The Burgeoning Importance of Nanomotion Sensors in Microbiology and Biology
Abstract
1. Introduction
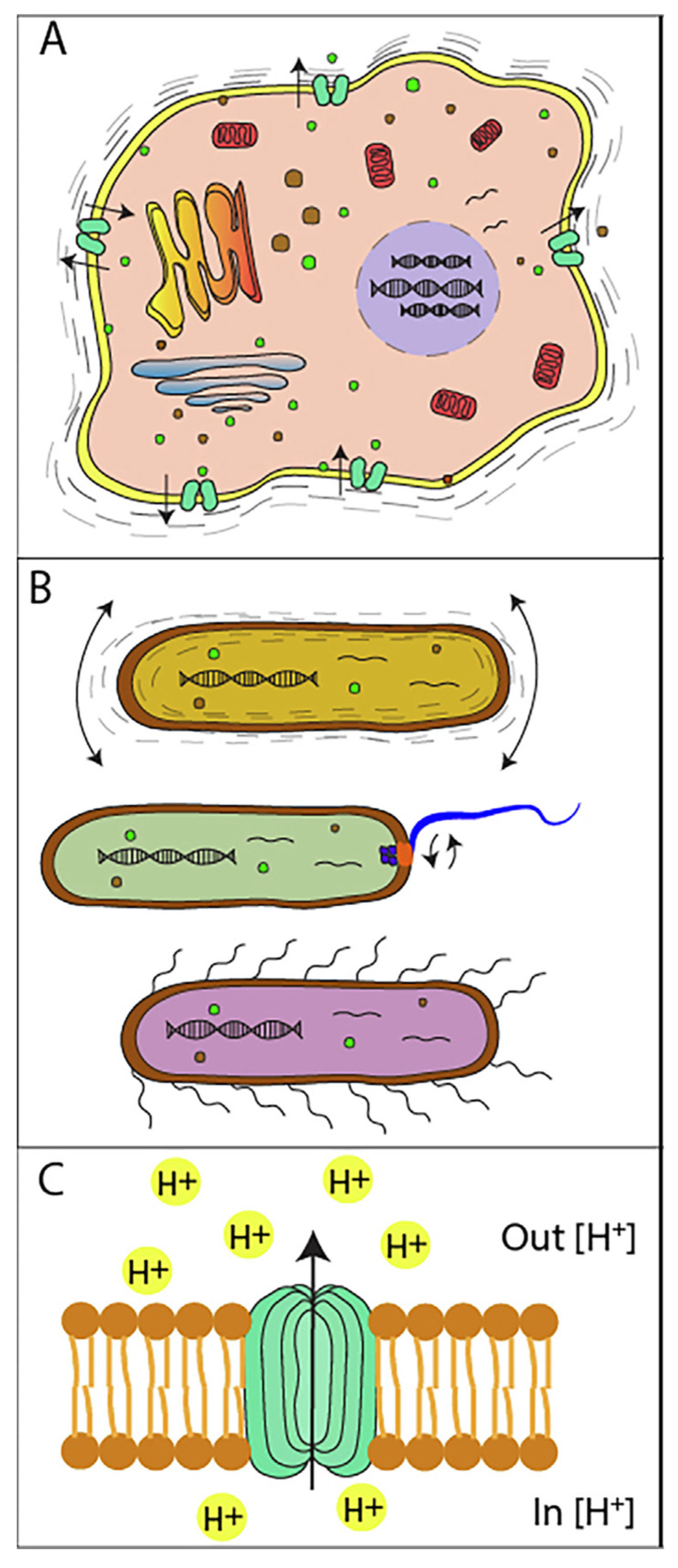
2. Nanomechanical Sensors
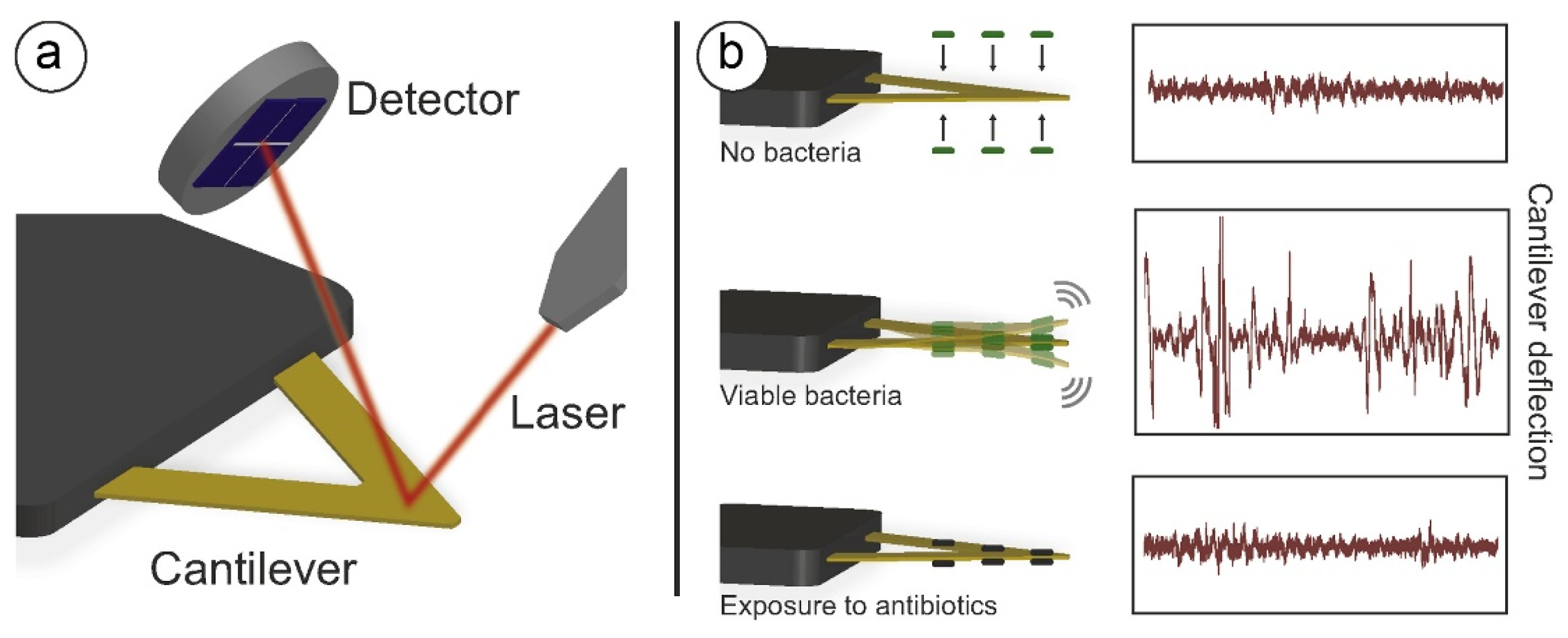
3. AFM-Based Nanomotion Sensor
3.1. The Principle of the AFM-Based Banomotion Sensor
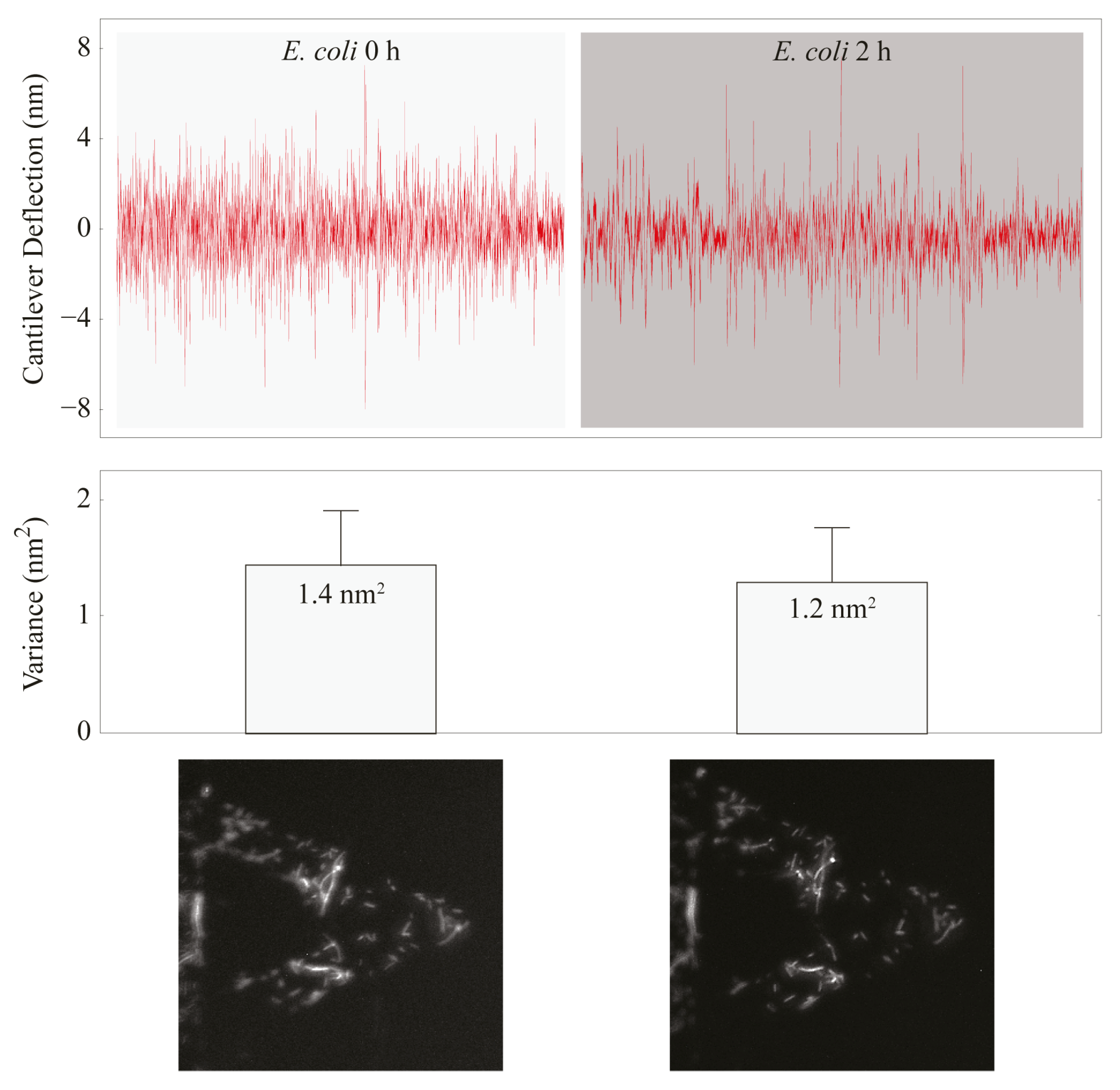
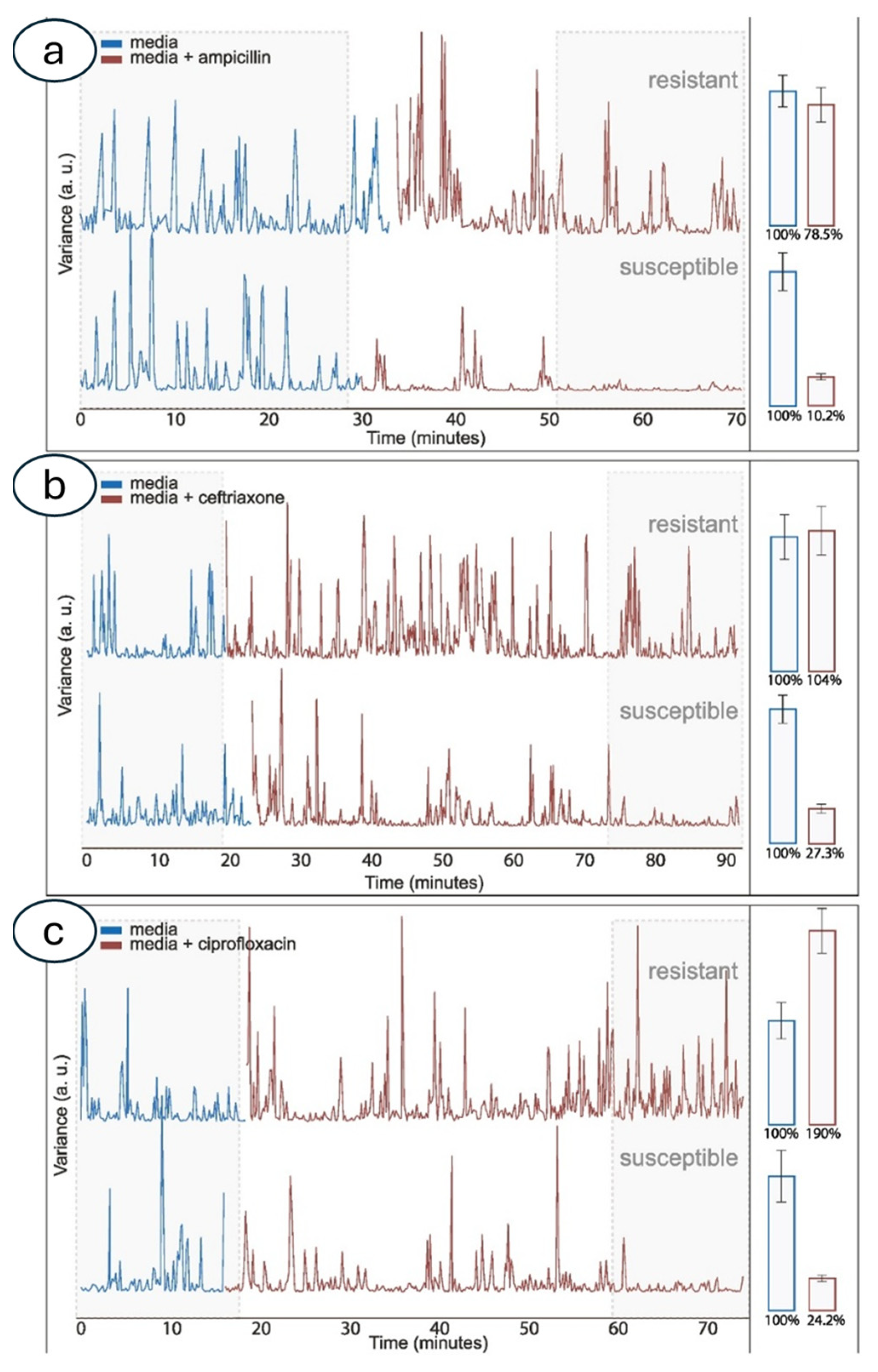
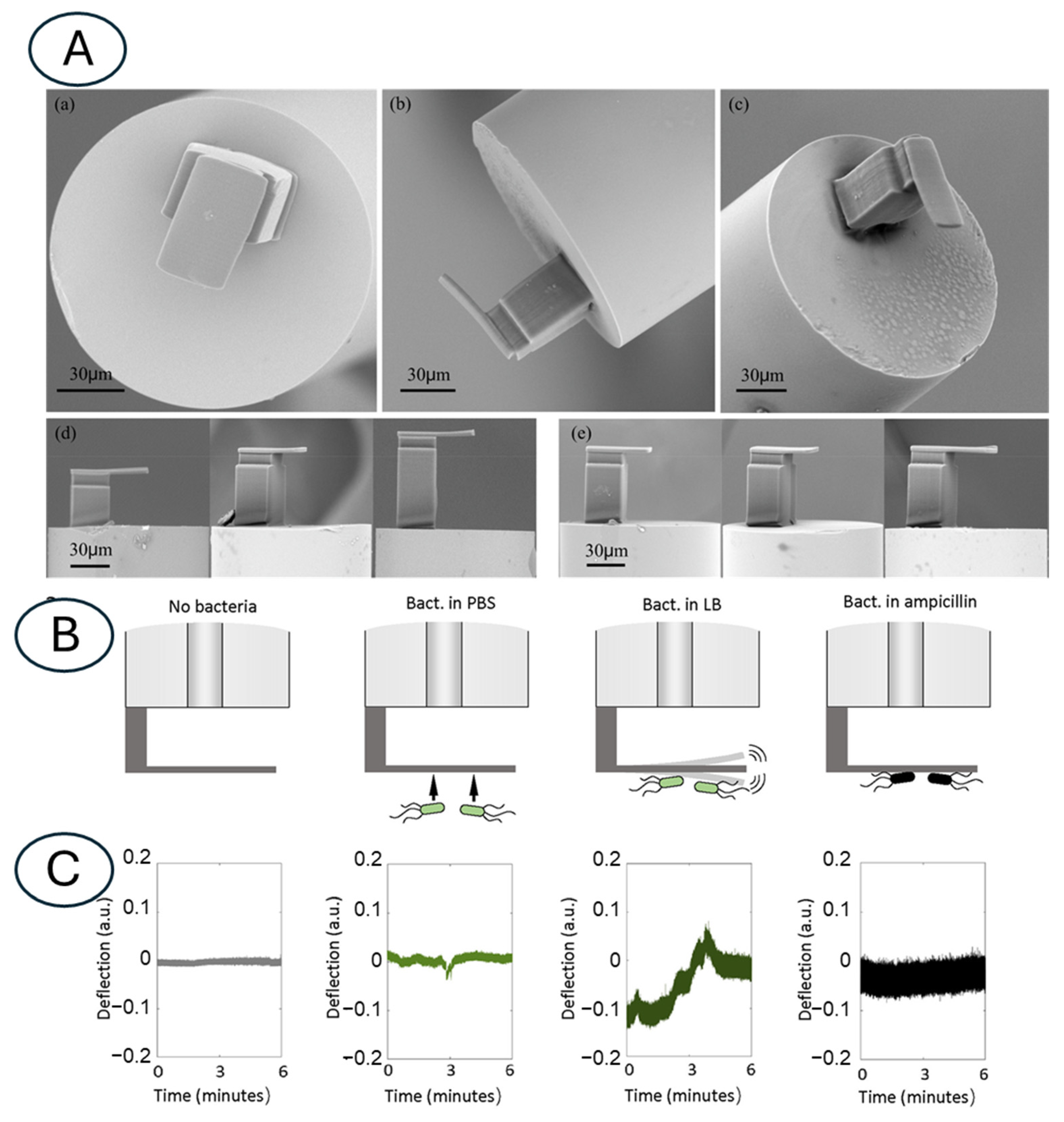
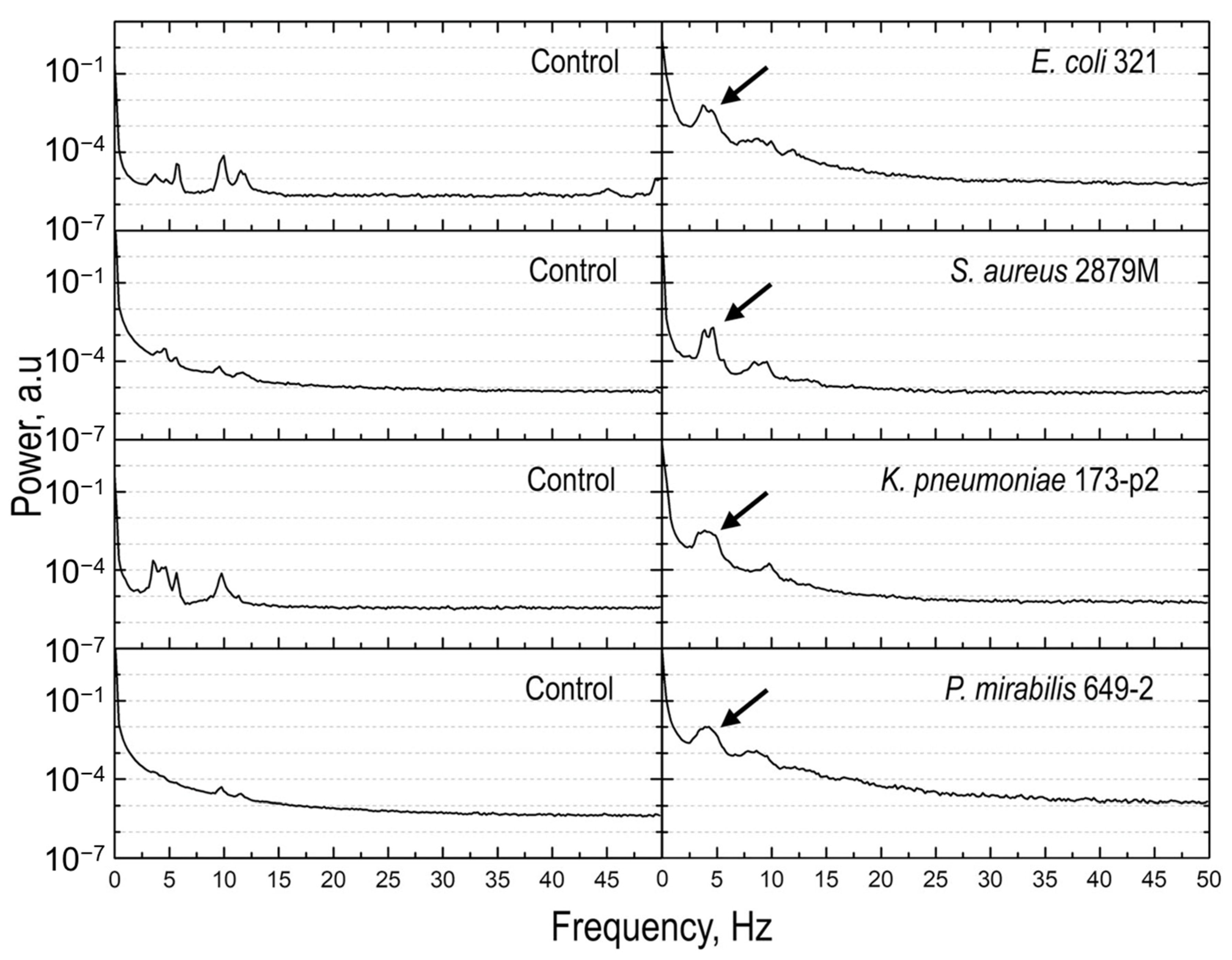
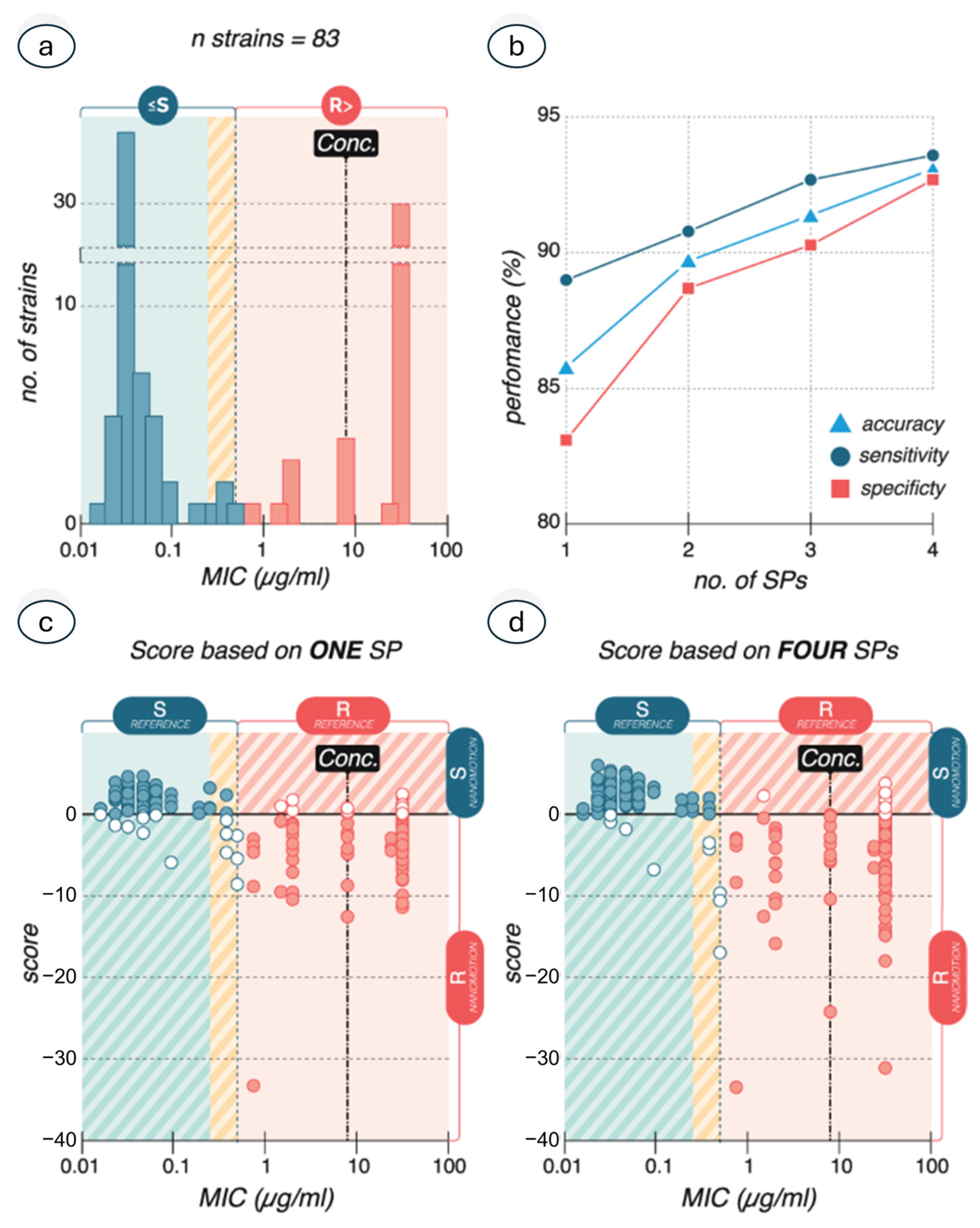
3.2. Atomic Force Microscopy-Based Nanomotion Antibiotic Susceptibility Test
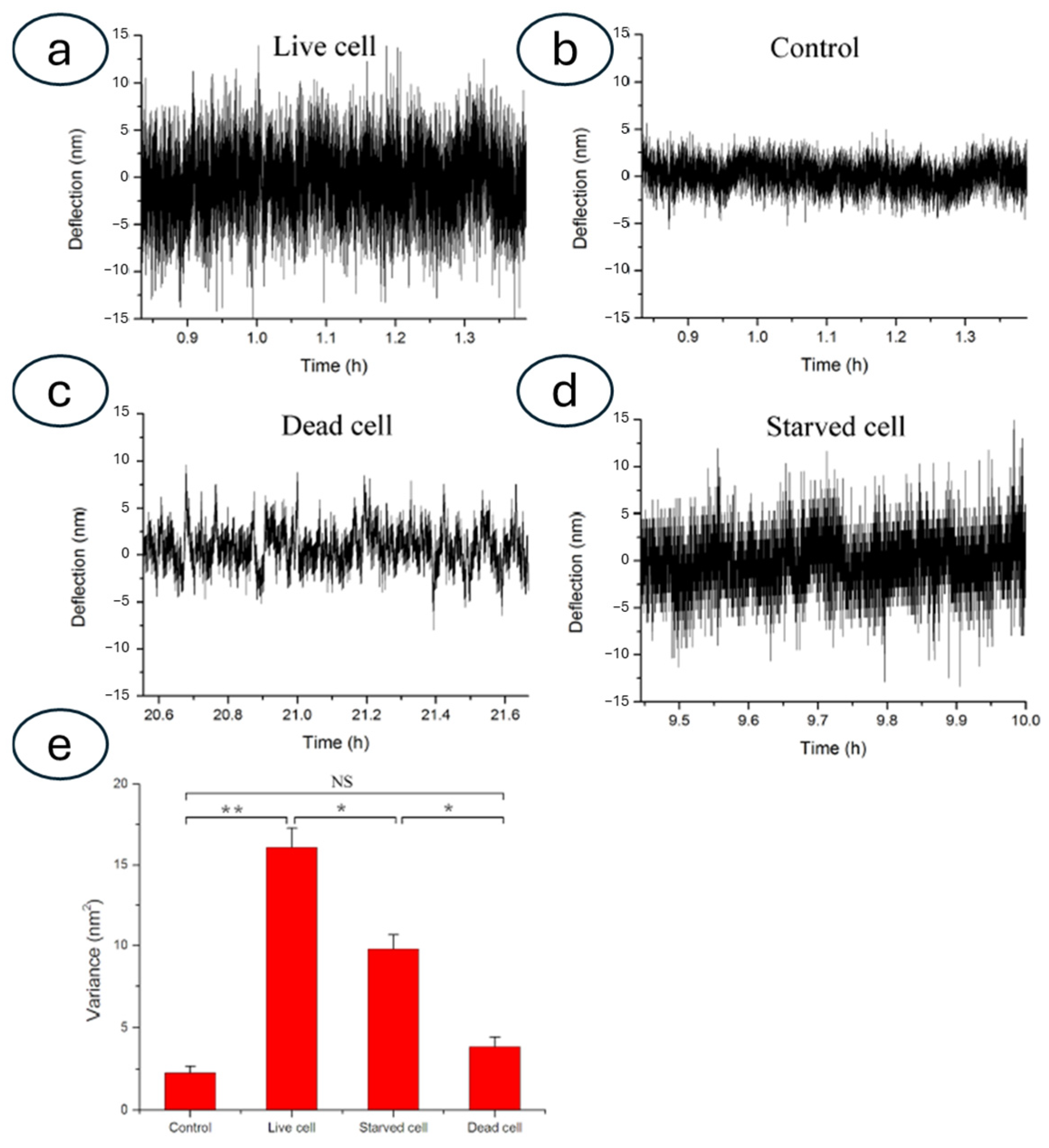
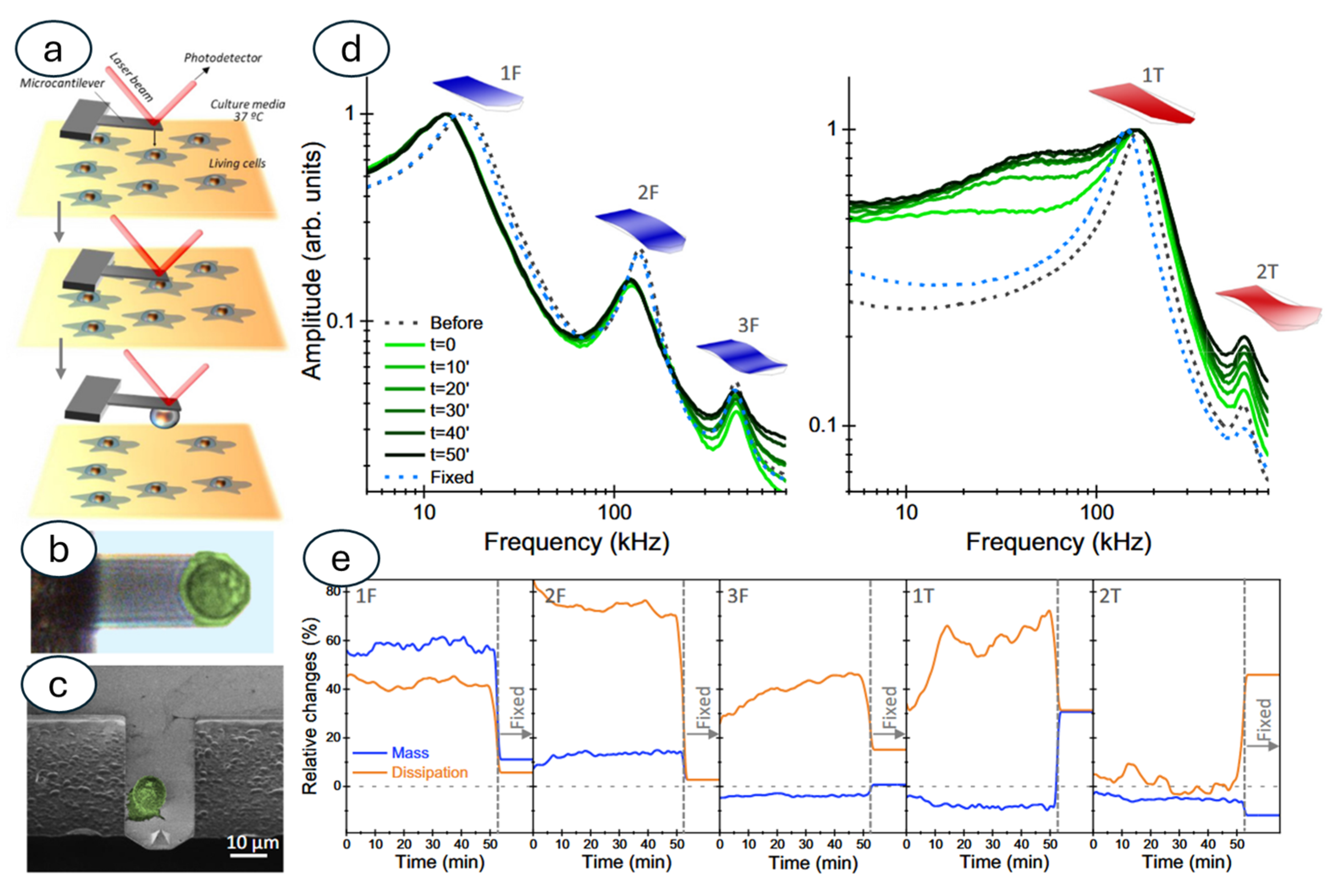
3.3. Nanomotion Sensor to Study Other Biological Specimens: Mammalian Cells and Sub-Cellular Organelles
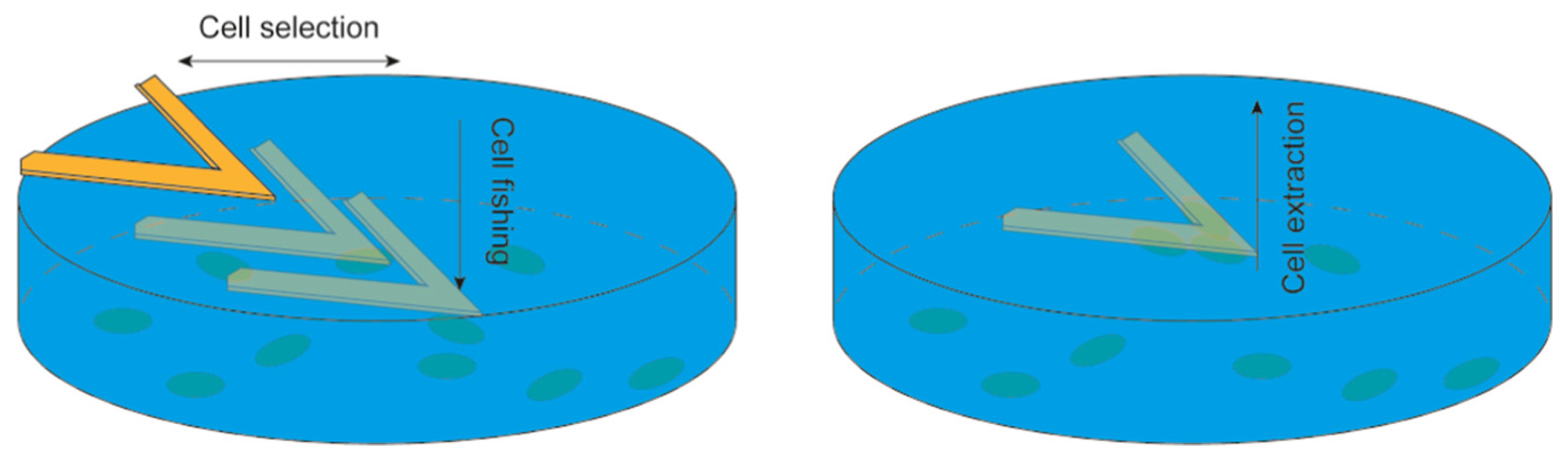
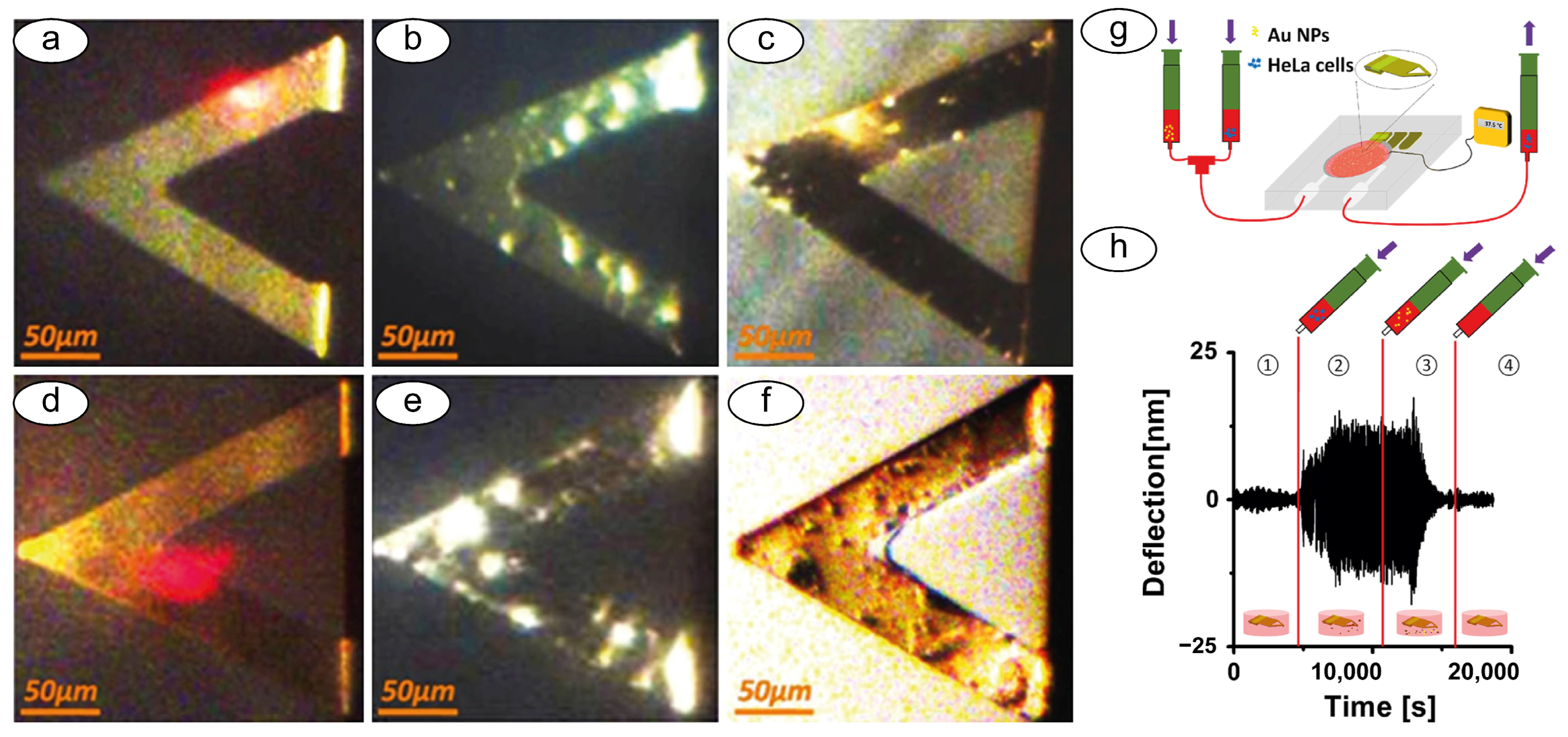
4. Novel Setups
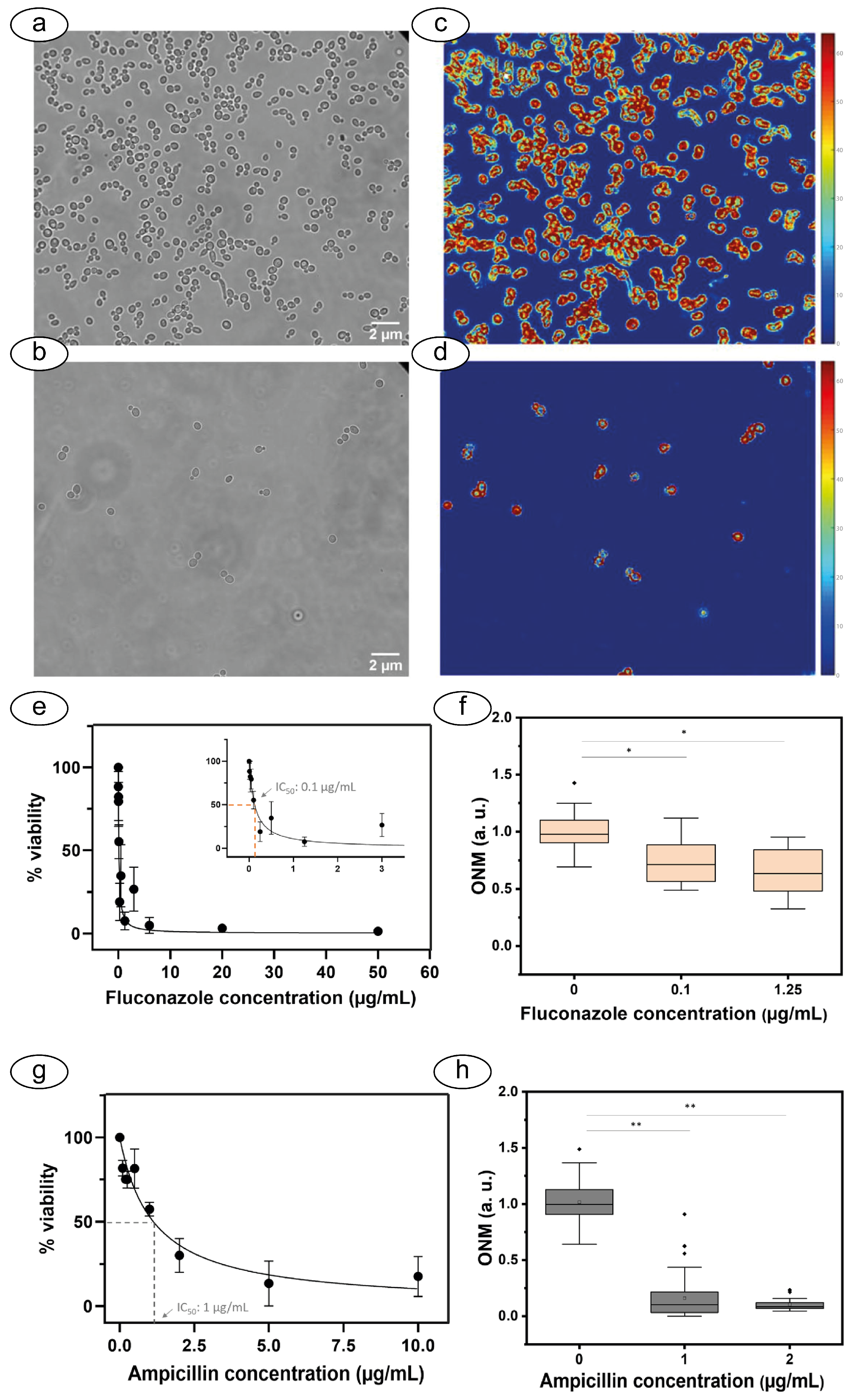
4.1. Optical Nanomotion
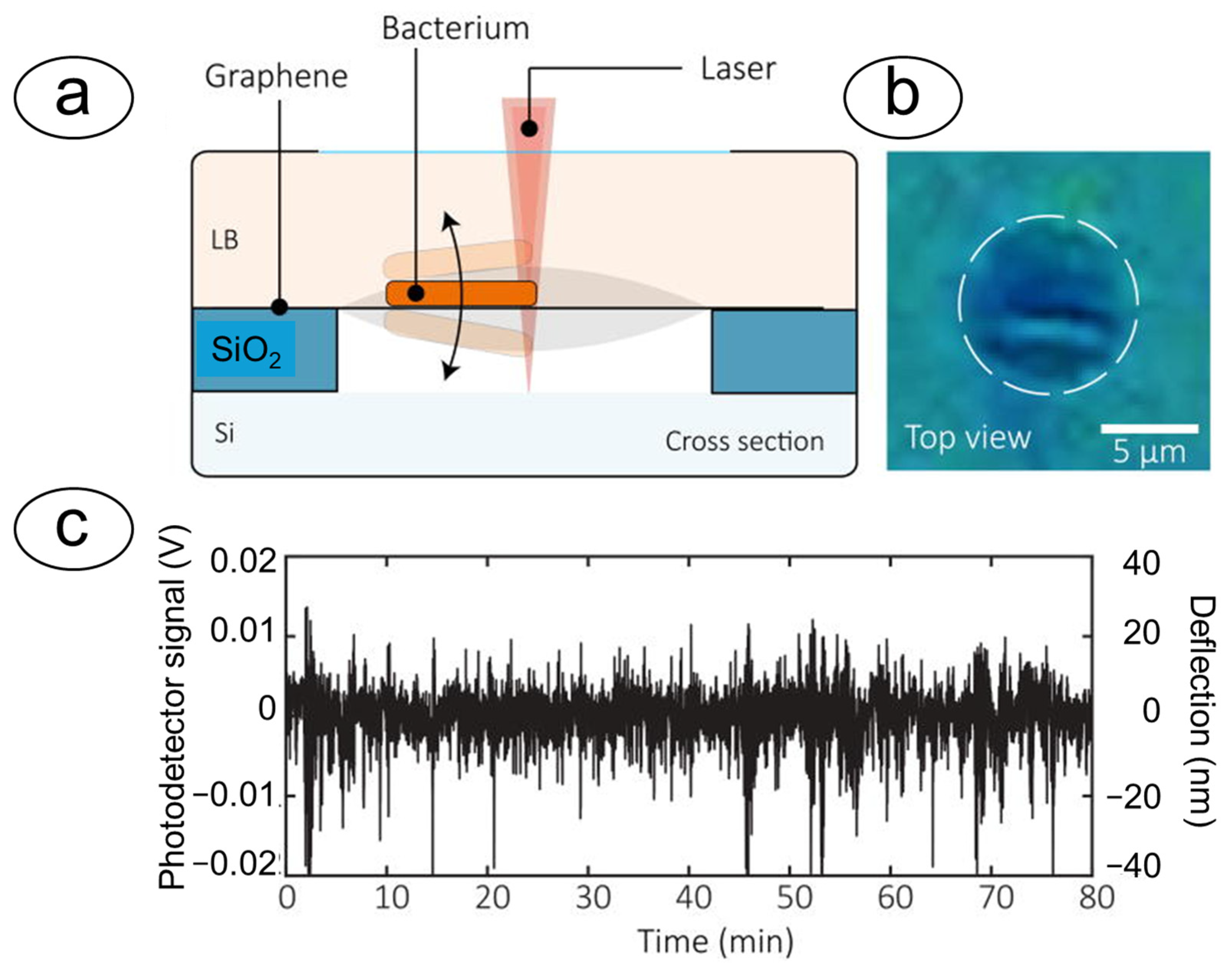
4.2. Optical Cavities and Graphene Membranes
5. Models, Big Data, and Integration with Artificial Intelligence
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | Atomic Force Microscope |
| AST | Antibiotic Susceptibility Test |
| ONMD | Optical Nano Motion Detection |
References
- Sarkar, A. Biosensing, Characterization of Biosensors, and Improved Drug Delivery Approaches Using Atomic Force Microscopy: A Review. Front. Nanotechnol. 2022, 3, 798928. [Google Scholar] [CrossRef]
- Lam, C.-D.; Park, S. Nanomechanical characterization of soft nanomaterial using atomic force microscopy. Mater. Today Bio 2025, 31, 101506. [Google Scholar] [CrossRef]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 2011, 6, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.-J.; Güntherodt, H.; Gerber, C.; Gimzewski, J.K. Translating Biomolecular Recognition into Nanomechanics. Science 2000, 288, 316–318. [Google Scholar] [CrossRef]
- Dorobantu, L.S.; Gray, M.R. Application of atomic force microscopy in bacterial research. Scanning 2010, 32, 74–96. [Google Scholar] [CrossRef] [PubMed]
- Beaussart, A.; El-Kirat-Chatel, S. Microbial adhesion and ultrastructure from the single-molecule to the single-cell levels by Atomic Force Microscopy. Cell Surf. 2019, 5, 100031. [Google Scholar] [CrossRef]
- Pujol-Vila, F.; Villa, R.; Alvarez, M. Nanomechanical Sensors as a Tool for Bacteria Detection and Antibiotic Susceptibility Testing. Front. Mech. Eng. 2020, 6, 44. [Google Scholar] [CrossRef]
- Helenius, J.; Heisenberg, C.-P.; Gaub, H.E.; Muller, D.J. Single-cell force spectroscopy. J. Cell Sci. 2008, 121, 1785. [Google Scholar] [CrossRef]
- Spitalieri, P.; Talarico, R.V.; Caioli, S.; Murdocca, M.; Serafino, A.; Girasole, M.; Dinarelli, S.; Longo, G.; Pucci, S.; Botta, A.; et al. Modelling the pathogenesis of Myotonic Dystrophy type 1 cardiac phenotype through human iPSC-derived cardiomyocytes. J. Mol. Cell. Cardiol. 2018, 118, 95–109. [Google Scholar] [CrossRef]
