Genomic Characterization of a Proteus sp. Strain of Animal Origin Co-Carrying blaNDM-1 and lnu(G)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Genetic Characterization of the blaNDM-1-Harboring Proteus sp. NMG38-2
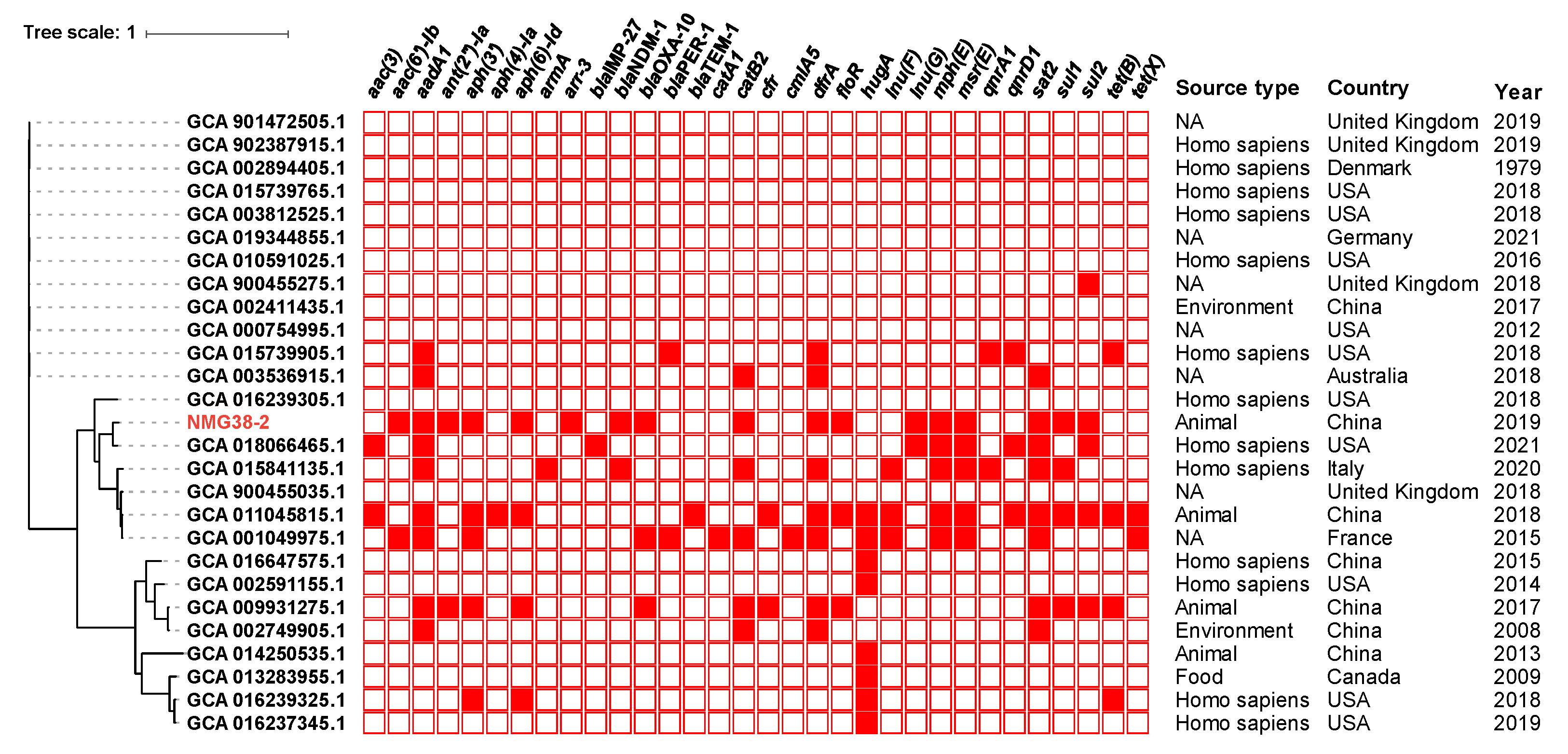
2.2. Phylogenetic Analysis of Proteus sp. NMG38-2
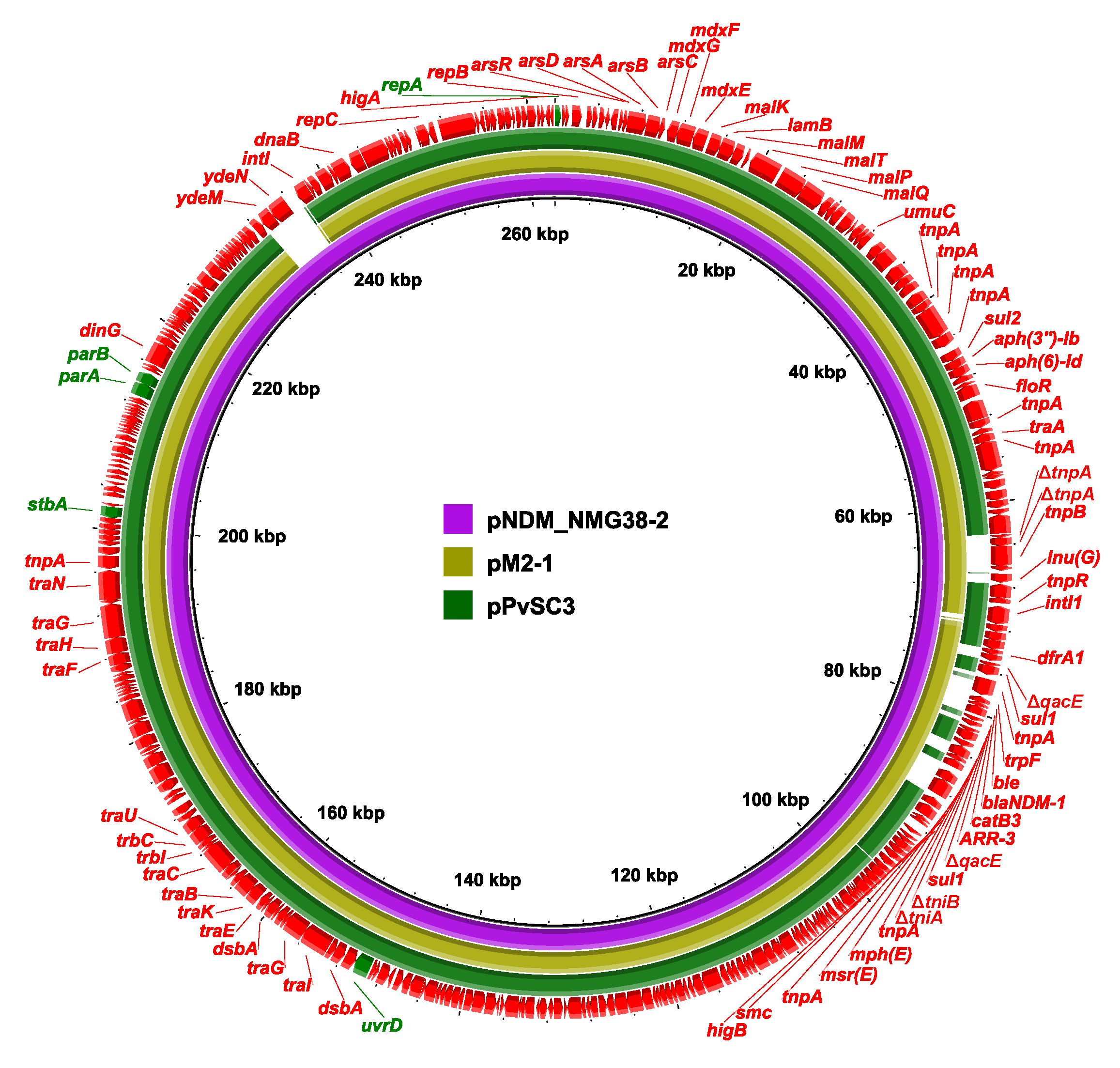
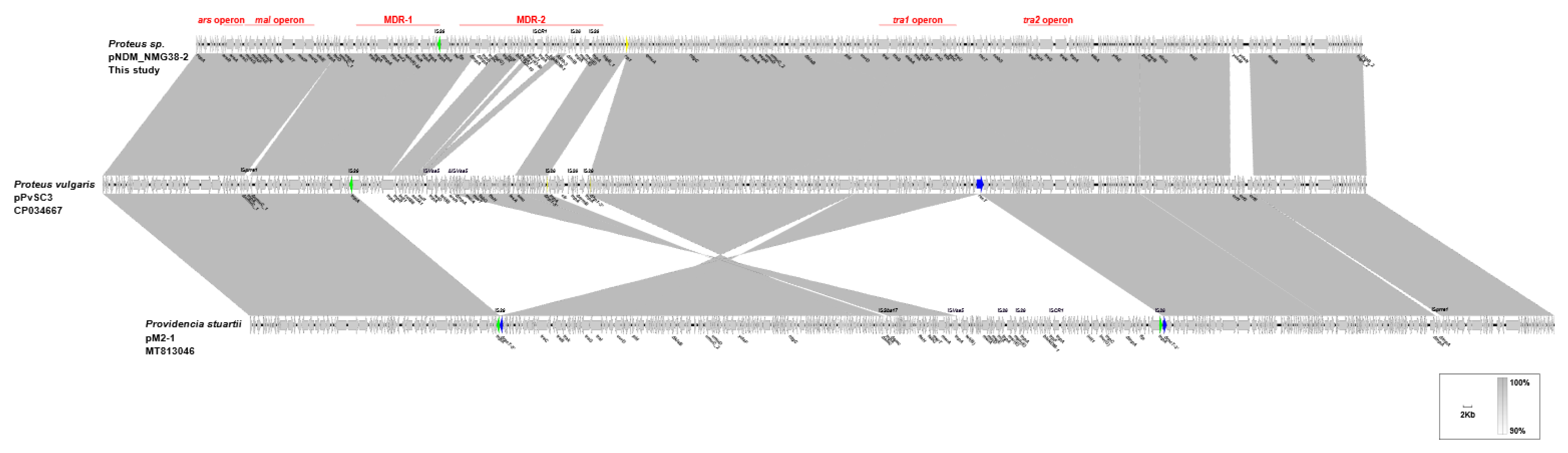
2.3. Genetic Structure of pPvSC3-like Plasmids
2.4. Transfer Ability of pNDM_NMG38-2 and Fitness Cost
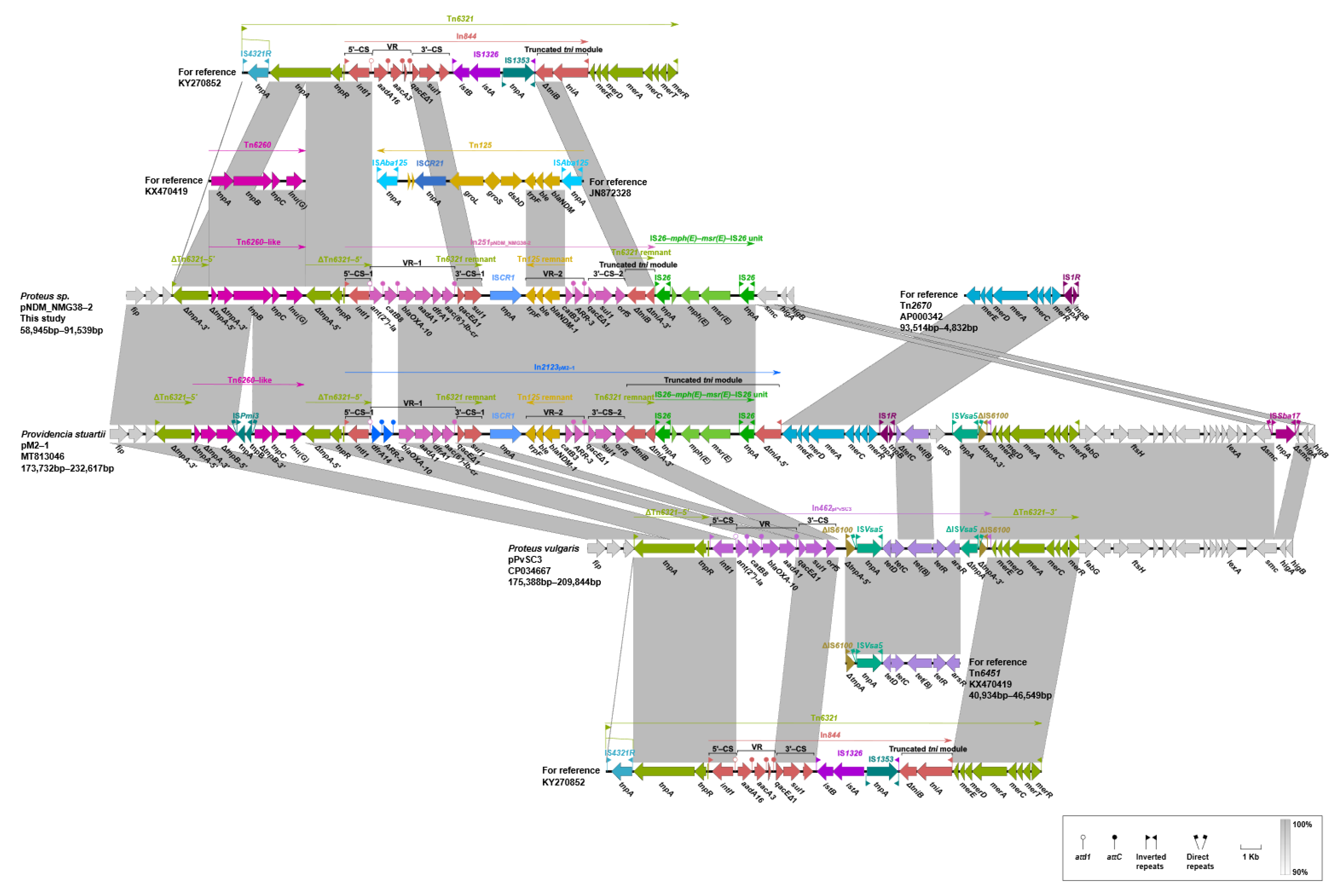
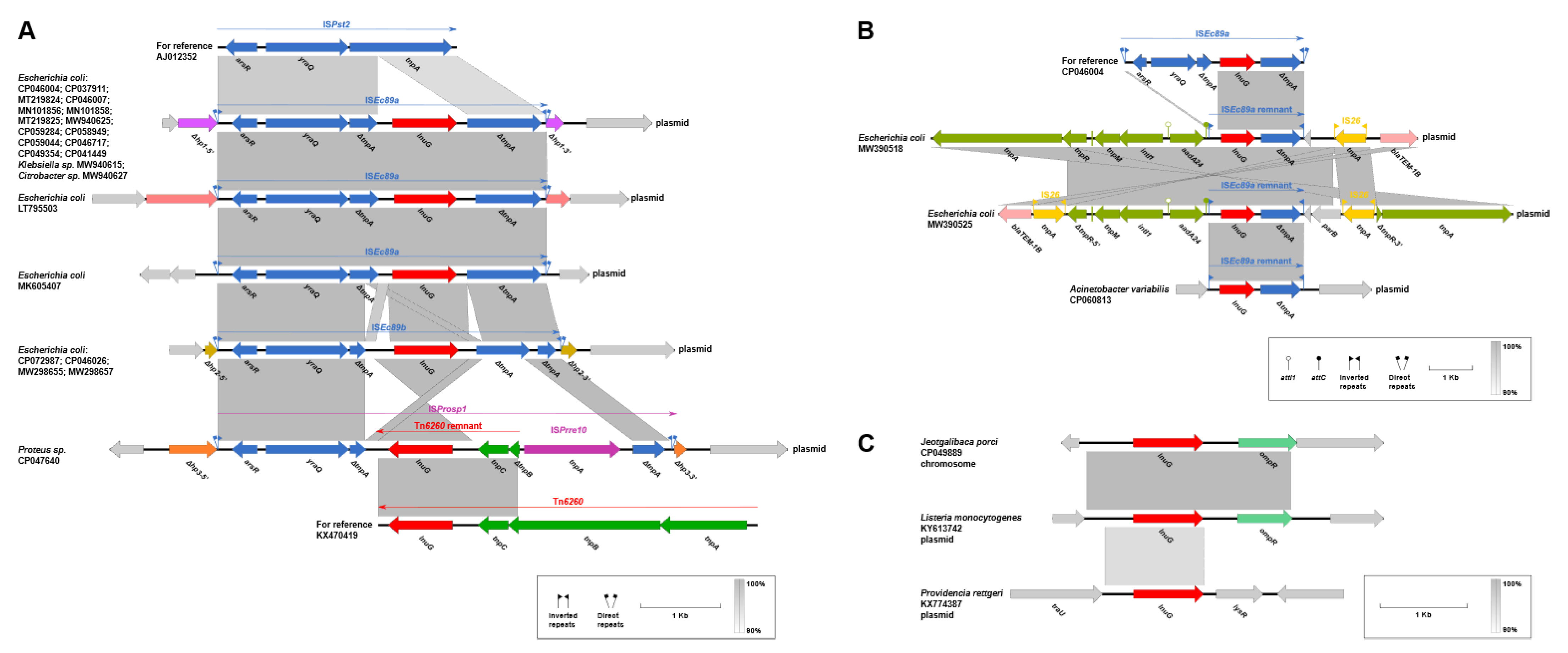
2.5. Genetic Contexts of lnu(G)
3. Materials and Methods
3.1. Bacterial Strain and In Vitro Susceptibility Testing
3.2. Conjugation Assay
3.3. In Vitro Growth Assays
3.4. Genome Sequencing and Data Analyses
3.5. Phylogenetic Analysis
3.6. Comparative Analysis of lnu(G)-Harboring Genome Sequences
3.7. Nucleotide Sequence Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar Trivedi, M. Phenotyping and genotyping characterization of Proteus vulgaris after biofield treatment. Int. J. Genet. Genom. 2015, 3, 66. [Google Scholar] [CrossRef]
- O’Hara, C.M.; Brenner, F.W.; Miller, J.M. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 2000, 13, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115–e00118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Liu, L.; McNally, A.; Zong, Z. Coexistence of two blaNDM-5 genes on an IncF plasmid as revealed by Nanopore sequencing. Antimicrob. Agents Chemother. 