Abstract
Tuberculosis, caused by the pathogen Mycobacterium tuberculosis, is a serious infectious disease worldwide. Multidrug-resistant TB (MDR-TB) remains a global problem, and the understanding of this resistance is incomplete. Studies suggested that DNA methylation promotes bacterial adaptability to antibiotic treatment, but the role of mycobacterial HsdM in drug susceptibility has not been explored. Here, we constructed an inactivated Mycobacterium bovis (BCG) strain, ΔhsdM. ΔhsdM shows growth advantages over wild-type BCG under isoniazid treatment and hypoxia-induced stress. Using high-precision PacBio single-molecule real-time sequencing to compare the ΔhsdM and BCG methylomes, we identified 219 methylated HsdM substrates. Bioinformatics analysis showed that most HsdM-modified genes were enriched in respiration- and energy-related pathways. qPCR showed that HsdM-modified genes directly affected their own transcription, indicating an altered redox regulation. The use of the latent Wayne model revealed that ΔhsdM had growth advantages over wild-type BCG and that HsdM regulated trcR mRNA levels, which may be crucial in regulating transition from latency to reactivation. We found that HsdM regulated corresponding transcription levels via gene methylation; thus, altering the mycobacterial redox status and decreasing the bacterial susceptibility to isoniazid, which is closely correlated with the redox status. Our results provide valuable insight into DNA methylation on drug susceptibility.
1. Introduction
Tuberculosis (TB) is a chronic wasting infectious disease that causes infections of various tissues and organs after Mycobacterium tuberculosis (Mtb) is inhaled, and then spreads through the respiratory tract. According to the Global Tuberculosis Report issued by the World Health Organization in 2019 [], the number of TB-infected patients has been relatively stable in recent years, with approximately 10 million new cases annually worldwide. In 2018, TB caused approximately 1.5 million deaths and is among the top ten causes of human deaths. TB causes most infection-related deaths primarily because of antimicrobial resistance. Understanding how Mtb mutates and evolves in the host to produce drug resistance will provide theoretical guidance for improving TB treatment and control strategies. Mtb is genetically static, with a low mutation rate, and evolves via single nucleotide polymorphisms [,]. However, under continued chemotherapy, a fraction of Mtb eventually develops complete antibiotic resistance []. It remains unclear how this genetically stable organism adapts so quickly to antibiotic treatments and infected host’s immune pressures such as reactive oxygen species, which are products of the host’s innate immune responses induced by infected Mtb [].
Recent studies have shown that bacterial epigenomics is an important and newly rising field for genetic and phenotypic analysis of microbial diversity, gene regulation and evolution. Modifying DNA to regulate gene expression facilitates physiological adaptation to new environments without needing to drastically alter the genome. Previous studies of bacterial methylation have shown that DNA methylation promotes bacterial adaptability in infected hosts [,] and to antibiotic treatments []. These studies suggested that environmental changes (such as pathogens infecting the host) lead to selection of a well-adjusted subgroup. That is, this phenotypic plasticity helps bacteria quickly adapt to changing environmental pressures. Studies have shown that the different evolutionary lineages of Mtb come from different geographical regions [], and genetic variations in these TB subgroups affect the evolution of drug resistance [].
We speculate that DNA methylation is a feasible but poorly understood mechanism of Mtb phenotypic variation. Mtb has three DNA methyltransferases (MTases), MamA, MamB and HsdM, which target different DNA sequence motifs of N6-adenine methylation [,,]. The bacterial methylome has been linked to antibiotic stress survival [], indicating that DNA methylation may play important roles in antibiotic susceptibility. Most methylation studies on Mtb have been conducted on strains collected from clinical samples, and the MTases in these strains have complex backgrounds with varying numbers of MTases and different degrees of mutated sites; thus, conclusions from these studies are conflicting [,].
Here, we characterize the biological function of an orphan MTase, HsdM, in Mycobacterium bovis BCG Pasteur (BCG). Using specialized transduction, we constructed ΔhsdM and showed that ΔhsdM decreased the bacterial susceptibility to isoniazid (INH) compared with that of its parental strain, BCG. We used high-precision PacBio single-molecule real-time (SMRT) sequencing technology to compare the ΔhsdM and BCG methylomes. We identified 219 methylated HsdM substrates, of which, 192 were located in open reading frames (ORFs), and 28 were located upstream of ORFs. Bioinformatics analysis showed that most HsdM-modified genes were enriched in respiration- and energy-related pathways. qPCR showed that HsdM modification regulated the transcription level of its modified DNA, suggesting that HsdM-mediated changes in mRNA levels may be related to redox regulation. Using the latent Wayne model, we show that ΔhsdM had a growth advantage over wild-type BCG under hypoxic conditions and that HsdM regulated trcR mRNA levels, which was predicted to be a key regulator of transition from latency to reactivation. Our results showed that HsdM regulated the corresponding transcription levels via gene methylation, leading to changes in the mycobacterial redox status, thereby affecting mycobacterial susceptibility to INH, which was closely correlated with the mycobacterial redox status.