- Radmacher, M.; Fritz, M.; Hansma, H.G.; Hansma, P.K. Direct Observation of Enzyme-Activity with the Atomic-Force Microscope. Science 1994, 265, 1577–1579. [Google Scholar] [CrossRef]
- Boisen, A.; Dohn, S.; Keller, S.S.; Schmid, S.; Tenje, M. Cantilever-like micromechanical sensors. Rep. Prog. Phys. 2011, 74, 036101. [Google Scholar] [CrossRef]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; San Paulo, A.; Calleja, M. Biosensors based on nanomechanical systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Lechuga, L.M. Microcantilever-based platforms as biosensing tools. Analyst 2010, 135, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.M.; Thundat, T. Microcantilever biosensors. Methods 2005, 37, 57–64. [Google Scholar] [CrossRef]
- Berger, R.; Delamarche, E.; Lang, H.P.; Gerber, C.; Gimzewski, J.K.; Meyer, E.; Guntherodt, H.J. Surface stress in the self-assembly of alkanethiols on gold. Science 1997, 276, 2021–2024. [Google Scholar] [CrossRef]
- Godin, M.; Tabard-Cossa, V.; Miyahara, Y.; Monga, T.; Williams, P.J.; Beaulieu, L.Y.; Lennox, R.B.; Grutter, P. Cantilever-based sensing: The origin of surface stress and optimization strategies. Nanotechnology 2010, 21, 075501. [Google Scholar] [CrossRef]
- Ndieyira, J.W.; Watari, M.; Barrera, A.D.; Zhou, D.; Vogtli, M.; Batchelor, M.; Cooper, M.A.; Strunz, T.; Horton, M.A.; Abell, C.; et al. Nanomechanical detection of antibiotic mucopeptide binding in a model for superbug drug resistance. Nat. Nanotechnol. 2008, 3, 691–696. [Google Scholar] [CrossRef]
- Braun, T.; Ghatkesar, M.K.; Backmann, N.; Grange, W.; Boulanger, P.; Letellier, L.; Lang, H.-P.; Bietsch, A.; Gerber, C.; Hegner, M. Quantitative time-resolved measurement of membrane protein-ligand interactions using microcantilever array sensors. Nat. Nanotechnol. 2009, 4, 179–185. [Google Scholar] [CrossRef]
- Ilic, B.; Czaplewski, D.; Craighead, H.G.; Neuzil, P.; Campagnolo, C.; Batt, C. Mechanical resonant immunospecific biological detector. Appl. Phys. Lett. 2000, 77, 450–452. [Google Scholar] [CrossRef]
- Kosaka, P.M.; Pini, V.; Ruz, J.J.; da Silva, A.; Gonzalez, M.U.; Ramos, D.; Calleja, M.; Tamayo, J. Detection of cancer biomarkers in serum using a hybrid mechanical and optoplasmonic nanosensor. Nat. Nanotechnol. 2014, 9, 1047–1053. [Google Scholar] [CrossRef]
- Longo, G. Cancer biomarkers: Detected twice for good measure. Nat. Nanotechnol. 2014, 9, 959–960. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, G.S.; Dravid, V.P. Nanomechanical Sensors: Bent on detecting cancer. Nat. Nanotechnol. 2013, 8, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Sato, R.H.; Kosaka, P.M.; Omori, Á.T.; Ferreira, E.A.; Petri, D.F.S.; Malvar, Ó.; Domínguez, C.M.; Pini, V.; Ahumada, Ó.; Tamayo, J.; et al. Development of a methodology for reversible chemical modification of silicon surfaces with application in nanomechanical biosensors. Biosens. Bioelectron. 2019, 137, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Ndieyira, J.W.; Kappeler, N.; Logan, S.; Cooper, M.A.; Abell, C.; McKendry, R.A.; Aeppli, G. Surface-stress sensors for rapid and ultrasensitive detection of active free drugs in human serum. Nat. Nanotechnol. 2014, 9, 225–232. [Google Scholar] [CrossRef]