2018, 62, e00110-18. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Sekizuka, T.; Matsui, M.; Yamane, K.; Takeuchi, F.; Ohnishi, M.; Hishinuma, A.; Arakawa, Y.; Kuroda, M. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE 2011, 6, e25334. [Google Scholar] [CrossRef]
- Wailan, A.M.; Sidjabat, H.E.; Yam, W.K.; Alikhan, N.F.; Petty, N.K.; Sartor, A.L.; Williamson, D.A.; Forde, B.M.; Schembri, M.A.; Beatson, S.A.; et al. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob. Agents Chemother. 2016, 60, 4082–4088. [Google Scholar] [CrossRef] [Green Version]
- Le-Vo, H.N.; Tran, P.T.; Le, L.; Matsumoto, Y.; Motooka, D.; Nakamura, S.; Jones, J.W.; Iida, T.; Cao, V. Complex class 1 integron in a clinical Escherichia coli strain from Vietnam carrying both mcr-1 and blaNDM-1. Front. Microbiol. 2019, 10, 2472. [Google Scholar] [CrossRef]
- Kong, L.-H.; Xiang, R.; Wang, Y.-L.; Wu, S.-K.; Lei, C.-W.; Kang, Z.-Z.; Chen, Y.-P.; Ye, X.-L.; Lai, Y.; Wang, H.-N. Integration of the blaNDM-1 carbapenemase gene into a novel SXT/R391 integrative and conjugative element in Proteus vulgaris. J. Antimicrob. Chemother. 2020, 75, 1439–1442. [Google Scholar] [CrossRef]