2. Results
2.1. DNA Methyltransferase HsdM Decreased INH Susceptibility in Mycobacteria
Previous studies have shown that MTases mediate various functions, including antibiotic stress responses [,,,]. Mycobacterial DNA methylation is complex. Mtb has three known MTases, MamA (Rv3263), HsdM (Rv2756c) and MamB (Rv2024c), which modify distinct DNA sequence motifs [,]. Mycobacteria from different lineages contain different numbers of DNA MTases from different sources with different mutation sites [,,]. These complexities make it difficult to identify the functions of these MTases. The clinical isolates containing different numbers of MTases and different degrees of mutated sites [,] are difficult to explore the biological functions of HsdM. Here, we used M. bovis BCG Pasteur (BCG) to assess how HsdM functioned in antibiotic susceptibility. HsdM in BCG is similar to HsdM in Mtb H37Rv. BCG_2772c from BCG is equivalent to Rv2756c from M. tuberculosis strain H37Rv with 99.8% identity (Mycobrowser website https://mycobrowser.epfl.ch/genes (accessed on 12 August 2021) and KEGG website https://www.kegg.jp (accessed on 12 August 2021)). Additionally, BCG contains three intact DNA MTases. We examined whether HsdM affected DNA methylation via MamA and MamB.
First, we constructed an hsdM knockout in BCG using specialized transduction []. We cloned approximately 1 kb fragments both upstream and downstream of hsdM via PCR and constructed an allelic exchange vector on phAE159 [,]. Colonies carrying hyg to replace hsdM were confirmed via PCR (Figure 1A,B). The corresponding complementary strain pMV361-hsdM/ΔhsdM was constructed. The in vitro growth kinetics of ΔhsdM in Middlebrook 7H9 supplemented with albumin–dextrose–saline (ADS) were comparable to those of wild-type BCG and the complementary strain for up to 14 days of incubation (Figure 1C). No differences were observed throughout the entire growth process, indicating that HsdM did not affect mycobacterial growth in vitro in ADS-supplemented Middlebrook 7H9.
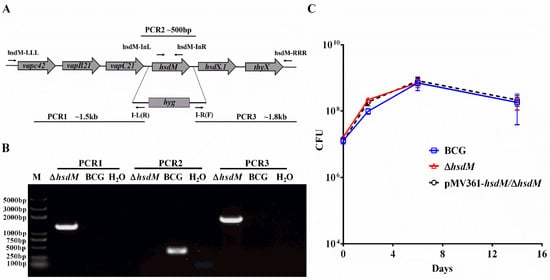
Figure 1.
HsdM did not impact mycobacterial growth in vitro. (A) Genomic organization of the hsdM gene locus. Large arrows represent coding genes in their orientation. Small arrows represent the primer pair used for corresponding PCR. (B) PCR to confirm the knockout hsdM (ΔhsdM) strains. The replacement gene, hygromycin-coding gene (hyg), is also shown. The PCR products of the upstream (PCR1) and downstream (PCR3) regions of hsdM using the primers pairs LLL/I-L(R) and I-R(F)/RRR, respectively. The PCR product of the gene fragment in hsdM using primers hsdM-InL and hsdM-InR (PCR2). (C) Growth curves of BCG, ΔhsdM and pMV361-hsdM/ΔhsdM strains in 7H9 medium. Data presented the means ± standard deviations (SD) from three independent experiments.
Next, we selected antibiotics used for the clinical treatment of TB, including isoniazid (INH), rifampicin (RFP), streptomycin (STR), ethambutol (EMB), ciprofloxacin (CIP) and ofloxacin (OFX), compared the susceptibility of ΔhsdM and wild-type BCG (Figure 2). Strikingly, ΔhsdM showed a marked resistance to INH compared with that of BCG (Figure 2A). The minimum inhibitory concentration (MIC) of ΔhsdM to INH was ~32-fold higher than that of the wild-type BCG exposed to INH. No differences in antibiotic susceptibility between ΔhsdM and wild-type BCG were detected in any other antibiotics (Figure 2B–F). We, at first, performed antibiotic susceptibility testing of three mycobacterial strains (BCG, ΔhsdM and pMV361-hsdM/ΔhsdM) treated with INH at the indicated concentrations and showed the growth advantage of ΔhsdM was partly reduced by complementation with an integrated copy of hsdM, pMV361-hsdM/ΔhsdM (Supplementary Material Figure S1). To further confirm the effect of HsdM on INH susceptibility, we performed the drug exposure experiments in the presence of 0.25 mg/L INH (8 × MIC of INH) to compare the growth rates among strains BCG, ΔhsdM and pMV361-hsdM/ΔhsdM. Consistent to previous studies [,,], we observed the typical biphasic killing curve of these three mycobacterial strains (Figure 2G). All three of these strains showed typical INH-killing growth curves after INH treatment; that is, the INH treatment arrested the mycobacterial growth for up to 6 days. After reaching the lowest point, the mycobacterial tolerance to INH began to increase (Figure 2G). On days 2–3 after INH treatment, ΔhsdM exhibited a significant growth advantage (Figure 2H), which was partly reduced by complementation with an integrated copy of hsdM constitutively expressed and driven by the mycobacterial promoter, pMV361-hsdM/ΔhsdM. Thus, HsdM affected the mycobacterial susceptibility to INH likely because HsdM methylation-modified DNA affected transcription, which in turn affected INH susceptibility.
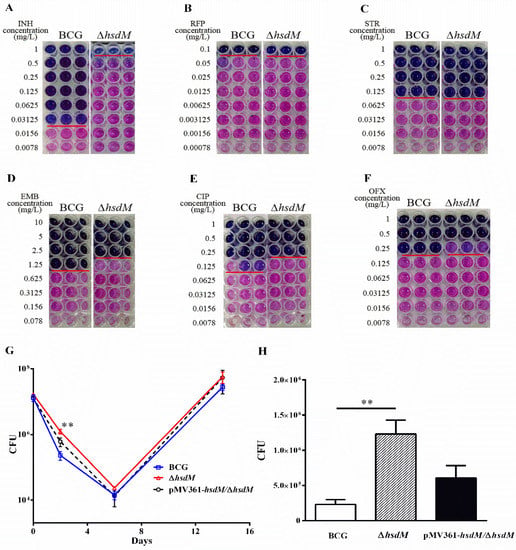
Figure 2.