- Huber, F.; Lang, H.P.; Backmann, N.; Rimoldi, D.; Gerber, C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nat. Nanotechnol. 2013, 8, 125–129. [Google Scholar] [CrossRef]
- Waggoner, P.S.; Craighead, H.G. Micro- and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip 2007, 7, 1238–1255. [Google Scholar] [CrossRef]
- Lang, H.P.; Gerber, C. Microcantilever sensors. In STM and AFM Studies on (Bio)molecular Systems: Unravelling the Nanoworld; Samori, P., Ed.; Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2008; Volume 285, pp. 1–27. [Google Scholar]
- Gil-Santos, E.; Ruz, J.J.; Malvar, O.; Favero, I.; Lemaître, A.; Kosaka, P.M.; García-López, S.; Calleja, M.; Tamayo, J. Optomechanical detection of vibration modes of a single bacterium. Nat. Nanotechnol. 2020, 15, 469–474. [Google Scholar] [CrossRef]
- Ramos, D.; Tamayo, J.; Mertens, J.; Calleja, M.; Villanueva, L.G.; Zaballos, A. Detection of bacteria based on the thermomechanical noise of a nanomechanical resonator: Origin of the response and detection limits. Nanotechnology 2008, 19, 035503. [Google Scholar] [CrossRef]
- Lang, H.P.; Baller, M.K.; Berger, R.; Gerber, C.; Gimzewski, J.K.; Battiston, F.M.; Fornaro, P.; Ramseyer, J.P.; Meyer, E.; Guntherodt, H.J. An artificial nose based on a micromechanical cantilever array. Anal. Chim. Acta 1999, 393, 59–65. [Google Scholar] [CrossRef]
- McKendry, R.; Zhang, J.; Arntz, Y.; Strunz, T.; Hegner, M.; Lang, H.P.; Baller, M.K.; Certa, U.; Meyer, E.; Güntherodt, H.-J.; et al. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc. Natl. Acad. Sci. USA 2002, 99, 9783–9788. [Google Scholar] [CrossRef]
- Gupta, A.; Akin, D.; Bashir, R. Detection of bacterial cells and antibodies using surface micromachined thin silicon cantilever resonators. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. 2004, 22, 2785–2791. [Google Scholar] [CrossRef]
- Djuric, Z.G.; Jokic, I.M. Thermomechanical noise of nanooscillators with time-dependent mass. Microelectron. Eng. 2007, 84, 1639–1642. [Google Scholar] [CrossRef]
- Barnes, J.R.; Stephenson, R.J.; Welland, M.E.; Gerber, C.; Gimzewski, J.K. Photothermal spectroscopy with femtojoule sensitivity using a micromechanical device. Nature 1994, 372, 79–81. [Google Scholar] [CrossRef]
- Berger, R.; Gerber, C.; Gimzewski, J.K.; Meyer, E.; Guntherodt, H.J. Thermal analysis using a micromechanical calorimeter. Appl. Phys. Lett. 1996, 69, 40–42. [Google Scholar] [CrossRef]
- Etayash, H.; Khan, M.F.; Kaur, K.; Thundat, T. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat. Commun. 2016, 7, 12947. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, R.; Adhikari, S.; Mitchell, J. Vibrating carbon nanotube based bio-sensors. Phys. E Low-Dimens. Syst. Nanostruct. 2009, 42, 104–109. [Google Scholar] [CrossRef]
