Rapid, Portable, Multiplexed Detection of Bacterial Pathogens Directly from Clinical Sample Matrices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Platform Development
2.2. Microfluidic Disc Design and Fabrication
2.3. Reagent Preparation
2.4. Immunoassay Protocol
2.5. ELISA Protocol
2.6. Safe Handling of Human and Animal Products
2.7. Multiplexed Analysis
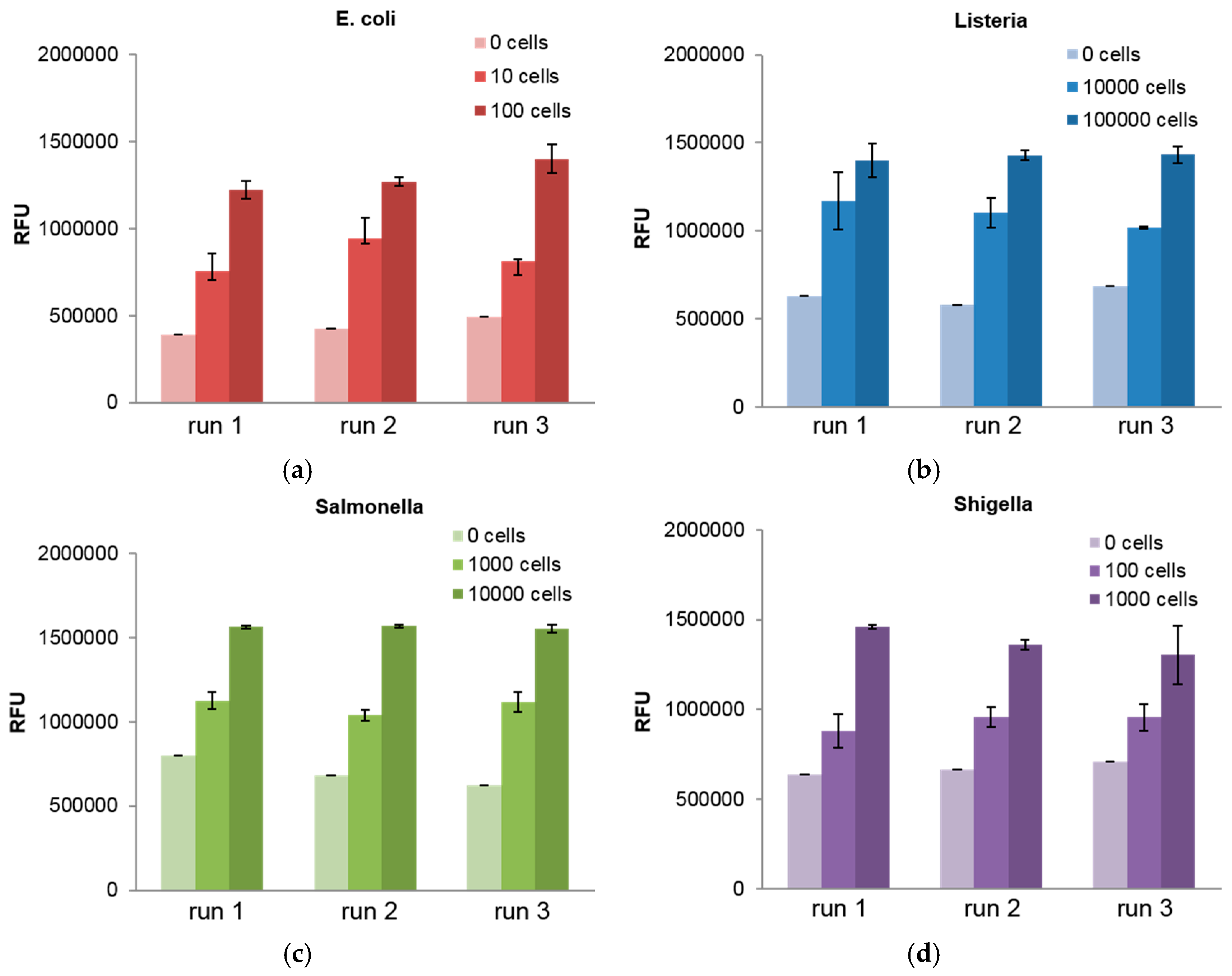
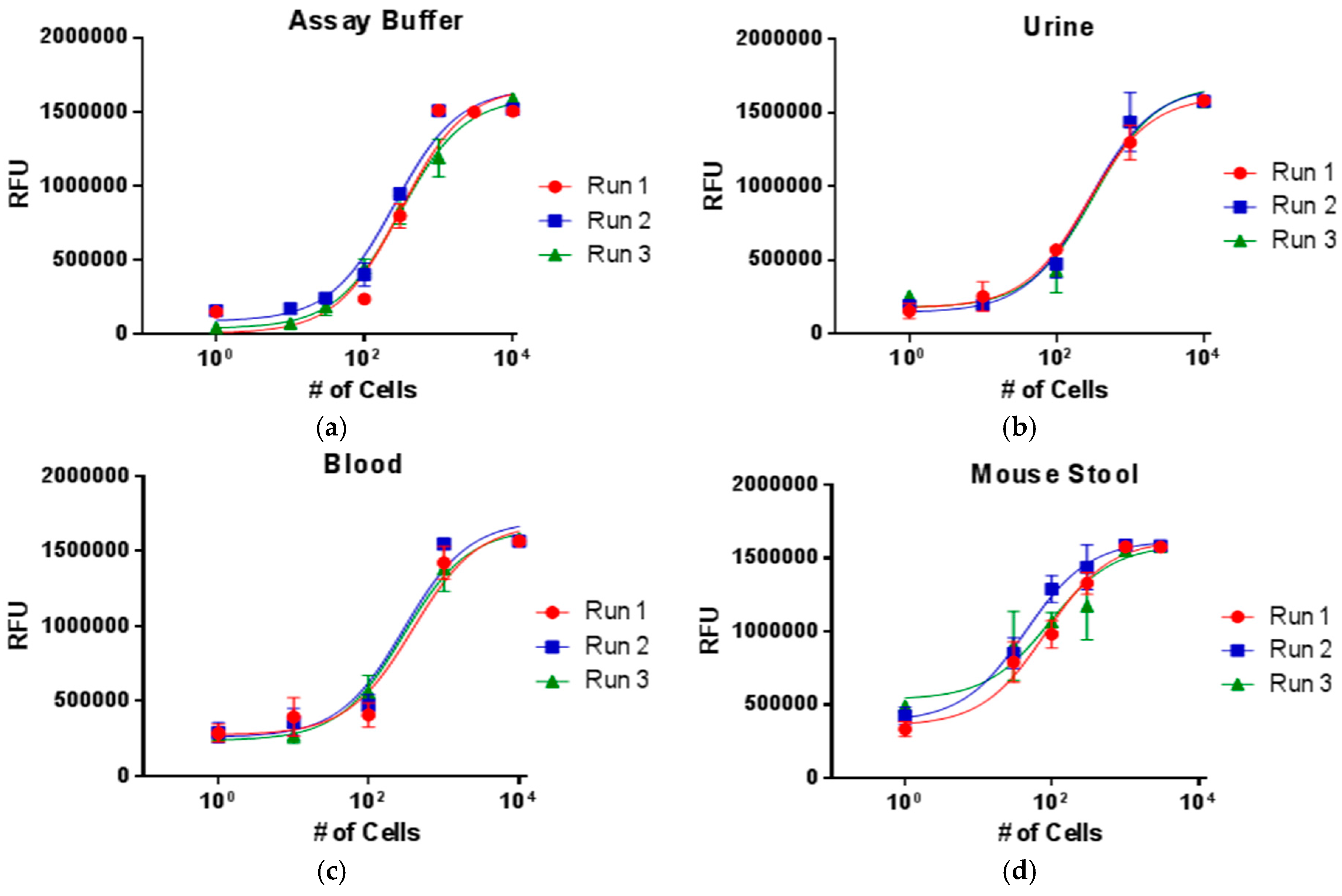
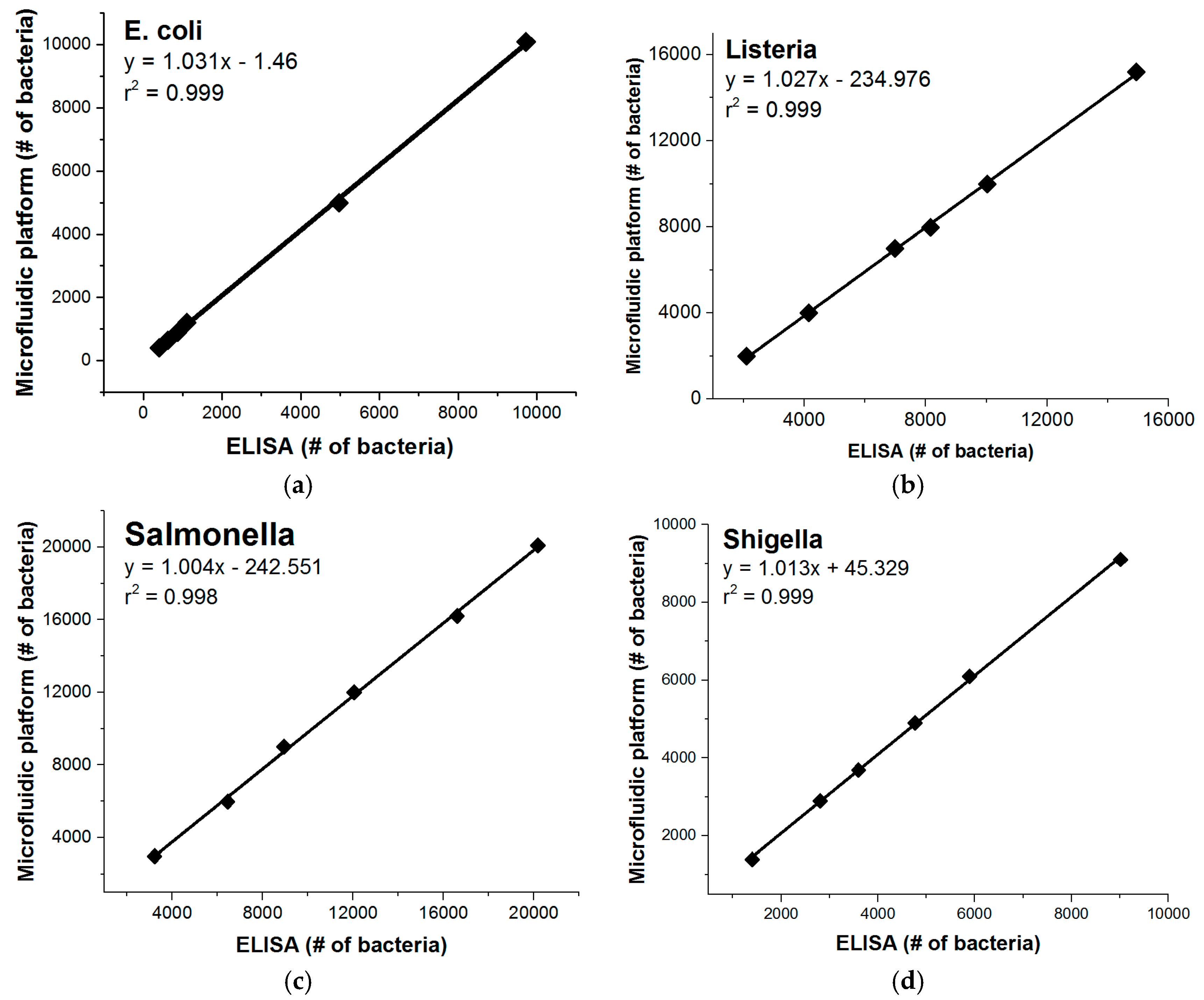
3. Results
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Black, R.E.; Morris, S.S.; Bryce, J. Where and why are 10 million children dying every year? Lancet 2003, 361, 2226–2234. [Google Scholar] [CrossRef]
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- Lanata, C.F.; Fischer-Walker, C.L.; Olascoaga, A.C.; Torres, C.X.; Aryee, M.J.; Black, R.E. Global Causes of Diarrheal Disease Mortality in Children <5 Years of Age: A Systematic Review. PLoS ONE 2013, 8, e72788. [Google Scholar]