HsdM affected drug susceptibility in BCG. (A–F) MICs of INH, RFP, EMB, STR, OFX and CIP on microplates in BCG and ΔhsdM. Blue wells indicate no growth owing to drug inhibition; pink wells indicate growth. (G) Growth curves of BCG, ΔhsdM and pMV361-hsdM/ΔhsdM strains treated with 0.25 mg/L INH. (H) The number of surviving cells was examined by monitoring the colony-forming units (CFUs) for 2 days. Data are presented as the mean ± standard deviation of three independent replicates. The figure presents the results of three biological replicates. ** p < 0.01.
2.2. Bioinformatic Analysis of the HsdM Substrate via Whole-Genome Sequencing
To verify our hypothesis that HsdM affected susceptibility to INH via its methylated DNA, the three mycobacterial strains BCG, ΔhsdM and pMV361-hsdM/ΔhsdM were sequenced via PacBio SMRT sequencing [], which enables directly detecting methylated DNA at the genomic level. A previous study showed that the GATC methylome was stable, and drug treatment did not affect the adenine methylome []; thus, we did not examine the mycobacterial methylome after drug treatment. Three strains were cultured on 7H10 plates supplemented with ADS for 10 days, and all colonies growing on the plates were collected. Genomic DNA was extracted and sequenced using PacBio SMRT sequencing. The average sequencing coverages of BCG, ΔhsdM and pMV361-hsdM/ΔhsdM were approximately 133.79×, 156.51× and 219.69×, respectively. Similar to a previous study [], the sequencing results provided general bioinformation, including the respective genomic sizes (~4.345–4.352 Mb), the number of ORFs (~4069–4102) and the GC% (65.6%). Adenine MTases have been shown to target distinctive DNA sequence motifs, of which, MamA, MamB and HsdM target the corresponding motifs CTCCAG, CACGCAG and GATN4RTAC, respectively [,]. The percentage of GATN4RTAC sites methylated by HsdM in BCG was 72.5%; no methylated GATN4RTAC sites were detected in ΔhsdM (Table 1). The percentage of the methylated GATN4RTAC sites was 58.7% in the complementary pMV361-hsdM/ΔhsdM strain, indicating that a single integrated copy of hsdM expressed from a mycobacterial promoter could at least partially complement the methylation function of HsdM. Most of the CTCCAG (99.3%) and CACGCAG (99.8%) were methylated by the corresponding MamA and MamB in BCG. Additionally, the motifs of both MamA and MamB were almost fully methylated (99.1% and 100%, respectively) in ΔhsdM. Similar to the methylation profiles modified by MamA and MamB in BCG and ΔhsdM, approximately 99–100% of the methylated motifs were modified in pMV361-hsdM/ΔhsdM. Hence, HsdM did not affect DNA methylation via MamA and MamB.

Table 1.
N6-methyl-adenine base modifications in sequenced BCG strains.
The SMRT sequencing results for BCG, ΔhsdM and pMV361-hsdM/ΔhsdM revealed 489 methylated sites, of which 430 were located in the 192 ORFs, and 59 were located in noncoding gene regions, including 28 downstream genes (Supplementary Material Tables S1 and S2). One substrate, Rv1522c, had modification sites both in and upstream of the gene.
We, then, investigated the Clusters of Orthologous Groups (COG) functional category of those substrates. The methylated substrates of HsdM included 219 genes (Tables S1 and S2), of which, 158 were successfully annotated by COG and fell into 19 classification categories, including replication, recombination and repair, transcription, translation, ribosomal structure and biogenesis, lipid transport and metabolism and carbohydrate transport metabolism (Figure 3). Manually searching the biofunctions of these substrates revealed that 56 methylated substrates of HsdM (~25%) were involved in the respiration pathway (Table S3), and the bioinformatic analysis implied that HsdM-mediated modification was linked to cellular redox regulation or energy metabolism. Previous studies have shown that bacterial antibiotic susceptibility is associated with the redox status [,]. Therefore, after combining the experimental results and bioinformatics analysis of the HsdM substrates, we speculated that methylation by HsdM affects the transcription level of the corresponding substrates; thus, affecting the redox state of the cell and affecting its susceptibility to INH, a prodrug with redox-mediated activation [,].
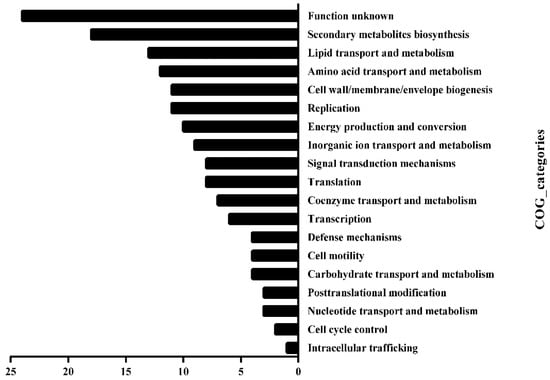
Figure 3.
Functional classification of HsdM substrates in BCG. Substrate gene functions were classified by comparing them in the COG database.