- Patil, S.B.; Al-Jehani, R.M.; Etayash, H.; Turbe, V.; Jiang, K.; Bailey, J.; Al-Akkad, W.; Soudy, R.; Kaur, K.; McKendry, R.A.; et al. Modified cantilever arrays improve sensitivity and reproducibility of nanomechanical sensing in living cells. Commun. Biol. 2018, 1, 175. [Google Scholar] [CrossRef]
- Han, Y.; Li, Y.; Wu, W.E.; Liu, Z. Noninvasive strategies of cell-tracking in vivo. TrAC Trends Anal. Chem. 2024, 172, 117616. [Google Scholar] [CrossRef]
- Han, S.; Kang, B.; Son, H.Y.; Choi, Y.; Shin, M.-K.; Park, J.; Min, J.-K.; Park, D.; Lim, E.-K.; Huh, Y.-M.; et al. In vivo monitoring platform of transplanted human stem cells using magnetic resonance imaging. Biosens. Bioelectron. 2021, 178, 113039. [Google Scholar] [CrossRef]
- Carniello, V.; Peterson, B.W.; Sjollema, J.; Busscher, H.J.; van der Mei, H.C. Surface enhanced fluorescence and nanoscopic cell wall deformation in adhering Staphylococcus aureus upon exposure to cell wall active and non-active antibiotics. Nanoscale 2018, 10, 11123–11133. [Google Scholar] [CrossRef]
- Marsden, N.V. Red blood cells, phase contrast, interference contrast microscopy and microspectrophotometry. Upsala J. Med. Sci. 1995, 100, 33–40. [Google Scholar] [CrossRef]
- Lam, V.K.; Nguyen, T.C.; Chung, B.M.; Nehmetallah, G.; Raub, C.B. Quantitative assessment of cancer cell morphology and motility using telecentric digital holographic microscopy and machine learning. Cytom. Part A 2018, 93, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Palma, V.; Gutiérrez, M.S.; Vargas, O.; Parthasarathy, R.; Navarrete, P. Methods to Evaluate Bacterial Motility and Its Role in Bacterial-Host Interactions. Microorganisms 2022, 10, 563. [Google Scholar] [CrossRef]
- Brzozowski, R.S.; White, M.L.; Eswara, P.J. Live-Cell Fluorescence Microscopy to Investigate Subcellular Protein Localization and Cell Morphology Changes in Bacteria. J. Vis. Exp. 2019, 153, 59905. [Google Scholar] [CrossRef]
- Johnson, M.B.; Criss, A.K. Fluorescence microscopy methods for determining the viability of bacteria in association with mammalian cells. J. Vis. Exp. 2013, 79, 50729. [Google Scholar] [CrossRef]
- Calleja, M.; Kosaka, P.M.; San Paulo, Á.; Tamayo, J. Challenges for nanomechanical sensors in biological detection. Nanoscale 2012, 4, 4925–4938. [Google Scholar] [CrossRef]
- Longo, G.; Alonso Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef]
- Stupar, P.; Opota, O.; Longo, G.; Prod’hom, G.; Dietler, G.; Greub, G.; Kasas, S. Nanomechanical sensor applied to blood culture pellets: A fast approach to determine the antibiotic susceptibility against agents of bloodstream infections. Clin. Microbiol. Infect. 2017, 23, 400–405. [Google Scholar] [CrossRef]
- Pleskova, S.N.; Lazarenko, E.V.; Bezrukov, N.A.; Bobyk, S.Z.; Boryakov, A.V.; Kriukov, R.N. Differences in bacteria nanomotion profiles and neutrophil nanomotion during phagocytosis. Front. Microbiol. 2023, 14, 1113353. [Google Scholar] [CrossRef]