- He, J.; Sun, L.; Zhang, L.; Leptihn, S.; Yu, Y.; Hua, X. A novel SXT/R391 integrative and conjugative element carries two copies of the blaNDM-1 gene in Proteus mirabilis. mSphere 2021, 6, e0058821. [Google Scholar] [CrossRef]
- Dong, D.; Li, M.; Liu, Z.; Feng, J.; Jia, N.; Zhao, H.; Zhao, B.; Zhou, T.; Zhang, X.; Tong, Y.; et al. Characterization of a NDM-1- encoding plasmid pHFK418-NDM from a clinical Proteus mirabilis isolate harboring two novel transposons, Tn6624 and Tn6625. Front. Microbiol. 2019, 10, 2030. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, Q.; Peng, K.; Liu, Y.; Li, R.; Wang, Z. Emergence of carbapenem- and tigecycline-resistant Proteus cibarius of animal origin. Front. Microbiol. 2020, 11, 1940. [Google Scholar] [CrossRef] [PubMed]
- Spizek, J.; Rezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 2017, 133, 20–28. [Google Scholar] [CrossRef]
- Zhu, X.-Q.; Wang, X.-M.; Li, H.; Shang, Y.-H.; Pan, Y.-S.; Wu, C.-M.; Wang, Y.; Du, X.-D.; Shen, J.-Z. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from Enterococcus faecalis E531. J. Antimicrob. Chemother. 2017, 72, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Spizek, J.; Novotna, J.; Rezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Adv. Appl. Microbiol. 2004, 56, 121–154. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brisson-Noel, A.; Delrieu, P.; Samain, D.; Courvalin, P. Inactivation of lincosaminide antibiotics in Staphylococcus. Identification of lincosaminide O-nucleotidyltransferases and comparison of the corresponding resistance genes. J. Biol. Chem. 1988, 263, 15880–15887. [Google Scholar] [CrossRef]
- Bozdogan, B.; Berrezouga, L.; Kuo, M.S.; Yurek, D.A.; Farley, K.A.; Stockman, B.J.; Leclercq, R. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 1999, 43, 925–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achard, A.; Villers, C.; Pichereau, V.; Leclercq, R. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 2005, 49, 2716–2719. [Google Scholar] [CrossRef] [Green Version]
- Petinaki, E.; Guerin-Faublee, V.; Pichereau, V.; Villers, C.; Achard, A.; Malbruny, B.; Leclercq, R. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob. Agents Chemother. 2008, 52, 626–630. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Wendlandt, S.; Li, H.; Li, J.; Wu, C.; Shen, J.; Schwarz, S.; Wang, Y. Identification of the novel lincosamide resistance gene lnu(E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: De novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 1785–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cen, D.-J.; Sun, R.-Y.; Mai, J.-L.; Jiang, Y.-W.; Wang, D.; Guo, W.-Y.; Jiang, Q.; Zhang, H.; Zhang, J.-F.; Zhang, R.