2.3. HsdM Regulated Gene Expression of Its Substrates
To verify whether HsdM methylation affected the transcriptional level of its substrates, resulting in different mycobacterial redox statuses and affecting the susceptibility to INH, we designed qPCR primers to compare the mRNA levels of all identified HsdM substrates between BCG and ∆hsdM. Under the detection conditions of all HsdM substrates, 17 ORFs were significantly downregulated, and 41 were significantly upregulated in ∆hsdM compared with those in BCG (Table S1). The qPCR examination of the cis-regulation of DNA methylation revealed that nine methylation sites upstream caused statistically different BCG and ∆hsdM expressions in the corresponding genes (Table S2). Consistent with a previous study [], hsdS.1 (equivalent to rv2755c in Mtb) expression was significantly upregulated in the knockout strain (Figure 4A), suggesting that its regulation was associated with hsdM (equivalent to rv2756c in Mtb). Conversely, hsdS (equivalent to rv2761c in Mtb) was unchanged (Figure 4A). Additionally, the two-component system response regulator, TrcR (equivalent to Rv1033c in Mtb), which is thought to help bacteria adapt to the infection environment in the host and switch from latency to reactivation [], was upregulated in ∆hsdM compared with that in the wild-type. This switch process is closely linked to redox regulation []. Moreover, a study showed that TrcR represses BCG_1115 (equivalent to Rv1057 in Mtb), a putative surface antigen []. The qPCR analysis showed that BCG_1115 expression levels were downregulated in ∆hsdM compared with those in the wild-type, which was consistent with the previously reported negative regulation of BCG_1115 (equivalent to rv1057 in Mtb) by TrcR (Figure 4B).
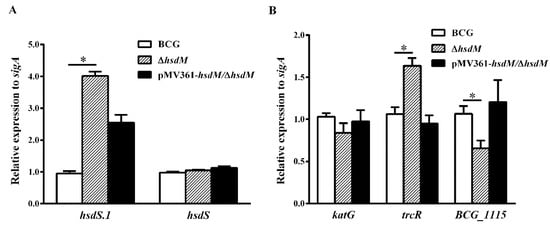
Figure 4.
Genes differentially expressed in ∆hsdM. Expression of each gene was determined by qPCR in the ∆hsdM strain and its parental strain. (A) Relative expressions of hsdS and hsdS.1. * p < 0.05. (B) qPCR analysis of the mRNA expression of katG, trcR and BCG_1115.
To rule out that the reduced susceptibility of ∆hsdM to INH compared with that of BCG was due to differential katG expression, we examined the expression levels of katG in ∆hsdM and BCG via qPCR. The qPCR analysis showed that katG mRNA levels in ∆hsdM were not significantly reduced (~0.84 ± 0.08-fold) compared with those of the parental strain, BCG (Figure 4B). These results suggested that HsdM methylation affected gene expression in the reduced INH susceptibility of ∆hsdM compared with that of BCG.
2.4. HsdM Deletion Increases Survival of BCG during Hypoxia
To further confirm the role of HsdM in oxidative stress responses, we constructed a latent Wayne model [] and examined the growth kinetics and expression levels of related genes of BCG, ∆hsdM and pMV361-hsdM/∆hsdM. At 10 days postinoculation, all strains were collected and plated on 7H10 media supplemented with ADS. When the methylene blue, an oxygen concentration indicator, became colorless, it indicated that the O2 concentration dropped to 0. ∆hsdM exhibited a significant growth advantage over BCG (Figure 5A). Growth kinetics of the complemented strains pMV361-hsdM/∆hsdM and BCG did not differ, indicating that ∆hsdM had a clear growth advantage relative to that of wild-type BCG under hypoxic conditions. Under these conditions, the relative hsdM mRNA levels decreased by 0.49 ± 0.06-fold, and compared with those under standard culture conditions in BCG, the relative trcR mRNA levels increased by 1.75 ± 0.11-fold. Additionally, BCG_1115 was decreased by 0.50 ± 0.05-fold (Figure 5B). Hence, HsdM-mediated modification was linked to redox regulation.
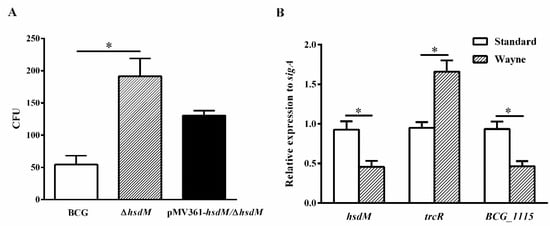
Figure 5.
HsdM deletion increased survival of BCG under hypoxia. (A) CFUs of the ∆hsdM, wild-type BCG and complementary strain pMV361-hsdM/∆hsdM were detected under the latent Wayne model. (B) qPCR analysis of the mRNA expressions of hsdM, trcR and BCG_1115 in BCG between the latent Wayne model and standard culture. * p < 0.05.
3. Discussion
Here, we showed that mycobacterial HsdM, an adenine methyltransferase, altered the transcription levels of the corresponding substrates by methylating its own substrates, thereby reducing the mycobacterial susceptibility to INH. Furthermore, HsdM deletion increased mycobacterial survival under hypoxia. Our results indicated that HsdM-mediated DNA methylation alters mycobacterial sensitivity to INH.
To explore the biological role of HsdM, we took advantage of antibiotics as chemical probes to screen the difference between the wild-type strain BCG and the mutant strain ∆hsdM (Figure 2A–F). Both previous studies and our studies linked isoniazid action with redox homeostatic [,,]. We observed that the hsdM knockout in BCG showed that ∆hsdM had remarkable resistance to INH compared with that of BCG (Figure 2A), and complementary HsdM partially abrogated INH resistance in ∆hsdM (Figure 2G,H). We hypothesized that HsdM affects the transcriptional levels of its substrates, which in turn affects the oxidation reduction in the bacteria. To prove our hypothesis, we compared the methylomes of those three strains (BCG, ΔhsdM and pMV361-hsdM/ΔhsdM) using high-precision PacBio single-molecule real-time (SMRT) sequencing technology.
