Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format
Abstract
1. Introduction
- cost of analysis should not be too high;
- analysis should be rapid, sensitive and specific;
- multiplex detection of a number of pathogens; and
- procedure of analysis should be simple and does not require special facilities and conditions;
- diagnostic kits should not fall under special storage and transportation limitations.
2. Materials and Methods
2.1. Fungal Isolates, Cultivation, and DNA Extraction
2.2. Oligonucleotide Design
2.3. Sample Preparation
2.4. Amplification Conditions and Data Analysis
2.5. Validation of Developed qPCR Assays
3. Results
3.1. Primer Design
3.2. Specificity Assay
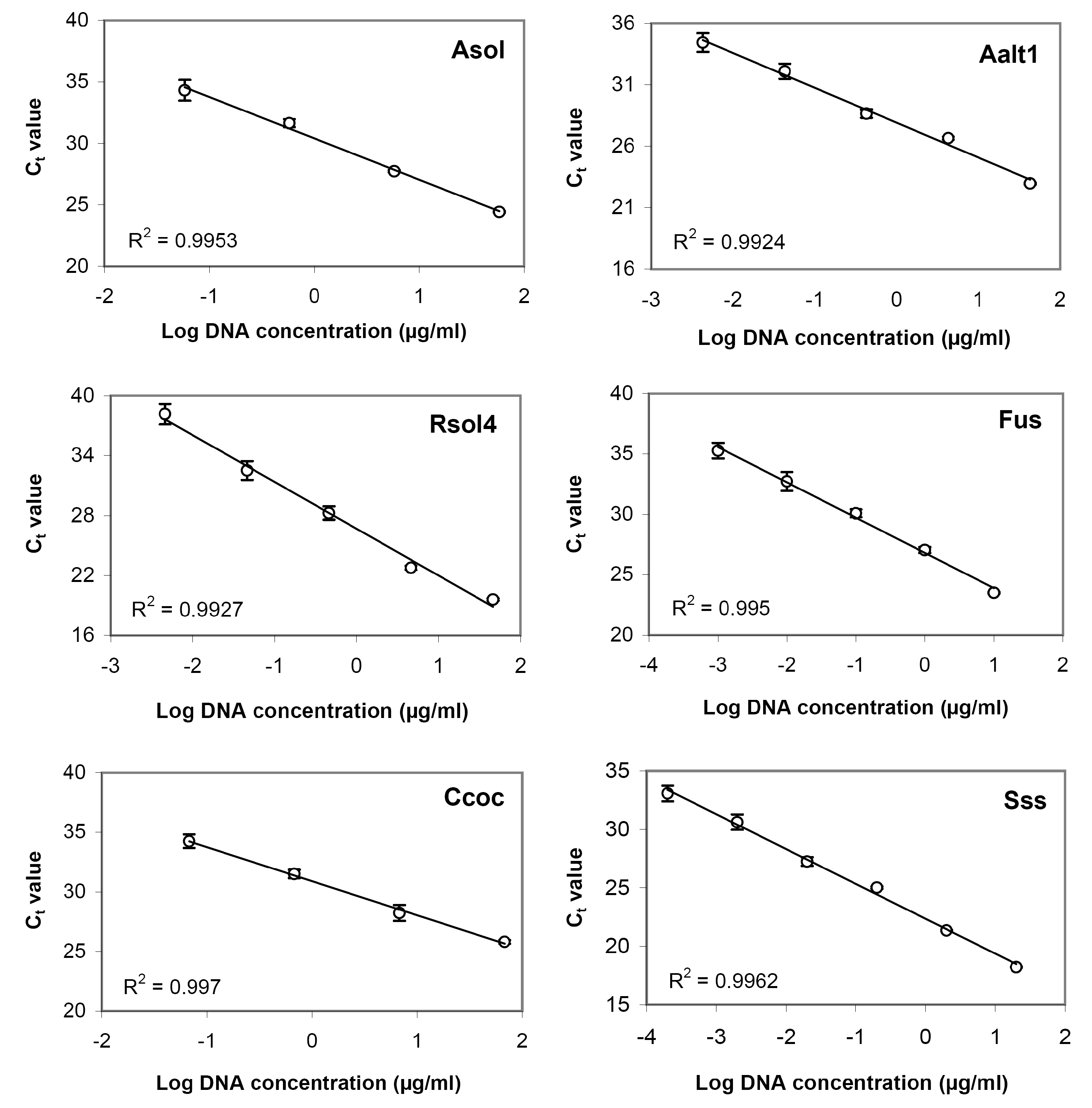
3.3. Sensitivity Assay and Regression Curves
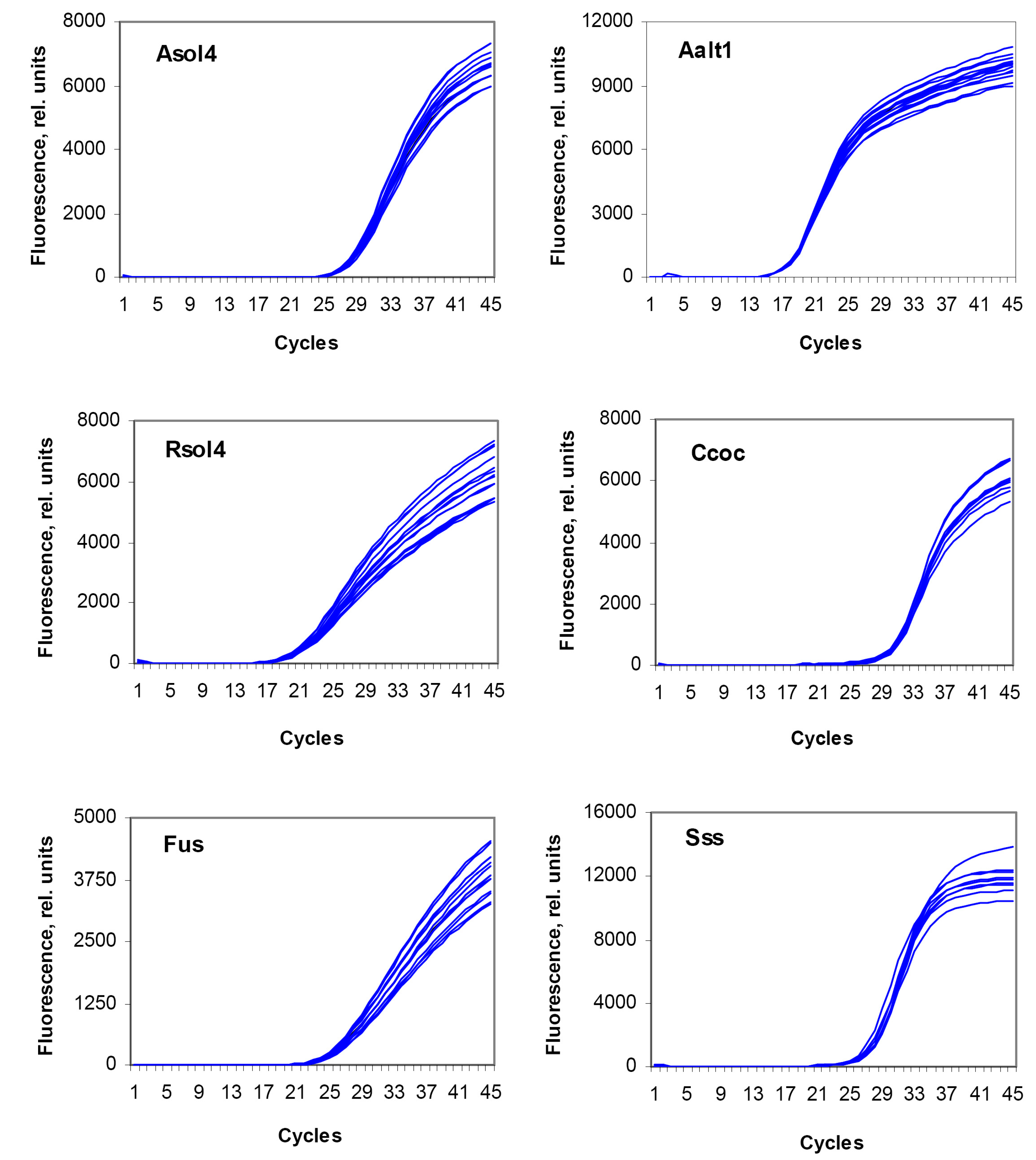
3.4. Reproducibility Assay
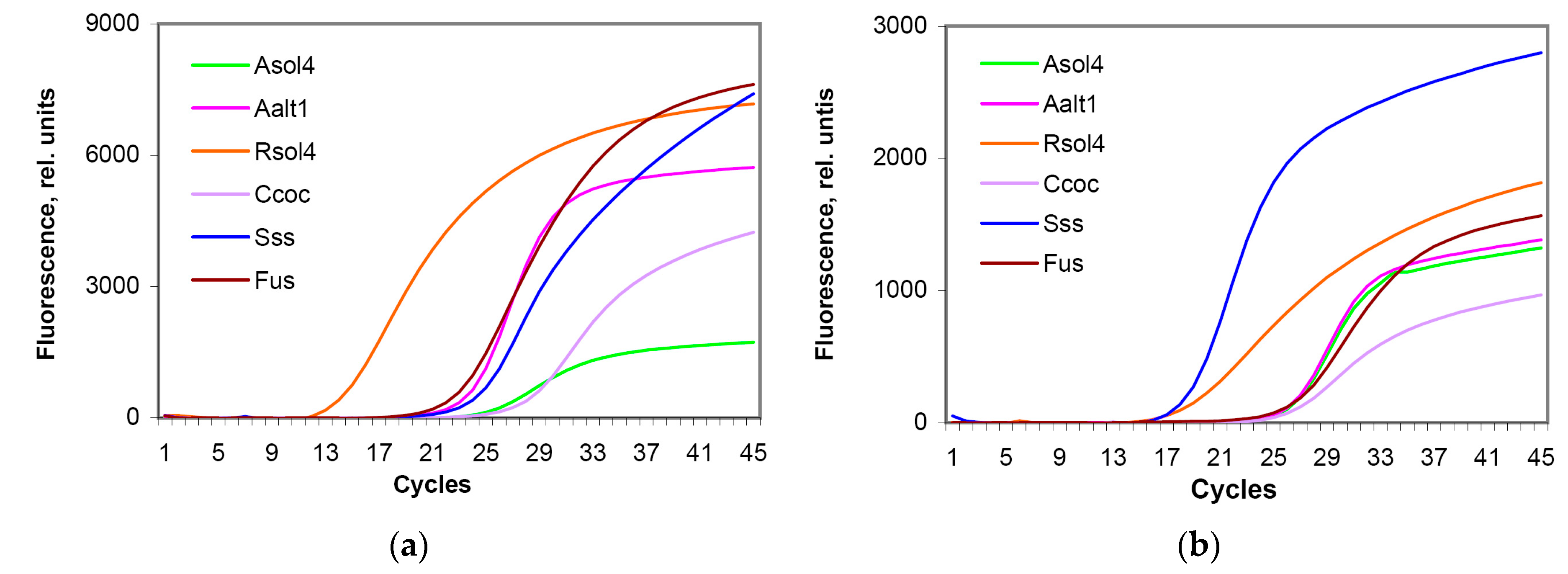
3.5. Comparison of the Working Efficiency of Fresh and Lyophilized Test Systems
3.6. Validation of Test Systems with Field Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Potato Statistics. Available online: https://www.potatopro.com/world/potato-statistics (accessed on 10 October 2018).