- Rosłoń, I.E.; Japaridze, A.; Steeneken, P.G.; Dekker, C.; Alijani, F. Probing nanomotion of single bacteria with graphene drums. Nat. Nanotechnol. 2022, 17, 637–642. [Google Scholar] [CrossRef]
- McNiven, M.A.; Ridley, A.J. Focus on membrane dynamics. Trends Cell Biol. 2006, 16, 485–486. [Google Scholar] [CrossRef]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. USA 2015, 112, 378–381. [Google Scholar] [CrossRef]
- Kohler, A.; Venturelli, L.; Longo, G.; Dietler, G.; Kasas, S. Nanomotion detection based on Atomic Force Microscopy cantilevers. Cell Surf. 2019, 5, 100021. [Google Scholar] [CrossRef] [PubMed]
- Aghayee, S.; Benadiba, C.; Notz, J.; Kasas, S.; Dietler, G.; Longo, G. Combination of fluorescence microscopy and nanomotion detection to characterize bacteria. J. Mol. Recognit. 2013, 26, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Hinnu, M.; Mets, T.; Kerkez, I.; Putrinš, M.; Kaldalu, N.; Cathomen, G.; Pla Verge, M.; Cichocka, D.; Sturm, A.; Tenson, T. Nanomotion technology for testing azithromycin susceptibility of Salmonella enterica. Microbiol. Spectr. 2025, 13, e02385-24. [Google Scholar] [CrossRef] [PubMed]
- Aubry, C.; Kebbi-Beghdadi, C.; Luraschi-Eggemann, A.; Cathomen, G.; Cichocka, D.; Sturm, A.; Greub, G.; Consortium, T.E. Nanomotion technology: An innovative method to study cell metabolism in Escherichia coli, as a potential indicator for tolerance. J. Med. Microbiol. 2024, 73, 001912. [Google Scholar] [CrossRef]
- Pleskova, S.N.; Bezrukov, N.A.; Nikolaeva, E.D.; Boryakov, A.V.; Kuzina, O.V. Rapid Detection of Acinetobacter baumannii Suspension and Biofilm Nanomotion and Antibiotic Resistance Estimation. Biomedicines 2024, 12, 2034. [Google Scholar] [CrossRef]
- Villalba, M.I.; Venturelli, L.; Willaert, R.; Vela, M.E.; Yantorno, O.; Dietler, G.; Longo, G.; Kasas, S. Nanomotion spectroscopy as a new approach to characterize bacterial virulence. Microorganisms 2021, 9, 1545. [Google Scholar] [CrossRef]
- Opota, O.; Croxatto, A.; Prod’hom, G.; Greub, G. Blood Culture-Based Diagnosis of Bacteremia: State of the Art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef]
- Mustazzolu, A.; Venturelli, L.; Dinarelli, S.; Brown, K.; Floto, R.A.; Dietler, G.; Fattorini, L.; Kasas, S.; Girasole, M.; Longo, G. A rapid unraveling of the activity and antibiotic susceptibility of mycobacteria. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef]
- Vocat, A.; Luraschi-Eggemann, A.; Antoni, C.; Cathomen, G.; Cichocka, D.; Greub, G.; Riabova, O.; Makarov, V.; Opota, O.; Mendoza, A.; et al. Real-time evaluation of macozinone activity against Mycobacterium tuberculosis through bacterial nanomotion analysis. Antimicrob. Agents Chemother. 2025, 69, e01318-24. [Google Scholar] [CrossRef]
- Alonso-Sarduy, L.; De Los Rios, P.; Benedetti, F.; Vobornik, D.; Dietler, G.; Kasas, S.; Longo, G. Real-time monitoring of protein conformational changes using a nano-mechanical sensor. PLoS ONE 2014, 9, e103674. [Google Scholar] [CrossRef]