SMRT technology has been used to identify methylated DNA sequences in bacteria, including Mycobacterium [,,,,]. In mycobacteria, HsdM is predicted to be an orphan that lacks cognate restriction enzymes []. This means that unmethylated HsdM-modified DNA sequences cannot be degraded; that is, the HsdM-modified motifs are stable regardless of whether they are methylated or unmethylated. Consistent with a previous study [,,], the percentage/ratio of methylated modification by HsdM was lower (67–69%), indicating that methylation by HsdM is limited to specific biological functions. As predicted, HsdM substrates were enriched in the respiration pathway (Table S3), whereas the MamA and MamB substrates were not enriched in specific pathways. The bioinformatic analysis of 25% of HsdM substrates presenting in the respiration pathway supported our hypothesis that biological function of HsdM links to redox homeostatic regulation.
Recent studies on the bacterial GATC methylome suggested that MTases regulate transcription [,,,,]. In Mtb, more HsdM motifs than MamA and SigA motifs are located in ORFs [], suggesting that the biological functions of HsdM modification differ from those of MamA and SigA. Here, we showed that HsdM methylated 88% of sites in ORFs. Further assays showed that the expression levels of most motifs tended to be upregulated in ∆hsdM (Table S1). Thus, HsdM-methylated encoding genes could directly affect their own transcription.
Through a comparison of different methylomes in BCG and ∆hsdM, we identified 219 HsdM substrates and confirmed via qPCR (Table S1). In particular, trcR, a two-component system member, was identified as a HsdM substrate (Table S1). The trcR mRNA levels were upregulated in ∆hsdM compared with those in BCG (Figure 4). Because TrcR is important in reactivation from latency [], a process being linked to the redox status change. Using a Wayne model of hypoxia, we also showed increased trcR mRNA levels under hypoxic conditions, compared with standard growth (growth in 7H9 medium) (Figure 5B). Additionally, ∆hsdM exhibited a significant growth advantage over BCG (Figure 5A). Thus, HsdM is important for fitness during hypoxia. Similar to a previous study [], we found a relationship between the latent gene, trcR, and drug resistance, which, thus, requires further study. MamA has also been associated with hypoxia []. Further studies are needed to determine how the synergistic effect of MamA and HsdM on genome methylation affects mycobacterial antibiotic susceptibility.
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
In this study, Mycobacterium bovis BCG Pasteur (BCG) was used to assess HsdM functions in antibiotic susceptibility. Mycobacterial strains were grown in 7H9 medium comprising Middlebrook 7H9 medium (Becton Dickinson, Sparks, MD, USA) supplemented with 10% ADS (5% w/v bovine serum albumin fraction V, 2% w/v D-dextrose, 8.1% w/v NaCl), 0.5% v/v glycerol and 0.05% v/v TWEEN 80. The ∆hsdM mutant strain was maintained in media supplemented with 50 mg/L hygromycin B (Roche, Indianapolis, IN, USA). BCG was used to explore the biological function of the methyltransferase, HsdM.
4.2. Generation of the hsdM Knockout Mutant Strain
Mycobacteriophage-based specialized transduction was used to replace hsdM as previously described [,]. The upstream and downstream sequences were amplified from BCG genomic DNA, using primer pairs hsdM-LL/hsdM-LR and hsdM-RL/hsdM-RR. Table S4 lists the corresponding primers for the ∆hsdM mutant strain, and Figure 1A shows their corresponding positions. The resulting cloned upstream and downstream regions of hdsM were ligated with plasmid p0004s (Hsu and Jacobs, unpublished data) digested with Van91I. The constructed plasmid p0004-hsdM was then linearized with PacI and inserted into the PacI-digested phAE159 (Hsu and Jacobs, unpublished data). A MaxPlax packaging extract (Epicentre Biotechnologies, Madison, WI, USA) was used for phage packaging and the resulting shuttle plasmids were transformed and amplified into E. coli HB101 cells. The selected plasmids were electroporated into M. smegmatis mc2155 for phage propagation. Transduction into individual BCG cells was performed and hsdM was replaced with hygromycin-coding gene (hyg) (Figure 1), and correct transformants were characterized by PCR using the primer pair, hsdM-InL/hsdM-InR (Table S4). The corresponding complementary strain was constructed as described previously. Briefly, the full-length sequence of hsdM was amplified from BCG genomic DNA using primer pair 361-hsdM-F/361-hsdM-R (Table S4), and the PCR product was cloned into the integrating vector, pMV361 [,]. The constructed plasmid was then electroporated into the knockout strain, ∆hsdM, yielding pMV361-hsdM/ΔhsdM.
4.3. Antibiotic Susceptibility Testing
Growth rates of the hsdM mutant strain ∆hsdM and its parent strain were compared in 7H9 medium by monitoring the OD600 at different time points. Early phase cultures (OD600~0.1) were treated with drugs at the indicated concentrations. The OD600 values and the number of CFUs were measured at the indicated point times. Experiments were performed in triplicate.