- Moore, D.; Robson, G.D.; Trinci, A.P. 21st Century Guidebook to Fungi; Cambridge University Press: Cambridge, UK, 2011; pp. 368–369. ISBN 978-1107419711. [Google Scholar]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Haverkort, A.; Struik, P.; Visser, R.; Jacobsen, E. Applied biotechnology to combat late blight in potato caused by Phytophthora Infestans. Potato Res. 2009, 52, 249–264. [Google Scholar] [CrossRef]
- Tsror, L. Biology, epidemiology and management of Rhizoctonia solani on potato. J. Phytopathol. 2010, 110, 111–118. [Google Scholar] [CrossRef]
- Thangavel, T.; Tegg, R.S.; Wilson, C.R. Monitoring Spongospora subterranea development in potato roots reveals distinct infection patterns and enables efficient assessment of disease control methods. PLoS ONE 2015, 10, e0137647. [Google Scholar] [CrossRef] [PubMed]
- Baayen, R.; Cochius, G.; Hendriks, H.; Meffert, J.; Bakker, J.; Bekker, M.; van den Boogert, P.; Stachewicz, H.; van Leeuwen, G. History of potato wart disease in Europe—A proposal for harmonisation in defining pathotypes. Eur. J. Plant Pathol. 2006, 116, 21–31. [Google Scholar] [CrossRef]
- Abbas, M.F.; Naz, F.; Irshad, G. Important fungal diseases of potato and their management—A brief review. Mycopath 2013, 11, 45–50. [Google Scholar]
- Tsror, L.; Erlich, O.; Hazanovsky, M. Effect of Colletotrichum coccodes on potato yield, tuber quality, and stem colonization during spring and autumn. Plant Dis. 1999, 83, 561–565. [Google Scholar] [CrossRef]
- Bojanowski, A.; Avis, T.J.; Pelletier, S.; Tweddell, R.J. Management of potato dry rot. Postharvest Boil. Technol. 2013, 84, 99–109. [Google Scholar] [CrossRef]
- De Haan, E.G.; Van Den Bovenkamp, G. Improved diagnosis of powdery scab (Spongospora subterranea f.sp. subterranea) symptoms on potato tubers (Solarium tuberosum L.). Potato Res. 2005, 48, 1–14. [Google Scholar] [CrossRef]
- Turkensteen, J.; Spoelder, J.; Mulder, A. Will the real Alternaria stand up please: Experiences with Alternaria-like diseases on potatoes during the 2009 season in the Netherlands. PPO-Spec. Rep. 2010, 14, 165–170. [Google Scholar]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Bonantz, P.J.M.; Shoen, C.D.; Szemes, M.; Speksnijder, A.; Klerks, M.M.; van den Boogert, P.H.J.F.; Waalwijk, C.; van der Wolf, J.M.; Zijltra, C. From single to multiple detection of plant pathogens: PUMA, a new concept of multiplex detection using microarrays. Phytopathol. Pol. 2005, 35, 29–47. [Google Scholar]
- Chen, W.; Djama, Z.R.; Coffey, M.D.; Martin, F.N.; Bilodeau, G.J.; Radmer, L.; Denton, G.; Lévesque, C.A. Membrane-based oligonucleotide array developed from multiple markers for the detection of many Phytophthora species. Phytopathology 2013, 103, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; McCarthy, M.L.; Smart, C.D. A macroarray system for the detection of fungal and oomycete pathogens of solanaceous crops. Plant Dis. 2008, 92, 953–960. [Google Scholar] [CrossRef]
- Vreeburg, R.A.; Bergsma-Vlami, M.; Bollema, R.M.; Haan, E.G.; Kooman-Gersmann, M.; Smits-Mastebroek, L.; Tameling, W.I.; Tjou-Tam-Sin, N.N.; Vossenberg, B.T.; Janse, J.D. Performance of real-time PCR and immunofluorescence for the detection of Clavibacter michiganensis subsp. sepedonicus and Ralstonia solanacearum in potato tubers in routine testing. EPPO Bull. 2016, 46, 112–121. [Google Scholar] [CrossRef]
- Pastuszewska, T.; Lewosz, J.; Sadoch, Z. Application of IFAs and PCR for Clavibacter michiganensis subsp. sepedonicus detection in potato plants during the growing season. Phytopathol. Pol. 2005, 35, 95–102. [Google Scholar]
- Elad, Y.; Pertot, I. Climate change impacts on plant pathogens and plant diseases. J. Crop. Improv. 2014, 28, 99–139. [Google Scholar] [CrossRef]
- Garrett, K.A.; Dendy, S.P.; Frank, E.E.; Rouse, M.N.; Travers, S.E. Climate change effects on plant disease: Genomes to ecosystems. Annu. Rev. Phytopathol. 2006, 44, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Sanati Nezhad, A. Future of portable devices for plant pathogen diagnosis. Lab Chip 2014, 14, 2887–2904. [Google Scholar] [CrossRef]
- Mirmajlessi, S.M.; Loit, E.; Mand, M.; Mansouripour, S.M. Real-time PCR applied to study on plant pathogens: Potential applications in diagnosis—A review. Plant Prot. Sci. 2015, 51, 177–190. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Pankratova, G.K.; Zaitsev, I.A.; Varitsev, Y.A.; Vengerov, Y.Y.; Zherdev, A.V.; Dzantiev, B.B. Multiarray on a test strip (MATS): Rapid multiplex immunodetection of priority potato pathogens. Anal. Bioanal. Chem. 2016, 408, 6009–6017. [Google Scholar] [CrossRef] [PubMed]
- DeShields, J.B.; Bomberger, R.A.; Woodhall, J.W.; Wheeler, D.L.; Moroz, N.; Johnson, D.A.; Tanaka, K. On-site molecular detection of soil-borne phytopathogens using a portable real-time PCR system. J. Vis. Exp. 2018, 132, e56891. [Google Scholar] [CrossRef] [PubMed]
- Khiyami, M.A.; Almoammar, H.; Awad, Y.M.; Alghuthaymi, M.A.; Abd-Elsalam, K.A. Plant pathogen nanodiagnostic techniques: Forthcoming changes? Biotechnol. Biotechnol. Equip. 2014, 28, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Huanca-Mamani, W.; Salvatierra Martínez, R.; Sepúlveda-Chavera, G. A fast and efficient method for total DNA extraction from soil filamentous fungi. Idesia 2014, 32, 75–78. [Google Scholar] [CrossRef]
- Grund, E.; Darissa, O.; Adam, G. Application of FTA® cards to sample microbial plant pathogens for PCR and RT-PCR. J. Phytopathol. 2010, 158, 750–757. [Google Scholar] [CrossRef]
- Le, D.T.; Vu, N.T. Progress of loop-mediated isothermal amplification technique in molecular diagnosis of plant diseases. Appl. Boil. Chem. 2017, 60, 169–180. [Google Scholar] [CrossRef]
- Khater, M.; Escosura-Muñiza, A.; Merkoçi, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 4, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Hashsham, S.A. Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: A review. Anal. Chim. Acta 2012, 733, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ryazantsev, D.; Zavriev, S. An efficient diagnostic method for the identification of potato viral pathogens. Mol. Boil. 2009, 43, 515–523. [Google Scholar] [CrossRef]
- Cating, R.A.; Funke, C.N.; Kaur, N.; Hamm, P.B.; Frost, K.E. A multiplex reverse transcription (RT) high-fidelity PCR protocol for the detection of six viruses that cause potato tuber necrosis. Am. J. Potato Res. 2015, 92, 536–540. [Google Scholar] [CrossRef]
- Ranjan, R.K.; Singh, D.; Baranwal, V.K. Simultaneous detection of brown rot- and soft rot-causing bacterial pathogens from potato tubers through multiplex PCR. Curr. Microbiol. 2016, 73, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, M.; Statsyuk, N.; Frantsuzov, P.; Dzhavakhiya, V.; Golikov, A. Matrix approach to the simultaneous detection of multiple potato pathogens by real-time PCR. J. Appl. Microbiol. 2018, 124, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, M.M.; Statsyuk, N.V.; Frantsuzov, P.A.; Pridannikov, M.V.; Golikov, A.G. Rapid and simple detection of two potato cyst nematode species by real-time multiplex PCR using preserved microarray-based test systems. Russ. J. Nematol. 2017, 25, 51–60. [Google Scholar]
- Deych, K.O.; Pridannikov, M.V.; Nikitin, M.M.; Statsyuk, N.V.; Dzhavakhiya, V.G.; Golikov, A.G. Simultaneous detection of three plant parasitic nematode species from the genus Ditylenchus using a real-time qPCR matrix-based technology. Russ. J. Nematol. accepted.