- Stupar, P.; Chomicki, W.; Maillard, C.; Mikeladze, D.; Kalauzi, A.; Radotić, K.; Dietler, G.; Kasas, S. Mitochondrial activity detected by cantilever based sensor. Mech. Sci. 2017, 8, 23–28. [Google Scholar] [CrossRef]
- Parmar, P.; Villalba, M.I.; Horii Huber, A.S.; Kalauzi, A.; Bartolić, D.; Radotić, K.; Willaert, R.G.; MacFabe, D.F.; Kasas, S. Mitochondrial nanomotion measured by optical microscopy. Front. Microbiol. 2023, 14, 1133773. [Google Scholar] [CrossRef]
- Friedrichs, J.; Legate, K.R.; Schubert, R.; Bharadwaj, M.; Werner, C.; Muller, D.J.; Benoit, M. A practical guide to quantify cell adhesion using single-cell force spectroscopy. Methods 2013, 60, 169–178. [Google Scholar] [CrossRef]
- Yang, F.; Riedel, R.; del Pino, P.; Pelaz, B.; Said, A.H.; Soliman, M.; Pinnapireddy, S.R.; Feliu, N.; Parak, W.J.; Bakowsky, U.; et al. Real-time, label-free monitoring of cell viability based on cell adhesion measurements with an atomic force microscope. J. Nanobiotech. 2017, 15, 23. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Mahul-Mellier, A.-L.; Kasas, S.; Lashuel, H.A.; Longo, G.; Dietler, G. Amyloid single-cell cytotoxicity assays by nanomotion detection. Cell Death Discov. 2017, 3, 17053. [Google Scholar] [CrossRef]
- Lupoli, F.; Vannocci, T.; Longo, G.; Niccolai, N.; Pastore, A. The role of oxidative stress in Friedreich’s ataxia. FEBS Lett. 2018, 592, 718–727. [Google Scholar] [CrossRef]
- Vannocci, T.; Dinarelli, S.; Girasole, M.; Pastore, A.; Longo, G. A new tool to determine the cellular metabolic landscape: Nanotechnology to the study of Friedreich’s ataxia. Sci. Rep. 2019, 9, 19282. [Google Scholar] [CrossRef]
- Girasole, M.; Dinarelli, S.; Longo, G. Correlating nanoscale motion and ATP production in healthy and favism erythrocytes: A real-time nanomotion sensor study. Front. Microbiol. 2023, 14, 1196764. [Google Scholar] [CrossRef]
- Girasole, M.; Moretti, P.F.; Di Giannatale, A.; Di Paolo, V.; Galardi, A.; Lampis, S.; Dinarelli, S.; Longo, G. Towards a role for the acoustic field in cells interaction. Front. Syst. Neurosci. 2025, 19, 1484769. [Google Scholar] [CrossRef]
- Starodubtseva, M.N.; Shkliarava, N.M.; Chelnokova, I.A.; Villalba, M.I.; Krylov, A.Y.; Nadyrov, E.A.; Kasas, S. Mechanical Properties and Nanomotion of BT-20 and ZR-75 Breast Cancer Cells Studied by Atomic Force Microscopy and Optical Nanomotion Detection Method. Cells 2023, 12, 2362. [Google Scholar] [CrossRef]
- Venturelli, L.; Kohler, A.C.; Stupar, P.; Villalba, M.I.; Kalauzi, A.; Radotic, K.; Bertacchi, M.; Dinarelli, S.; Girasole, M.; Pešić, M. A perspective view on the nanomotion detection of living organisms and its features. J. Mol. Recognit. 2020, 33, e2849. [Google Scholar] [CrossRef]
- Stupar, P.; Podolski-Renić, A.; Villalba, M.I.; Dragoj, M.; Jovanović Stojanov, S.; Pešić, M.; Kasas, S. Nano-Motion Analysis for Rapid and Label Free Assessing of Cancer Cell Sensitivity to Chemotherapeutics. Medicina 2021, 57, 446. [Google Scholar] [CrossRef]
