---|---|---|---|
| Non-XDR | preXDR/XDR | ||
| Rifampicin | 6.9% (7/102) | 2.4% (1/42) | 0.505 |
| rpoB 511 + rpoB 516 | 3.9% (4/102) | / | / |
| rpoB 511 + rpoB 526 | 1.0% (1/102) | / | / |
| rpoB 516 + rpoB 526 | 1.0% (1/102) | / | / |
| rpoB 526 + rpoB 531 | 1.0% (1/102) | / | / |
| rpoB 516 + rpoB 531 | / | 2.4% (1/42) | / |
| Isoniazid | 4.9% (4/82) | 7.5% (3/40) | 0.891 |
| katG 315 + inhA | 4.9% (4/82) | 7.5% (3/40) | 0.891 |
| Ethambutol | 6.2% (10/161) | 3.8% (1/26) | 1.000 |
| embB 306 + embB 328 | 1.2% (2/161) | / | / |
| embB 306 + embB 354 | 1.2% (2/161) | / | / |
| embB 306 + embB 406 | 3.1% (5/161) | 3.8% (1/26) | 1.000 |
| embB 306 + embB 497 | 0.6% (1/161) | / | / |
| Ofloxacin | 26.7% (48/180) | 9.1% (2/22) | 0.072 |
| gyrA 90 + gyrA 91 | 5.0% (9/180) | 4.5% (1/22) | 1.000 |
| gyrA 90 + gyrA 94 | 21.7% (39/180) | 4.5% (1/22) | 0.053 |
| Isolate No. | INH | RIF | EMB | OFX | STR | |
|---|---|---|---|---|---|---|
| Total DR | DR and MM(+) | 40 | 42 | 26 | 22 | 27 |
| DR and MM(−) | 6 | 4 | 6 | 3 | 6 | |
| Total no. | 46 | 46 | 32 | 25 | 33 | |
| Total DS | DS and MM(+) | 0 | 0 | 7 | 11 | 2 |
| DS and MM(−) | 0 | 0 | 7 | 10 | 11 | |
| Total no. | 0 | 0 | 14 | 21 | 13 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, H.; Xu, Y.; Sun, Z. Analysis on Drug-Resistance-Associated Mutations among Multidrug-Resistant Mycobacterium tuberculosis Isolates in China. Antibiotics 2021, 10, 1367. https://doi.org/10.3390/antibiotics10111367
Jia H, Xu Y, Sun Z. Analysis on Drug-Resistance-Associated Mutations among Multidrug-Resistant Mycobacterium tuberculosis Isolates in China. Antibiotics. 2021; 10(11):1367. https://doi.org/10.3390/antibiotics10111367
Chicago/Turabian StyleJia, Hongbing, Yuhui Xu, and Zhaogang Sun. 2021. "Analysis on Drug-Resistance-Associated Mutations among Multidrug-Resistant Mycobacterium tuberculosis Isolates in China" Antibiotics 10, no. 11: 1367. https://doi.org/10.3390/antibiotics10111367
APA StyleJia, H., Xu, Y., & Sun, Z. (2021). Analysis on Drug-Resistance-Associated Mutations among Multidrug-Resistant Mycobacterium tuberculosis Isolates in China. Antibiotics, 10(11), 1367. https://doi.org/10.3390/antibiotics10111367