- Qu, X.; Christ, B.J. Single cystosorus isolate and restriction fragment length polymorphism characterization of the obligate biotroph Spongospora subterranea f. sp. subterranea. Phytopathology 2006, 96, 1157–1163. [Google Scholar] [CrossRef]
- GenBank Geneti Sequence Database. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 10 December 2018).
- OLIGO Primer Analysis Software. Available online: http://www.oligo.net/ (accessed on 10 December 2018).
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/ (accessed on 10 December 2018).
- State Standard R 55329-2012. Seed Potatoes. Acceptance Rules and Methods of Analysis; Standartinform: Moscow, Russia, 2013; pp. 8–10. (In Russian) [Google Scholar]
- Pidoplichko, N.M. Parasitic Fungi of Agricultural Crops. Identification Guide; Naukova Dumka: Kiev, Ukraine, 1977; Volume 2. (In Russian) [Google Scholar]
- Kokaeva, L.Y.; Belosokhov, A.F.; Doeva, L.Y.; Skolotneva, E.S.; Elansky, S.N. Distribution of Alternaria species on blighted potato and tomato leaves in Russia. J. Plant Dis. Prot. 2018, 125, 205–212. [Google Scholar] [CrossRef]
- Du, M.; Ren, X.; Sun, Q.; Wang, Y.; Zhang, R. Characterization of Fusarium spp. causing potato dry rot in China and susceptibility evaluation of Chinese potato germplasm to the pathogen. Potato Res. 2012, 55, 175–184. [Google Scholar] [CrossRef]
- Stefańczyk, E.; Sobkowiak, S.; Brylińska, M.; Śliwka, J. Diversity of Fusarium spp. associated with dry rot of potato tubers in Poland. Eur. J. Plant Pathol. 2016, 145, 871–884. [Google Scholar] [CrossRef]
- Gachango, E.; Hanson, L.E.; Rojas, A.; Hao, J.J.; Kirk, W.W. Fusarium spp. causing dry rot of seed potato tubers in Michigan and their sensitivity to fungicides. Plant Dis. 2012, 96, 1767–1774. [Google Scholar] [CrossRef]
- Konstantinova, P.; Bonants, P.J.M.; van Gent-Pelzer, M.P.E.; van der Zouwen, P.; van den Bulk, R. Development of specific primers for detection and identification of Alternaria spp. in carrot material by PCR and comparison with blotter and plating assays. Mycol. Res. 2002, 106, 23–33. [Google Scholar] [CrossRef]
- Van de Graaf, P.; Lees, A.K.; Cullen, D.W.; Duncan, J.M. Detection and quantification of Spongospora subterranea in soil, water and plant tissue samples using real-time PCR. Eur. J. Plant Pathol. 2003, 109, 589–597. [Google Scholar] [CrossRef]
- Qu, X.; Wanner, L.; Christ, B. Multiplex real-time PCR (TaqMan) assay for the simultaneous detection and discrimination of potato powdery and common scab diseases and pathogens. J. Appl. Microbiol. 2011, 110, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Leiminger, J.; Bäßler, E.; Knappe, C.; Bahnweg, G.; Hausladen, H. Quantification of disease progression of Alternaria spp. on potato using real-time PCR. Eur. J. Plant Pathol. 2015, 141, 295–309. [Google Scholar] [CrossRef]
- Grube, S.; Schönling, J.; Prange, A. Evaluation of a triplex real-time PCR system to detect the plant-pathogenic molds Alternaria spp., Fusarium spp. and C. Purpurea. J. Microbiol. Methods 2015, 119, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Graça, M.G.; van der Heijden, I.M.; Perdigão, L.; Taira, C.; Costa, S.F.; Levin, A.S. Evaluation of two methods for direct detection of Fusarium spp. in water. J. Microbiol. Methods 2016, 123, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Muraosa, Y.; Schreiber, A.Z.; Trabasso, P.; Matsuzawa, T.; Taguchi, H.; Moretti, M.L.; Mikami, Y.; Kamei, K. Development of cycling probe-based real-time PCR system to detect Fusarium species and Fusarium solani species complex (FSSC). Int. J. Med Microbiol. 2014, 304, 505–511. [Google Scholar] [CrossRef]
- Atallah, Z.K.; Stevenson, W.R. A methodology to detect and quantify five pathogens causing potato tuber decay using real-time quantitative polymerase chain reaction. Phytopathology 2006, 96, 1037–1045. [Google Scholar] [CrossRef]
- Edin, E. Species specific primers for identification of Alternaria solani, in combination with analysis of the F129L substitution associates with loss of sensitivity toward strobilurins. Crop. Prot. 2012, 38, 72–73. [Google Scholar] [CrossRef]
- Cullen, D.W.; Lees, A.K.; Toth, I.K.; Duncan, J.M. Detection of Colletotrichum coccodes from soil and potato tubers by conventional and quantitative real-time PCR. Plant Pathol. 2002, 51, 281–292. [Google Scholar] [CrossRef]