-M.; et al. Occurrence and transmission of blaNDM -carrying Enterobacteriaceae from geese and the surrounding environment on a commercial goose farm. Appl. Environ. Microb. 2021, 87, e00087-21. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Karageorgopoulos, D.E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: Need for international harmonization in terminology. Clin. Infect. Dis. 2008, 46, 1121–1122, author reply 1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Zhang, Y.; Lei, C.W.; Wang, H.N. Identification of a novel conjugative plasmid carrying the multiresistance gene cfr in Proteus vulgaris isolated from swine origin in China. Plasmid 2019, 105, 102440. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006, 70, 296–316. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.; Klima, C.L.; Ha, R.; Gruninger, R.J.; Zaheer, R.; McAllister, T.A. A novel aadA aminoglycoside resistance gene in bovine and porcine pathogens. mSphere 2018, 3, e00568-17. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ma, X.; Li, C.; Dai, X.; Zhang, L. Occurrence and genomic characterization of ESBL-producing Escherichia coli ST29 strains from swine with abundant virulence genes. Microb. Pathog. 2020, 148, 104483. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, Y.; Wang, G.; Li, C.; Chang, Y.F.; Chen, W.; Nian, S.; Mao, Y.; Zhang, J.; Zhong, F.; et al. blaNDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob. Resist. Infect. Control 2019, 8, 140. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Li, Y.; Luo, L.; Xiao, Z.; Wang, G.; Li, C.; Zhang, Z.; Zhou, Y.; Zhang, L. Characterization of a carbapenem-resistant Kluyvera Cryocrescens isolate carrying blaNDM-1 from hospital sewage. Antibiotics 2019, 8, 149. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, A.; Soares, M.; Pereira, C.; Leitao, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009, 25, 1096–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strains a | Species’ Name | MIC (μg/mL) b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | CST | MEM | IMP | CFT | AZT | CIP | CTX | CLN | CHL | ||
| NMG38-2 | Proteus sp. | 128 | 256 | 512 | 512 | >512 | 512 | 256 | ≤0.5 | 256 | >512 | 256 |
| JM-1 | E. coli | 64 | 32 | 256 | 512 | >512 | 512 | 256 | 1 | 256 | >512 | 256 |
| JM-2 | E. coli | 64 | 32 | 512 | 512 | >512 | 512 | 256 | 1 | 512 | 512 | 256 |
| JM-3 | E. coli | 64 | 64 | 256 | 512 | >512 | 512 | 128 | 2 | 256 | 512 | 256 |
| J53 | E. coli | 16 | 2 | ≤0.5 | ≤0.5 | ≤0.5 | 16 | 8 | ≤0.5 | ≤0.5 | 256 | 8 |
| ATCC25922 | E. coli | 4 | 4 | ≤0.5 | ≤0.5 | ≤0.5 | 8 | 2 | ≤0.5 | ≤0.5 | 256 | 16 |
| Chromosome /Plasmid | Length (bp) | GC% | No. of Predicted Open Reading Frames | Inc Type | Drug Resistance Gene |
|---|---|---|---|---|---|
| Chromosome | 4,433,346 | 37.62 | 4231 | - | aadA1, dfrA1, catB2 |