- Wazny, K.; Zipursky, A.; Black, R.; Curtis, V.; Duggan, C.; Guerrant, R.; Levine, M.; Petri, W.A., Jr.; Santosham, M.; Scharf, R.; et al. Setting Research Priorities to Reduce Mortality and Morbidity of Childhood Diarrhoeal Disease in the Next 15 Years. PLOS Med. 2013, 10, e1001446. [Google Scholar] [CrossRef] [PubMed]
- Wardlaw, T.; Salama, P.; Brocklehurst, C.; Chopra, M.; Mason, E. Diarrhoea: Why children are still dying and what can be done. Lancet 2010, 375, 870–872. [Google Scholar] [CrossRef]
- Bunyakul, N.; Baeumner, A.J. Combining Electrochemical Sensors with Miniaturized Sample Preparation for Rapid Detection in Clinical Samples. Sensors 2014, 14, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, J.A.; Operario, D.J.; Houpt, E.R. Molecular Diagnosis of Diarrhea: Current Status and Future Potential. Curr. Infect. Dis. Rep. 2011, 14, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Harrington, S.M.; Buchan, B.W.; Doern, C.; Fader, R.; Ferraro, M.J.; Pillai, D.R.; Rychert, J.; Doyle, L.; Lainesse, A.; Karchmer, T.; et al. Multicenter Evaluation of the BD Max Enteric Bacterial Panel PCR Assay for Rapid Detection of Salmonella spp., Shigella spp., Campylobacter spp. (C. jejuni and C. coli), and Shiga Toxin 1 and 2 Genes. J. Clin. Microbiol. 2015, 53, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Yager, P.; Edwards, T.; Fu, E.; Helton, K.; Nelson, K.; Tam, M.R.; Weigl, B.H. Microfluidic diagnostic technologies for global public health. Nature 2006, 442, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Ravikumar, A.; Seubert, J.; Figueira, S. Detection of bacterial pathogens through microfluidic DNA sensors and mobile interface toward rapid, affordable, and point-of-care water monitoring. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 1–4.
- Ghosh Dastider, S.; Barizuddin, S.; Yuksek, N.S.; Dweik, M.; Almasri, M.F. Efficient and Rapid Detection of Salmonella Using Microfluidic Impedance Based Sensing. J. Sens. 2015, 2015, e293461. [Google Scholar] [CrossRef]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Song, X.C.; Mrázek, J.; Lamačová, J.; Scott Lynn, N., Jr.; Šedivák, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Czilwik, G.; Messinger, T.; Strohmeier, O.; Wadle, S.; von Stetten, F.; Paust, N.; Roth, G.; Zengerle, R.; Saarinen, P.; Niittymäki, J.; et al. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic LabDisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip 2015, 15, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Weigl, B.H.; Gerdes, J.; Tarr, P.; Yager, P.; Dillman, L.; Peck, R.; Ramachandran, S.; Lemba, M.; Kokoris, M.; Nabavi, M.; et al. Fully integrated multiplexed lab-on-a-card assay for enteric pathogens. In Proceedings of the Microfluidics, BioMEMS, and Medical Microsystems IV, San Jose, CA, USA, 21 January 2006; Volume 6112.