- Dauch, A.L.; Watson, A.K.; Seguin, P.; Jabaji-Hare, S.H. Real-time PCR quantification of Colletotrichum coccodes DNA in soils from bioherbicide field-release assays, with normalization for PCR inhibition. Can. J. Plant Pathol. 2006, 28, 42–51. [Google Scholar] [CrossRef]
- Dubey, S.C.; Tripathi, A.; Upadhyay, B.K.; Kumar, A. Development of conventional and real time PCR assay for detection and quantification of Rhizoctonia solani infecting pulse crops. Biologia 2016, 71, 133–138. [Google Scholar] [CrossRef]
- Woodhall, J.W.; Adams, I.P.; Peters, J.C.; Harper, G.; Boonham, N. A new quantitative real-time PCR assay for Rhizoctonia solani AG3-PT and the detection of AGs of Rhizoctonia solani associated with potato in soil and tuber samples in Great Britain. Eur. J. Plant Pathol. 2013, 136, 273–280. [Google Scholar] [CrossRef]
- Lees, A.K.; Cullen, D.W.; Sullivan, L.; Nicolson, M.J. Development of conventional and quantitative real-time PCR assays for the detection and identification of Rhizoctonia solani AG-3 in potato and soil. Plant Pathol. 2002, 51, 293–302. [Google Scholar] [CrossRef]
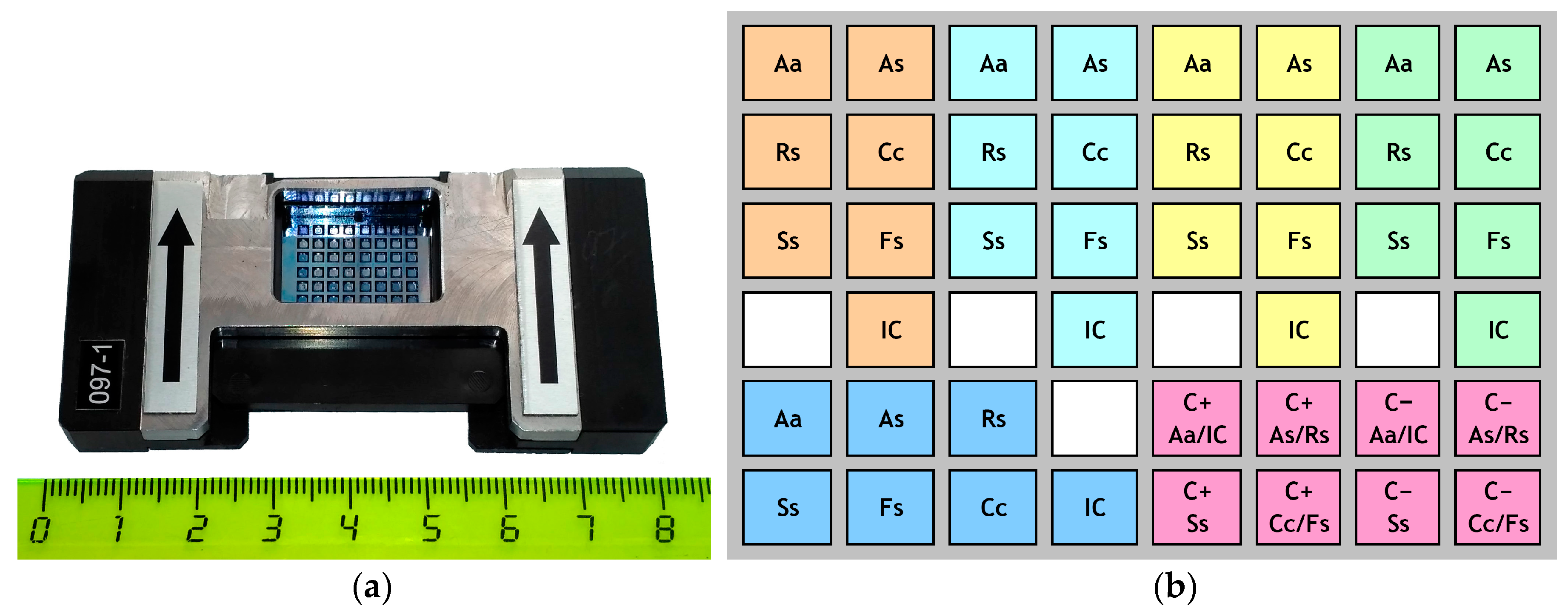
| Species | Isolate Code | Year and Region of Collection | Host Plant | Source 1 |
|---|---|---|---|---|
| Alternaria solani | 100053 | 2009, Mariy El Republic, Russia | Potato | SCPPM |
| A. solani | 043-021 | – 2 | – | MF |
| A. solani | 044-051 | – | – | MF |
| A. alternata | 4l/6 | 2011, Republic of Mordovia, Russia | Barley | SCPPM |
| A. alternata | 4k/2 | 2011, Republic of Mordovia, Russia | Barley | SCPPM |
| A. alternata | AK2 | – | – | MF |
| A. alternata | HKK1 | – | – | MF |
| A. alternata | 12RKL9 | 2007, Ryazan region, Russia | Potato | MF |
| A. alternata | 12RKL10 | 2007, Ryazan region, Russia | Potato | MF |
| A. infectoria | K100088 | 2009, Krasnodar region, Russia | Bindweed | SCPPM |
| A. radicina | K100011 | 2009, Moscow region, Russia | Carrot | SCPPM |
| A. longipes | K100055 | 2000, Moscow region, Russia | Tomato | SCPPM |
| Rhizoctonia solani | 100006 | 2005, Moscow region, Russia | Potato | SCPPM |
| R. solani | 100063 | 2011, Moscow region, Russia | Potato | SCPPM |
| R. solani | 100106 | 2013, Leningrad region, Russia | Potato | SCPPM |
| R. solani | R15BKK nev25 | 2015, Vladimir region, Russia | Potato | SCPPM |
| R. solani | 14BM rs2 | 2014, Vladimir region, Russia | Potato | SCPPM |
| R. solani | 14KC man1 | 2014, Kostroma region, Russia | Potato | SCPPM |
| R. cerealis | 1000025 | 2009, Ryazan region, Russia | Spring wheat | SCPPM |
| Colletotrichum coccodes | 100004 | 2009, Moscow region, Russia | Potato | SCPPM |
| C. coccodes | 100123(3) | 2012, Voronezh region, Russia | Potato | SCPPM |
| C. coccodes | 100124(3) | 2013, Tula region, Russia | Potato | SCPPM |
| C. coccodes | 100119(3) | 2015, Orenburg region, Russia | Potato | SCPPM |
| C. coccodes | 100120(3) | 2015, Moscow region, Russia | Potato | SCPPM |
| C. coffeanum | 100003 | 2008, Moscow region, Russia | Ornamental plants | SCPPM |
| C. gloeosporioides | 100002 | 2009, Bryansk region, Russia | Blue lupine | SCPPM |
| C. gloeosporioides | 100094 | 2013, Moscow region, Russia | Tatarian honeysuckle | SCPPM |
| C. lilii | 100001 | 2009, Moscow region, Russia | Lily | SCPPM |
| C. dematium | 100158 | 2017, Republic of Mordovia, Russia | Barley | SCPPM |
| Fusarium avenaceum | MOK-16-3 | 2016, Moscow region, Russia | Potato | SCPPM |
| F. avenaceum | 110501 | 2003, Moscow region, Russia | Barley | SCPPM |
| F. avenaceum | U-08-8-1 | 2008, Ulyanovsk region, Russia | Wheat | SCPPM |
| F. culmorum | MKRS-15-3 | 2015, Moscow region, Russia | Potato | SCPPM |
| F. culmorum | 100135(6-1) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. culmorum | 100136(6-2) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. gibbosum | 100130(7-2) | 2014, Moscow region, Russia | Potato | SCPPM |
| F. gibbosum | 100131(7-2) | 2014, Moscow region, Russia | Potato | SCPPM |
| F. gibbosum | 100132(7) | 2009, Voronezh region, Russia | Potato | SCPPM |
| F. heterosporum | MOK-16-1 | 2016, Moscow region, Russia | Potato | SCPPM |
| F. heterosporum | 100133(8) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. oxysporum | FO-1 | 1995, Moscow region, Russia | Potato | SCPPM |