| pNDM_NMG38-2 | 262,556 | 46.65 | 369 | UT | aac(6′)-Ib-cr, ant(2″)-Ia, aph(6)-Id, aac(6′)-Ib3, aph(3″)-Ib, aac(6′)-Ib-cr, aadA1, ARR-3, sul1 a, dfrA1, sul2, catB8, floR, catB3, blaOXA-10, blaNDM-1, mph(E), msr(E), lnu(G) |
| P1_SCLZS62 | 5500 | 34.11 | 10 | UT | |
| P2_SCLZS62 | 2683 | 41.78 | 2 | Col3M | qnrD1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Qiu, Y.; She, J.; Wang, X.; Dai, X.; Zhang, L. Genomic Characterization of a Proteus sp. Strain of Animal Origin Co-Carrying blaNDM-1 and lnu(G). Antibiotics 2021, 10, 1411. https://doi.org/10.3390/antibiotics10111411
Li Y, Qiu Y, She J, Wang X, Dai X, Zhang L. Genomic Characterization of a Proteus sp. Strain of Animal Origin Co-Carrying blaNDM-1 and lnu(G). Antibiotics. 2021; 10(11):1411. https://doi.org/10.3390/antibiotics10111411
Chicago/Turabian StyleLi, Ying, Yichuan Qiu, Junping She, Xu Wang, Xiaoyi Dai, and Luhua Zhang. 2021. "Genomic Characterization of a Proteus sp. Strain of Animal Origin Co-Carrying blaNDM-1 and lnu(G)" Antibiotics 10, no. 11: 1411. https://doi.org/10.3390/antibiotics10111411
APA StyleLi, Y., Qiu, Y., She, J., Wang, X., Dai, X., & Zhang, L. (2021). Genomic Characterization of a Proteus sp. Strain of Animal Origin Co-Carrying blaNDM-1 and lnu(G). Antibiotics, 10(11), 1411. https://doi.org/10.3390/antibiotics10111411