Isoniazid (INH), rifampicin (RFP), streptomycin (STR), ethambutol (EMB), ciprofloxacin (CIP) and ofloxacin (OFX) were obtained from Sigma-Aldrich (Saint Louis, MO, USA). Susceptibility of the mycobacterial strains to these antibiotic drugs was determined on microplates. A modified microplate Alamar Blue assay was performed to examine the mycobacterial susceptibility as previously described []. Briefly, approximately 105 cells/well were incubated for 7 days with different drug concentrations at 37°C. The indicator, 0.02% resazurin, was then added to individual samples, and color changes (from blue to pink) were recorded after 48 h. Blue indicated no growth; pink indicated growth. The MIC was defined as the lowest antibiotic drug concentration that prevented the color change from blue to pink. A difference of 4-fold or more indicated a significant difference in the antibiotic susceptibility of bacterial strains.
4.4. SMRT Sequencing and Bioinformatics Analysis
Genomic DNA was extracted from M. bovis BCG strains using a previously described method. Briefly, pellets were scraped from plates containing each strain into 4 mL DNA buffer (0.3 M Tris, pH 8.0; 0.1 M NaCl; 6 mM EDTA) and vortexed vigorously with 3 mm glass beads. The supernatant was treated with lysozyme solution, extracted in phenol/chloroform/isoamyl alcohol and precipitated to obtain the genomic DNA. Whole-genome sequencing was used, combining the Illumina HiSeq2000 (Illumina Inc., San Diego, CA, USA) and Pacific Biosciences Sequel II (Pacific Biosciences, Menlo Park, CA, USA) sequencing platforms, and the sequence data from the Illumina platform were used to proofread the PacBio assembly sequence. An Illumina paired-end sequencing library was prepared using TruSeq DNA sample prep kits (Illumina Inc.) as per the manufacturer’s instructions. A 20-kb SMRT bell library was prepared from sheared genomic DNA using a 20 kb template library preparation workflow. SMRT sequencing was conducted on a PacBio Sequel II sequencing platform.
The Hierarchical Genome Assembly Process (HGAP.4) algorithm in the SMRT Link (version 9.0.0) was used to assemble the genome. Errors in assembly of the raw sequence reads were corrected with the Quiver algorithm included in the SMRT software package []. Standard settings (QV > 30) in the “Base Modification Analysis” protocol included in SMRT Link, version 9.0.0, were used to detect base modifications and sequence motifs. The genomic sequence was uploaded into Rapid Annotation using Subsystem Technology for genome annotation. Functions of the predicted protein-coding genes were then annotated via comparisons with the NCBI-NR and COG databases. Functional gene categories were obtained by searching the Mycobrowser website (https://mycobrowser.epfl.ch/genes, accessed on 12 August 2021).
4.5. Hypoxia Survival Experiments
BCG strains were cultured in 7H9 media to OD600~0.5. Cultures were inoculated at 1 × 106 CFU/mL in anaerobic bottles and tightly sealed. The headspace ratio of the samples was 0.5, as defined by the latent Wayne model [], and methylene blue (1.5 mg/L) was used as an indicator of reduced oxygen tension. All samples were prepared in triplicate. The cultures were collected when the indicator turned from blue to colorless. Because the OD600 reached 0.2 when the indicator became colorless, we used samples grown aerobically to an OD600 of 0.2 as controls.
4.6. RNA Isolation and Quantitative Real-Time PCR
Mycobacterial cells were collected by centrifugation at 12,000× g, and bacterial pellets were resuspended in TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was then purified following the manufacturer’s instructions. cDNA was synthesized using 5× All-In One RT MasterMix (ABM, Richmond, BC, Canada). The EvaGreen 2× qPCR Master Mix (ABM) was used for quantitative real-time PCR in a Bio-Rad CFX Connect Real-Time System. The amplification conditions were set as follows: initial denaturation at 95 °C for 3 min; then, 40 cycles of 95 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s, ending with a melting curve step of 65 °C to 95 °C. The BCG RNA polymerase sigma factor, sigA, was used as a control to normalize gene expression. The 2–∆∆CT method [] was used to calculate the relative mycobacterial gene expression. Table S4 lists the qRT-PCR primers.
4.7. Statistical Analysis
The results were those of three biological replicates. Statistical analyses were performed using unpaired two-tailed t-tests in GraphPad Prism 6 or Microsoft Excel. ** p < 0.01 and * p < 0.05.
5. Conclusions
In summary, we explored the biological functions of the MTase, HsdM, in BCG and showed that deleting hsdM decreased mycobacterial susceptibility to INH and increased mycobacterial survival under hypoxia in BCG. Using SMRT sequencing, we identified 219 HsdM-methylated genes, including trcR, a regulator in mycobacterial latency and reactivation. Our study indicated that HsdM methylase genes alter the corresponding gene transcription and, thereby, alter the cellular redox status, resulting in mycobacterial susceptibility to INH. Our work provides insights into the relationship between DNA methylation and antibiotic susceptibility. Further studies are necessary to confirm whether the observed phenotypes related to HsdM are not just restricted to the used BCG model organism, but also to other M. bovis BCG and to virulent M. tuberculosis strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10111323/s1, Figure S1: The MIC of INH on microplates in BCG, ΔhsdM and pMV361-hsdM/ΔhsdM, Table S1: Methylated HsdM substrates: modification sites located in the gene, Table S2: Methylated HsdM substrates: modification sites located upstream of the gene, Table S3: HsdM-methylated substrates involved in intermediary metabolism and the respiration pathway, Table S4: Primers used in this study.
Author Contributions
K.M. conceived and designed the experiments; X.H., X.Z. and T.Y. performed the experiments; X.H., K.C., X.Z. and K.M. analyzed the data; K.M. and X.H. wrote the paper; Y.H. and B.Z. conducted the editing and proofreading. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology of China (2018YFC1603900, 2017YFA0505901 to K.M.), National Natural Science Foundation of China (31970136 to K.M.), International Joint Research Project of the Institute of Medical Science, University of Tokyo (Extension-2019-K3006 to K.M.) and Open Research Projects of the Key Laboratory of Major Diseases in Children, Ministry of Education.