| F. oxysporum | 100139(9) | 2014, Moscow region, Russia | Potato | SCPPM |
| F. oxysporum | 100140(9) | 2013, Bryansk region, Russia | Potato | SCPPM |
| F. oxysporum | RAM-14 | 2014, Moscow region, Russia | Potato | SCPPM |
| F. sambucinum | P-2-02 | 2002, Ryazan region, Russia | Barley | SCPPM |
| F. sambucinum | KRT 11-1 kch | 2012, Krasnodar region, Russia | Wheat | SCPPM |
| F. sambucinum | 100134(10) | 2009, Lipetsk region, Russia | Potato | SCPPM |
| F. solani | FSL-9 | 2002, Moscow region, Russia | Potato | SCPPM |
| F. solani | 100021 | 2001, Moscow region, Russia | Potato | SCPPM |
| F. solani | 100137(11-1) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. solani | 100138(11-2) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. sporotrichioides | KRT12-1kch | 2012, Krasnodar region, Russia | Wheat | SCPPM |
| F. sporotrichioides | 100141(12) | 2009, Moscow region, Russia | Potato | SCPPM |
| F. javanicum | MKRS-15-1 | 2015, Moscow region, Russia | Potato | SCPPM |
| F. sacchari | RAM-16 | 2016, Moscow region, Russia | Potato | SCPPM |
| Phytophthora infestans | MVK 118a-07 | 2007, Moscow region, Russia | Potato | SCPPM |
| P. infestans | ATP-3.08 | 2008, Astrakhan region, Russia | Tomato | SCPPM |
| Test System | Primers and Probe Sequences (5′–3′) | Length, Bases | Tm *, °C | Amplicon Size, bp |
|---|---|---|---|---|
| Asol4 Alternaria solani | F: GGTCAGCGACGAGTAAGTT | 19 | 59.4 | 71 |
| R: CAGATATACTAACGCTTTTCCA | 22 | 60 | ||
| Probe: ROX-CACGCTTTTCACCACCTTTTAC-BHQ2 | 22 | 66.7 | ||
| Aalt1 Alternaria alternata | F: AGGAACCCTCGACTTCACCT | 20 | 62.0 | 75 |
| R: TTCTCGCCACAGGAGTACCA | 20 | 62.0 | ||
| Probe: FAM- CTCTGCTCAGGCCGATAAGCT-BHQ1 | 21 | 66.0 | ||
| Rsol4 Rhizoctonia solani | F: TTCACACCTGCTCCTCTTT | 19 | 59.5 | 128 |
| R: TTCATCTGCATTTACCTTGG | 20 | 60.8 | ||
| Probe: FAM-TGCTTGGTTCCACTCAGCG-BHQ1 | 19 | 67.2 | ||
| Ccoc Colletotrichum coccodes | F: ACTTGTTCGAATAGGGTAACC | 21 | 60.5 | 115 |
| R: TAGGGCACAGTCAGTAATTCA | 21 | 60.5 | ||
| Probe: FAM-AACCAGACAGACGCCAACGA-BHQ1 | 20 | 68.5 | ||
| Sss Spongospora subterranea | F: GCCTCTTTGAGTGTCGGTT | 19 | 62 | 124 |
| R: AATCAGAAGCCAGAGACGC | 19 | 62 | ||
| Probe: FAM-TGTGCGTGGAAGGGGACTA-BHQ1 | 19 | 66 | ||
| Fus Fusarium spp. | F: TTGATCTACCAGTGCGGTG | 19 | 60 | 369 |
| R: GATACCACGCTCACGCTC | 18 | 60 | ||
| Probe: FAM-TGAGCTTGTCAAGAACCCAGG-BHQ1 | 21 | 67 |
| Species | Isolate | Ct Values (Ct ± SE) Obtained for the Pathogens Included into the Specificity Test 1 | |||||
|---|---|---|---|---|---|---|---|
| Asol4 | Aalt1 | Rsol4 | Fus | Ccoc | Sss | ||
| Alternaria solani | 100053 | 27.23 ± 0.15 | – | – | – | – | – |
| A. solani | 043-021 | 24.79 ± 0.16 | – | – | – | – | – |
| A. solani | 044-051 | 25.23 ± 0.05 | – | ||||
| Alternaria alternata | 4l/6 | – | 20.62 ± 0.47 | – | – | – | – |
| A. alternata | 4k/2 | – | 21.13 ± 0.90 | ||||
| A. alternata | AK2 | – | 21.52 ± 0.71 | ||||
| A. alternata | HKK1 | – | 22.02 ± 0.96 | ||||
| A. alternata | 12RKL9 | – | 24.86 ± 0.01 | – | – | – | – |
| A. alternata | 12RKL10 | – | 22.93 ± 0.95 | ||||
| A. infectoria | K100088 | – | – | – | – | – | – |
| A. longipes | K100055 | – | – | – | – | – | – |
| Rhizoctonia solani | 100006 | – | – | 16.53 ± 0.32 | – | – | – |
| R. solani | 100063 | – | – | 12.61 ± 0.08 | – | – | – |
| R. solani | 100106 | 16.29 ± 1.73 | |||||
| R. solani | R15BKK nev25 | 17.37 ± 0.24 | |||||
| R. solani | 14BM rs2 | 17.54 ± 0.46 | |||||
| R. solani | 14KC man1 | 20.29 ± 0.48 | |||||
| R. cerealis | 1000025 | – | |||||
| Colletotrichum coccodes | 100004 | – | – | – | – | 27.85 ± 0.37 | – |
| C. coccodes | 100123(3) | 28.14 ± 0.15 | |||||
| C. coccodes | 100124(3) | – | – | – | – | 25.24 ± 0.12 | – |
| C. coccodes | 100119(3) | 25.94 ± 0.39 | |||||
| C. coccodes | 100120(3) | 24.25 ± 0.65 | |||||
| C. coffeanum | 100003 | – | – | – | – | – | – |
| C. gloeosporioides | 100002 | – | – | – | – | – | – |
| C. gloeosporioides | 100094 | – | |||||
| C. lilii | 100001 | – | |||||
| C. dematium | 100158 | – | |||||
| Fusarium avenaceum | MOK-16-3 | – | – | – | 25.13 ± 0.10 | – | – |
| F. avenaceum | 110501 | 32.32 ± 0.29 | |||||
| F. avenaceum | U-08-8-1 | 23.86 ± 1.02 | |||||