- Wu, S.; Liu, X.; Zhou, X.; Liang, X.M.; Gao, D.; Liu, H.; Zhao, G.; Zhang, Q.; Wu, X. Quantification of cell viability and rapid screening anti-cancer drug utilizing nanomechanical fluctuation. Biosens. Bioelectron. 2016, 77, 164–173. [Google Scholar] [CrossRef]
- Villalba, M.I.; Rossetti, E.; Bonvallat, A.; Yvanoff, C.; Radonicic, V.; Willaert, R.G.; Kasas, S. Simple optical nanomotion method for single-bacterium viability and antibiotic response testing. Proc. Natl. Acad. Sci. USA 2023, 120, e2221284120. [Google Scholar] [CrossRef]
- Villalba, M.I.; Gligorovski, V.; Rahi, S.J.; Willaert, R.G.; Kasas, S. A simplified version of rapid susceptibility testing of bacteria and yeasts using optical nanomotion detection. Front. Microbiol. 2024, 15, 1328923. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, J.; Huang, H.; Zhao, C.; Zou, M.; Liu, D.; Weng, X.; Liu, L.; Qu, J.; Liu, L.; et al. Fiber-integrated cantilever-based nanomechanical biosensors as a tool for rapid antibiotic susceptibility testing. Biomed. Opt. Express 2023, 14, 1862–1873. [Google Scholar] [CrossRef]
- Xiong, C.; Zhou, J.; Liao, C.; Zhu, M.; Wang, Y.; Liu, S.; Li, C.; Zhang, Y.; Zhao, Y.; Gan, Z.; et al. Fiber-Tip Polymer Microcantilever for Fast and Highly Sensitive Hydrogen Measurement. ACS Appl. Mater. Interfaces 2020, 12, 33163–33172. [Google Scholar] [CrossRef]
- Zhou, J.; Liao, C.; Zou, M.; Villalba, M.I.; Xiong, C.; Zhao, C.; Venturelli, L.; Liu, D.; Kohler, A.-C.; Sekatskii, S.K.; et al. An optical fiber-based nanomotion sensor for rapid antibiotic and antifungal susceptibility tests. Nano Lett. 2024, 24, 2980–2988. [Google Scholar] [CrossRef]
- Rosłoń, I.E.; Japaridze, A.; Naarden, L.; Smeets, L.; Dekker, C.; van Belkum, A.; Steeneken, P.G.; Alijani, F. Prospects and challenges for graphene drums as sensors of individual bacteria. Appl. Phys. Lett. 2024, 124, 010501. [Google Scholar] [CrossRef]
- Therisod, R.; Tardif, M.; Villa, N.; Marcoux, P.R.; Picard, E.; Hadji, E.; Peyrade, D.; Houdré, R. Bacterial Gram-Type Differentiation Accomplished with Hollow Photonic Crystal Cavities. In Proceedings of the Light-Matter Interactions Towards the Nanoscale, Dordrecht, The Netherlands, 26 November 2022; pp. 357–359. [Google Scholar]
- Villa, N.; Tartari, E.; Glicenstein, S.; de Villiers de la Noue, H.; Picard, E.; Marcoux, P.R.; Zelsmann, M.; Resch, G.; Hadji, E.; Houdré, R. Optical Trapping and Fast Discrimination of Label-Free Bacteriophages at the Single Virion Level. Small 2024, 20, 2308814. [Google Scholar] [CrossRef]
- Tartari, E.; Villa, N.; de Villiers de la Noue, H.; Glicenstein, S.; Picard, E.; Marcoux, P.R.; Zelsmann, M.; Hadji, E.; Resch, G.; Houdré, R. Monitoring of Single-Cell Bacterial Lysis by Phages Within Integrated Optical Traps. Adv. Opt. Mater. 2025, 13, 2402586. [Google Scholar] [CrossRef]