Data Availability Statement
The data presented in this study are openly available in China National Microbiology Data Center (NMDC) with accession numbers NMDC40013959 (https://nmdc.cn/resource/genomics/sra/detail/NMDC40013959, accessed on 12 August 2021) and NMDC40013960 (https://nmdc.cn/resource/genomics/sra/detail/NMDC40013960, accessed on 12 August 2021).
Acknowledgments
We thank Chenyue Wu for her help in preparing the experimental materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Global Tuberculosis Report 2019; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Cohen, K.A.; Abeel, T.; McGuire, A.M.; Desjardins, C.A.; Munsamy, V.; Shea, T.P.; Walker, B.J.; Bantubani, N.; Almeida, D.V.; Alvarado, L.; et al. Evolution of extensively drug-resistant tuberculosis over four decades: Whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015, 12, e1001880. [Google Scholar] [CrossRef]
- Zhang, H.; Li, D.; Zhao, L.; Fleming, J.; Lin, N.; Wang, T.; Liu, Z.; Li, C.; Galwey, N.; Deng, J.; et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 2013, 45, 1255–1260. [Google Scholar] [CrossRef]
- Jain, P.; Weinrick, B.C.; Kalivoda, E.J.; Yang, H.; Munsamy, V.; Vilcheze, C.; Weisbrod, T.R.; Larsen, M.H.; O’Donnell, M.R.; Pym, A.; et al. Dual-reporter mycobacteriophages (Φ2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 2016, 7, e01023-16. [Google Scholar] [CrossRef]
- Roca, F.J.; Whitworth, L.J.; Redmond, S.; Jones, A.A.; Ramakrishnan, L. TNF induces pathogenic programmed macrophage necrosis in tuberculosis through a mitochondrial-lysosomal-endoplasmic reticulum circuit. Cell 2019, 178, 1344–1361. [Google Scholar] [CrossRef]
- Huang, X.T.; Wang, J.J.; Li, J.; Liu, Y.N.; Liu, X.; Li, Z.Y.; Kurniyati, K.; Deng, Y.J.; Wang, G.L.; Ralph, J.D.; et al. Prevalence of phase variable epigenetic invertons among host-associated bacteria. Nucleic Acids Res. 2020, 48, 11468–11485. [Google Scholar] [CrossRef] [PubMed]
- Estibariz, I.; Ailloud, F.; Woltemate, S.; Bunk, B.; Sproer, C.; Overmann, J.; Aebischer, T.; Meyer, T.F.; Josenhans, C.; Suerbaum, S. In Vivo genome and methylome adaptation of cag-negative helicobacter pylori during experimental human infection. mBio 2020, 11, e01803-20. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.R.; Ross, C.A.; Jain, S.; Shapiro, R.S.; Gutierrez, A.; Belenky, P.; Li, H.; Collins, J.J. A role for the bacterial GATC methylome in antibiotic stress survival. Nat. Genet. 2016, 48, 581–586. [Google Scholar] [CrossRef]
- Comas, I.; Coscolla, M.; Luo, T.; Borrell, S.; Holt, K.E.; Kato-Maeda, M.; Parkhill, J.; Malla, B.; Berg, S.; Thwaites, G.; et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 2013, 45, 1176–1182. [Google Scholar] [CrossRef]
- Castro, R.A.D.; Ross, A.; Kamwela, L.; Reinhard, M.; Loiseau, C.; Feldmann, J.; Borrell, S.; Trauner, A.; Gagneux, S. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol. Biol. Evol. 2020, 37, 195–207. [Google Scholar] [CrossRef]
- Shell, S.S.; Prestwich, E.G.; Baek, S.H.; Shah, R.R.; Sassetti, C.M.; Dedon, P.C.; Fortune, S.M. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013, 9, e1003419. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhong, J.; Jia, X.; Liu, G.; Kang, Y.; Dong, M.; Zhang, X.; Li, Q.; Yue, L.; Li, C.; et al. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 2016, 44, 730–743. [Google Scholar] [CrossRef]
- Modlin, S.J.; Conkle-Gutierrez, D.; Kim, C.; Mitchell, S.N.; Morrissey, C.; Weinrick, B.C.; Jacobs, W.R.; Ramirez-Busby, S.M.; Hoffner, S.E.; Valafar, F. Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates. eLife 2020, 9, e58542. [Google Scholar] [CrossRef] [PubMed]
- Chiner-Oms, A.; Berney, M.; Boinett, C.; Gonzalez-Candelas, F.; Young, D.B.; Gagneux, S.; Jacobs, W.R., Jr.; Parkhill, J.; Cortes, T.; Comas, I. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nat. Commun. 2019, 10, 3994. [Google Scholar] [CrossRef]
- Hernday, A.; Krabbe, M.; Braaten, B.; Low, D. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 16470–16476. [Google Scholar] [CrossRef]
- Ringquist, S.; Smith, C.L. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl. Acad. Sci. USA 1992, 89, 4539–4543. [Google Scholar] [CrossRef]
- Wright, R.; Stephens, C.; Shapiro, L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 1997, 179, 5869–5877. [Google Scholar] [CrossRef]
- Bardarov, S.; Bardarov, S.; Pavelka, M.S.; Sambandamurthy, V.; Larsen, M.; Tufariello, J.; Chan, J.; Hatfull, G.; Jacobs, W.R. Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 2002, 148, 3007–3017. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tao, J.; Hu, X.; Chan, J.; Xiao, J.; Mi, K. A bacterial hemerythrin-like protein MsmHr inhibits the SigF-dependent hydrogen peroxide response in mycobacteria. Front. Microbiol. 2014, 5, 800. [Google Scholar] [CrossRef][Green Version]