| F. culmorum | MKRS-15-3 | 31.60 ± 0.59 | |||||
| F. culmorum | 100135(6-1) | 23.30 ± 0.12 | |||||
| F. culmorum | 100136(6-2) | – | – | – | 30.68 ± 1.09 | – | – |
| F. gibbosum | 100130(7-2) | – | – | – | 32.87 ± 2.13 | – | – |
| F. gibbosum | 100131(7-2) | 26.11 ± 0.78 | |||||
| F. gibbosum | 100132(7) | 26.44 ± 0.74 | |||||
| F. heterosporum | MOK-16-1 | 31.06 ± 0.89 | |||||
| F. heterosporum | 100133(8) | 32.70 ± 1.56 | |||||
| F. oxysporum | FO-1 | 31.72 ± 0.29 | |||||
| F. oxysporum | 100139(9) | 26.04 ± 0.45 | |||||
| F. oxysporum | 100140(9) | – | – | – | 25.09 ± 0.52 | ||
| F. oxysporum | RAM-14 | 34.02 ± 0.59 | |||||
| F. sambucinum | P-2-02 | 28.05 ± 0.16 | |||||
| F. sambucinum | KRT 11-1 kch | – | – | – | 24.04 ± 0.08 | – | – |
| F. sambucinum | 100134(10) | 26.05 ± 0.49 | |||||
| F. solani | FSL-9 | 33.39 ± 0.78 | |||||
| F. solani | 100021 | – | – | – | 33.72 ± 0.69 | – | – |
| F. solani | 100137(11-1) | 32.40 ± 0.52 | |||||
| F. solani | 100138(11-2) | 30.62 ± 1.29 | |||||
| F. sporotrichioides | KRT12-1 kch | 31.96 ± 1.25 | |||||
| F. sporotrichioides | 100141(12) | 32.78 ± 0.96 | |||||
| F. javanicum | MKRS-15-1 | 33.35 ± 0.84 | |||||
| F. sacchari | RAM-16 | 33.65 ± 0.88 | |||||
| Phytophthora infestans | MVK 118a-07 | – | – | – | – | – | – |
| P. infestans | ATP-3.08 | – | – | – | – | – | – |
| Spongospora subterranea | Field sample, Sss-RS | – | – | – | – | – | 21.36 ± 0.05 |
| S. subterranea | Field sample, Sss-C | – | – | – | – | – | 20.53 ± 0.08 |
| Target Species | DNA Concentration, μg/mL | Threshold Cycle (Ct) 1 | Target Species | DNA Concentration, μg/mL | Threshold Cycle (Ct) |
|---|---|---|---|---|---|
| Alternaria solani | 58 | 24.42 ± 0.06 | Colletotrichum coccodes | 68 | 25.82 ± 0.15 |
| 5.8 | 27.73 ± 0.08 | 6.8 | 28.23 ± 0.67 | ||
| 0.58 | 31.64 ± 0.32 | 0.68 | 31.50 ± 0.35 | ||
| 0.058 | 34.32 ± 0.84 | 0.068 | 34.24 ± 0.57 | ||
| Alternaria alternata | 43 | 22.97 ± 0.08 | Rhizoctonia solani | 46 | 19.59 ± 0.12 |
| 4.3 | 26.66 ± 0.12 | 4.6 | 22.77 ± 0.20 | ||
| 0.43 | 28.65 ± 0.34 | 0.46 | 28.25 ± 0.66 | ||
| 0.043 | 32.09 ± 0.62 | 0.046 | 32.51 ± 0.94 | ||
| 0.0043 | 34.43 ± 0.76 | 0.0046 | 38.16 ± 1.04 | ||
| Fusarium sp. | 10 | 23.5 ± 0.07 | Spongospora subterranea | 20 | 18.22 ± 0.05 |
| 1 | 27.04 ± 0.24 | 2 | 21.36 ± 0.05 | ||
| 0.1 | 30.08 ± 0.33 | 0.2 | 25.02 ± 0.10 | ||
| 0.01 | 32.72 ± 0.78 | 0.02 | 27.25 ± 0.37 | ||
| 0.001 | 35.26 ±0.64 | 0.002 | 30.62 ± 0.66 | ||
| 0.0002 | 33.07 ± 0.64 |
| Test System | Number of Repeats | Ct Value (M ± SE) | SE (%) |
|---|---|---|---|
| Asol4 (Alternaria solani) | 14 | 27.23 ± 0.10 | 0.4 |
| Aalt1 (Alternaria alternata) | 14 | 16.20 ± 0.06 | 0.3 |
| Rsol4 (Rhizoctonia solani) | 14 | 20.24 ± 0.08 | 0.4 |
| Ccoc (Colletotrichum coccodes) | 9 | 27.39 ± 0.19 | 0.7 |
| Fus (Fusarium spp.) | 14 | 22.66 ± 0.09 | 0.4 |
| Sss (Spongospora subterranea) | 9 | 23.32 ± 0.17 | 0.7 |
| Test | Freshly Prepared | Lyophilized | ||
|---|---|---|---|---|
| System 1 | Ct (M ± SE) | Fluorescence Level at the End of Analysis | Ct (M ± SE) | Fluorescence Level at the End of Analysis |
| Asol4 | 24.06 ± 0.42 | 3000 | 26.50 ± 2.25 | 1300 |
| Aalt1 | 25.70 ± 0.05 | 5700 | 24.56 ± 0.42 | 1400 |
| Rsol4 | 19.54 ± 0.38 | 5600 | 18.80 ± 0.25 | 1900 |
| Ccoc | 27.25 ± 0.39 | 3500 | 26.61 ± 1.02 | 980 |
| Fus | 25.40 ± 0.25 | 7500 | 24.49 ± 0.88 | 1500 |
| Sss | 26.79 ±0.03 | 6800 | 26.56 ± 0.09 | 2200 |
| Target Species | DNA Concentration, μg/mL | Threshold Cycle (Ct)1 | Target Species | DNA Concentration, μg/mL | Threshold Cycle (Ct) |
|---|---|---|---|---|---|
| Alternaria solani | 136.4 | 26.50 ± 0.24 | Alternaria alternata | 43.5 | 20.64 ± 0.39 |
| 13.64 | 29.23 ± 0.22 | 4.35 | 23.74 ± 0.32 | ||
| 1.364 | 32.87 ± 0.41 | 0.435 | 27.73 ± 0.38 | ||
| 0.1364 | 34.08 ± 0.86 | 0.0435 | 31.85 ± 0.59 | ||
| 0.01364 | 37.18 ± 0.96 | 0.00435 | 35.51 ± 0.86 | ||
| Rhizoctonia solani | 18.32 | 18.40 ± 0.40 | Colletotrichum coccodes | 19.3 | 24.47 ± 0.38 |
| 1.832 | 24.41 ± 0.70 | 1.93 | 29.84 ± 0.47 | ||
| 0.1832 | 31.57 ± 0.98 | 0.193 | 34.09 ±0.55 | ||
| 0.01832 | 34.33 ± 1.14 | 0.0193 | 37.10 ± 0.85 | ||
| Fusarium sp. | 44.6 | 22.42 ± 0.24 | Spongospora subterranea | 23.2 | 23.35 ± 0.35 |
| 4.46 | 24.50 ± 0.33 | 2.32 | 26.46 ± 0.27 | ||