- Amasia, M.; Cozzens, M.; Madou, M.J. Centrifugal microfluidic platform for rapid PCR amplification using integrated thermoelectric heating and ice-valving. Sens. Actuators B Chem. 2012, 161, 1191–1197. [Google Scholar] [CrossRef]
- Oh, S.J.; Park, B.H.; Choi, G.; Seo, J.H.; Jung, J.H.; Choi, J.S.; Kim, D.H.; Seo, T.S. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip 2016, 16, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, Y.T.; Lee, J.W.; Kim, D.H.; Seo, T.S. An integrated direct loop-mediated isothermal amplification microdevice incorporated with an immunochromatographic strip for bacteria detection in human whole blood and milk without a sample preparation step. Biosens. Bioelectron. 2016, 79, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Park, B.H.; Jung, J.H.; Choi, G.; Lee, D.C.; Kim, D.H.; Seo, T.S. Centrifugal loop-mediated isothermal amplification microdevice for rapid, multiplex and colorimetric foodborne pathogen detection. Biosens. Bioelectron. 2016, 75, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Felipe, S.; Tortajada-Genaro, L.A.; Carrascosa, J.; Puchades, R.; Maquieira, Á. Real-time loop-mediated isothermal DNA amplification in compact disc micro-reactors. Biosens. Bioelectron. 2016, 79, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Sayad, A.A.; Ibrahim, F.; Uddin, S.M.; Pei, K.X.; Mohktar, M.S.; Madou, M.; Thong, K.L. A microfluidic lab-on-a-disc integrated loop mediated isothermal amplification for foodborne pathogen detection. Sens. Actuators B Chem. 2016, 227, 600–609. [Google Scholar] [CrossRef]
- Sayad, A.A.; Ibrahim, F.; Mohktar, M.S.; Thong, K.L. Salmonella Detection on Microfluidic CD Using Loop Mediated Isothermal Amplification. In Proceedings of the 7th WACBE World Congress on Bioengineering 2015, Singapore, 6–8 July 2015; Goh, J., Lim, C.T., Eds.; Springer: Cham, Switzerland, 2015; pp. 43–46. [Google Scholar]
- Xia, Y.; Liu, Z.; Yan, S.; Yin, F.; Feng, X.; Liu, B.-F. Identifying multiple bacterial pathogens by loop-mediated isothermal amplification on a rotate & react slipchip. Sens. Actuators B Chem. 2016, 228, 491–499. [Google Scholar]
- Madou, M.J.; Kellogg, G.J. LabCD: A centrifuge-based microfluidic platform for diagnostics. In Proceedings of Systems and Technologies for Clinical Diagnostics and Drug Discovery, San Jose, CA, USA, 27 January 1998; Volume 3259, pp. 80–93.