- Vilcheze, C.; Jacobs, W.R., Jr. The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J. Mol. Biol. 2019, 431, 3450–3461. [Google Scholar] [CrossRef] [PubMed]
- Vilcheze, C.; Hartman, T.; Weinrick, B.; Jain, P.; Weisbrod, T.R.; Leung, L.W.; Freundlich, J.S.; Jacobs, W.R., Jr. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, 4495–4500. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Gu, J.; Wang, T.; Bi, L.J.; Zhang, Z.P.; Cui, Z.Q.; Wei, H.P.; Deng, J.Y.; Zhang, X.E. Comparative analysis of mycobacterial NADH pyrophosphatase isoforms reveals a novel mechanism for isoniazid and ethionamide inactivation. Mol. Microbiol. 2011, 82, 1375–1391. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Lobritz, M.A.; Belenky, P.; Porter, C.B.M.; Gutierrez, A.; Yang, J.H.; Schwarz, E.G.; Dwyer, D.J.; Khalil, A.S.; Collins, J.J. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tao, J.; Han, J.; Hu, X.; Chen, Y.; Deng, H.; Zhang, G.; Hu, X.; Mi, K. The gain of hydrogen peroxide resistance benefits growth fitness in mycobacteria under stress. Protein Cell 2014, 5, 182–185. [Google Scholar] [CrossRef]
- Bardou, F.; Raynaud, C.; Ramos, C.; Laneelle, M.A.; Lanrelle, G. Mechanism of isoniazid uptake in Mycobacterium tuberculosis. Microbiology 1998, 144, 2539–2544. [Google Scholar] [CrossRef] [PubMed]
- Shoeb, H.A.; Bowman, B.U., Jr.; Ottolenghi, A.C.; Merola, A.J. Peroxidase-mediated oxidation of isoniazid. Antimicrob. Agents Chemother. 1985, 27, 399–403. [Google Scholar] [CrossRef]
- Haydel, S.E.; Benjamin, W.H., Jr.; Dunlap, N.E.; Clark-Curtiss, J.E. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J. Bacteriol. 2002, 184, 2192–2203. [Google Scholar] [CrossRef]
- Haydel, S.E.; Clark-Curtiss, J.E. The Mycobacterium tuberculosis TrcR response regulator represses transcription of the intracellularly expressed Rv1057 gene, encoding a seven-bladed beta-propeller. J. Bacteriol. 2006, 188, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.G. In vitro model of hypoxically induced nonreplicating persistence of Mycobacterium tuberculosis. Methods Mol. Med. 2001, 54, 247–269. [Google Scholar]
- Hu, X.L.; Li, X.J.; Huang, L.G.; Chan, J.; Chen, Y.L.; Deng, H.T.; Mi, K.X. Quantitative proteomics reveals novel insights into isoniazid susceptibility in sycobacteria sediated by a universal stress protein. J. Proteome Res. 2015, 14, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, A.; Chawla, M.; Mehta, M.; Parikh, P.; Chandra, P.; Bhave, D.; Kumar, D.; Carroll, K.S.; Singh, A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014, 10, e1003902. [Google Scholar] [CrossRef]
- Hale, W.B.; van der Woude, M.W.; Low, D.A. Analysis of nonmethylated GATC sites in the Escherichia coli chromosome and identification of sites that are differentially methylated in response to environmental stimuli. J. Bacteriol. 1994, 176, 3438–3441. [Google Scholar] [CrossRef]
- Blow, M.J.; Clark, T.A.; Daum, C.G.; Deutschbauer, A.M.; Fomenkov, A.; Fries, R.; Froula, J.; Kang, D.D.; Malmstrom, R.R.; Morgan, R.D.; et al. The epigenomic landscape of prokaryotes. PLoS Genet. 2016, 12, e1005854. [Google Scholar] [CrossRef]
- Ardissone, S.; Redder, P.; Russo, G.; Frandi, A.; Fumeaux, C.; Patrignani, A.; Schlapbach, R.; Falquet, L.; Viollier, P.H. Cell cycle constraints and environmental control of local DNA hypomethylation in alpha-proteobacteria. PLoS Genet. 2016, 12, e1006499. [Google Scholar] [CrossRef]
- Low, D.A.; Casadesus, J. Clocks and switches: Bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 2008, 11, 106–112. [Google Scholar] [CrossRef]
- Stover, C.K.; Delacruz, V.F.; Fuerst, T.R.; Burlein, J.E.; Benson, L.A.; Bennett, L.T.; Bansal, G.P.; Young, J.F.; Lee, M.H.; Hatfull, G.F.; et al. New use of BCG for recombinant vaccines. Nature 1991, 351, 456–460. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Florio, W.; Freer, G.; Campa, M.; Senesi, S. Analysis of the Mycobacterium bovis hsp60 promoter activity in recombinant Mycobacterium avium. FEMS Microbiol. Lett. 1998, 169, 117–124. [Google Scholar] [CrossRef]
- Franzblau, S.G.; Witzig, R.S.; McLaughlin, J.C.; Torres, P.; Madico, G.; Hernandez, A.; Degnan, M.T.; Cook, M.B.; Quenzer, V.K.; Ferguson, R.M.; et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 1998, 36, 362–366. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).