| 0.446 | 28.54 ± 0.48 | 0.232 | 29.43 ± 0.39 | ||
| 0.0446 | 31.73 ± 0.64 | 0.0232 | 32.73 ± 0.62 | ||
| 0.00446 | 34.73 ± 0.62 | 0.00232 | 35.37 ± 0.76 |
| Sample No. 1 | Origin 2 | Visual Symptoms | Microscopic Examination 3 | Real-Time PCR Analysis 4 |
|---|---|---|---|---|
| T1 | Leningrad region, Russia | Dry and dark surface lesions | Aalt Sss | Aalt (25.2) Sss (30.4) |
| T2 | Leningrad region, Russia | Dry and dark surface lesions | Sss | Sss (31.5) |
| T3 | Moscow region, Russia (Red Scarlett) | Black scurf (Rhizoctonia solani) | Rsol | Rsol (25.6) |
| T4 | Tambov region, Russia | Superficial scabs (powdery scab suspected) | No spore balls of S. subterranea | – |
| T5 | Omsk region, Russia | Superficial scabs (powdery or common scab suspected) | No spore balls of S. subterranea | – |
| T6 | Unknown (bought in a supermarket) | Black scurf, superficial scabs and dry lesions | Sss Rsol Aalt (single conidia) | Sss (26.7) Ccoc (29.2) Rsol (21.4) Aalt (34.3) |
| T7 | Leningrad region (Red Scarlett) | Black scurf, superficial dry lesions | Rsol Aalt Ccoc | Ccoc (31.4) Aalt (27.3) Rsol (20.4) |
| T8 | Moscow region (Red Scarlett) | Dry rot | Fus (F. oxysporum) | Fus (17.6) |
| T9 | Moscow region (Red Scarlett) | Dry rot | Fus (F. oxysporum) | Fus (21.4) |
| L1 | Republic of Tatarstan, Russia | Brown spot disease | Aalt | Aalt (29.4) |
| L2 | Republic of Kabardino-Balkaria, Russia 5 | Brown spot disease | Aalt | Aalt (33.7) |
| L3 | Moscow region, Russia (Red Scarlett) | Brown spot disease | Aalt | Aalt (27.7) |
| L4 | Moscow region, Russia (Alpha) | Early blight/brown spot | Aalt | Aalt (34.1) |
| L5 | Moscow region, Russia (Udacha) | Early blight/brown spot | Asol Aalt (single conidia) | Asol (26.8) Aalt (36.1) |
| L6 | Tambov region (Alouette) | Early blight/brown spot | Asol (single conidia) Aalt | Aalt (28.3) |
| L7 | Tambov region (Alouette) | Early blight/brown spot | – | – |
| L8 | Tambov region (Alouette) | Early blight/brown spot | Aalt | Aalt (32.5) |
| S1 | Moscow region, Russia (Red Scarlett) | Stem canker (R. solani) | Rsol | Rsol |
| S2 | Moscow region, Russia (Red Scarlett) | Black dot (C. coccodes) | Ccoc | Ccoc (32.1) |
| S3 | Tyumen region, Russia (Red Scarlett) | Stem canker | Rsol | Rsol (30.1) |
| S4 | Tyumen region, Russia (Evolution) | Stem canker | Rsol | Rsol (26.1) |
| S5 | Tyumen region, Russia (Red Scarlett) | Black dot suspected | – | – |
| S6 | Sverdlovsk region, Russia (Rozara) | Stem canker, black dot | Rsol, Ccoc, | Rsol (32.0) Ccoc (30.0) |
| S7 | Sverdlovsk region, Russia (Rozara) | Stem canker, black dot (suspected) | Rsol Ccoc | Rsol (33.2) Ccoc (34.0) |
| S8 | Sverdlovsk region, Russia (Gala) | Stem canker | Rsol | Rsol (24.3) |
| S9 | Kurgan region, Russia (Rozara) | Stem canker | Rsol | Rsol (34.0) |
| S10 | Chelyabinsk region, Russia (Red Scarlett) | Stem canker | Rsol | Rsol (34.2) |
| S11 | Chelyabinsk region, Russia (Red Scarlett) | Black dot | Ccoc | Ccoc (35.0) |
| S12 | Chelyabinsk region, Russia (Red Scarlett) | Stem canker | Rsol | Rsol (32.7) |
| S13 | Chelyabinsk region, Russia (Red Scarlett) | Stem canker suspected | – | – |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikitin, M.; Deych, K.; Grevtseva, I.; Girsova, N.; Kuznetsova, M.; Pridannikov, M.; Dzhavakhiya, V.; Statsyuk, N.; Golikov, A. Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format. Biosensors 2018, 8, 129. https://doi.org/10.3390/bios8040129
Nikitin M, Deych K, Grevtseva I, Girsova N, Kuznetsova M, Pridannikov M, Dzhavakhiya V, Statsyuk N, Golikov A. Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format. Biosensors. 2018; 8(4):129. https://doi.org/10.3390/bios8040129
Chicago/Turabian StyleNikitin, Maksim, Ksenia Deych, Inessa Grevtseva, Natalya Girsova, Maria Kuznetsova, Mikhail Pridannikov, Vitaly Dzhavakhiya, Natalia Statsyuk, and Alexander Golikov. 2018. "Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format" Biosensors 8, no. 4: 129. https://doi.org/10.3390/bios8040129
APA StyleNikitin, M., Deych, K., Grevtseva, I., Girsova, N., Kuznetsova, M., Pridannikov, M., Dzhavakhiya, V., Statsyuk, N., & Golikov, A. (2018). Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format. Biosensors, 8(4), 129. https://doi.org/10.3390/bios8040129