- Smith, S.; Mager, D.; Perebikovsky, A.; Shamloo, E.; Kinahan, D.; Mishra, R.; Torres Delgado, S.M.; Kido, H.; Saha, S.; Ducrée, J.; et al. CD-Based Microfluidics for Primary Care in Extreme Point-of-Care Settings. Micromachines 2016, 7, 22. [Google Scholar] [CrossRef]
- Gilmore, J.; Islam, M.; Martinez-Duarte, R. Challenges in the Use of Compact Disc-Based Centrifugal Microfluidics for Healthcare Diagnostics at the Extreme Point of Care. Micromachines 2016, 7, 52. [Google Scholar] [CrossRef]
- Gorkin, R.; Park, J.; Siegrist, J.; Amasia, M.; Lee, B.S.; Park, J.-M.; Kim, J.; Kim, H.; Madou, M.; Cho, Y.-K. Centrifugal microfluidics for biomedical applications. Lab Chip 2010, 10, 1758–1773. [Google Scholar] [CrossRef] [PubMed]
- Strohmeier, O.; Keller, M.; Schwemmer, F.; Zehnle, S.; Mark, D.; von Stetten, F.; Zengerle, R.; Paust, N. Centrifugal microfluidic platforms: Advanced unit operations and applications. Chem. Soc. Rev. 2015, 44, 6187–6229. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Wang, G.; Kong, S.-K.; Ho, H.-P. A Review of Biomedical Centrifugal Microfluidic Platforms. Micromachines 2016, 7, 26. [Google Scholar] [CrossRef]
- Schaff, U.Y.; Sommer, G.J. Whole Blood Immunoassay Based on Centrifugal Bead Sedimentation. Clin. Chem. 2011, 57, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Iii, D.I.W.; Sommer, G.J.; Schaff, U.Y.; Hahn, P.S.; Jaffe, G.J.; Murthy, S.K. A centrifugal fluidic immunoassay for ocular diagnostics with an enzymatically hydrolyzed fluorogenic substrate. Lab Chip 2014, 14, 2673–2680. [Google Scholar]
- Koh, C.-Y.; Schaff, U.Y.; Piccini, M.E.; Stanker, L.H.; Cheng, L.W.; Ravichandran, E.; Singh, B.-R.; Sommer, G.J.; Singh, A.K. Centrifugal Microfluidic Platform for Ultrasensitive Detection of Botulinum Toxin. Anal. Chem. 2015, 87, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, J.; Moen, S.T.; Koh, C.-Y.; Singh, A.K. Centrifugal sedimentation immunoassays for multiplexed detection of enteric bacteria in ground water. Biomicrofluidics 2016, 10, 14103. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Xu, K.; Qin, S.; Zhou, H.-X. Effect of Macromolecular Crowding on Protein Binding Stability: Modest Stabilization and Significant Biological Consequences. Biophys. J. 2009, 97, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Best, R.B.; Mittal, J. Macromolecular crowding effects on protein–protein binding affinity and specificity. J. Chem. Phys. 2010, 133, 205101. [Google Scholar] [CrossRef] [PubMed]


| LOD (# of Cells) | |||||||
|---|---|---|---|---|---|---|---|
| Microfluidic Singleplex | Microfluidic Multiplex | Conventional Singleplex | |||||
| Buffer | Urine | Blood | Stool | Buffer | Stool | Buffer | |
| E. coli | 11 | 34 | 33 | 9 | 51 | 31 | 38 |
| Listeria | 999 | 796 | 1668 | 320 | 2849 | 238 | 1745 |
| Salmonella | 2416 | 703 | 1200 | 974 | 1154 | 328 | 2648 |
| Shigella | 53 | 33 | 61 | 20 | 94 | 12 | 1236 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phaneuf, C.R.; Mangadu, B.; Piccini, M.E.; Singh, A.K.; Koh, C.-Y. Rapid, Portable, Multiplexed Detection of Bacterial Pathogens Directly from Clinical Sample Matrices. Biosensors 2016, 6, 49. https://doi.org/10.3390/bios6040049
Phaneuf CR, Mangadu B, Piccini ME, Singh AK, Koh C-Y. Rapid, Portable, Multiplexed Detection of Bacterial Pathogens Directly from Clinical Sample Matrices. Biosensors. 2016; 6(4):49. https://doi.org/10.3390/bios6040049
Chicago/Turabian StylePhaneuf, Christopher R., Betty Mangadu, Matthew E. Piccini, Anup K. Singh, and Chung-Yan Koh. 2016. "Rapid, Portable, Multiplexed Detection of Bacterial Pathogens Directly from Clinical Sample Matrices" Biosensors 6, no. 4: 49. https://doi.org/10.3390/bios6040049
APA StylePhaneuf, C. R., Mangadu, B., Piccini, M. E., Singh, A. K., & Koh, C.-Y. (2016). Rapid, Portable, Multiplexed Detection of Bacterial Pathogens Directly from Clinical Sample Matrices. Biosensors, 6(4), 49. https://doi.org/10.3390/bios6040049