- Katta, M.; Sandanalakshmi, R.; Veerraju, V. Microcantilever geometry analysis for array sensor design. Mater. Today Proc. 2022, 50, 1496–1501. [Google Scholar] [CrossRef]
- Ansari, M.Z.; Bisen, M.; Bera, U.N.; Kumar, S. Simulation of surface stress-induced deflections in microcantilever biochemical sensor. J. Mech. Eng. Autom 2015, 5, 23–26. [Google Scholar] [CrossRef]
- Tamayo, J.; Pini, V.; Kosaka, P.; Martinez, N.F.; Ahumada, O.; Calleja, M. Imaging the surface stress and vibration modes of a microcantilever by laser beam deflection microscopy. Nanotechnology 2012, 23, 315501. [Google Scholar] [CrossRef]
- Ramos, D.; Tamayo, J.; Mertens, J.; Calleja, M.; Zaballos, A. Origin of the response of nanomechanical resonators to bacteria adsorption. J. Appl. Phys. 2006, 100, 106105. [Google Scholar] [CrossRef]
- Allenstein, U.; Mayr, S.G.; Zink, M. Contractile cell forces deform macroscopic cantilevers and quantify biomaterial performance. Soft Matter 2015, 11, 5053–5059. [Google Scholar] [CrossRef]
- Puerto-Belda, V.; Ruz, J.J.; Millá, C.; Cano, Á.; Yubero, M.L.; García, S.; Kosaka, P.M.; Calleja, M.; Tamayo, J. Measuring Vibrational Modes in Living Human Cells. PRX Life 2024, 2, 013003. [Google Scholar] [CrossRef]
- Lissandrello, C.; Inci, F.; Francom, M.; Paul, M.R.; Demirci, U.; Ekinci, K.L. Nanomechanical motion of Escherichia coli adhered to a surface. Appl. Phys. Lett. 2014, 105, 113701. [Google Scholar] [CrossRef]
- Pleskova, S.N.; Lazarenko, E.V.; Bezrukov, N.A.; Kriukov, R.N.; Boryakov, A.V.; Dokukin, M.E.; Surodin, S.I. Nanomotion of bacteria to determine metabolic profile. Nanotechnol. Precis. Eng. 2023, 7, 013001. [Google Scholar] [CrossRef]
- Sturm, A.; Jóźwiak, G.; Verge, M.P.; Munch, L.; Cathomen, G.; Vocat, A.; Luraschi-Eggemann, A.; Orlando, C.; Fromm, K.; Delarze, E.; et al. Accurate and rapid antibiotic susceptibility testing using a machine learning-assisted nanomotion technology platform. Nat. Commun. 2024, 15, 2037. [Google Scholar] [CrossRef]
- Vocat, A.; Sturm, A.; Jóźwiak, G.; Cathomen, G.; Świątkowski, M.; Buga, R.; Wielgoszewski, G.; Cichocka, D.; Greub, G.; Opota, O. Nanomotion technology in combination with machine learning: A new approach for a rapid antibiotic susceptibility test for Mycobacterium tuberculosis. Microbes Infect. 2023, 25, 105151. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girasole, M.; Longo, G. The Burgeoning Importance of Nanomotion Sensors in Microbiology and Biology. Biosensors 2025, 15, 455. https://doi.org/10.3390/bios15070455
Girasole M, Longo G. The Burgeoning Importance of Nanomotion Sensors in Microbiology and Biology. Biosensors. 2025; 15(7):455. https://doi.org/10.3390/bios15070455
Chicago/Turabian StyleGirasole, Marco, and Giovanni Longo. 2025. "The Burgeoning Importance of Nanomotion Sensors in Microbiology and Biology" Biosensors 15, no. 7: 455. https://doi.org/10.3390/bios15070455
APA StyleGirasole, M., & Longo, G. (2025). The Burgeoning Importance of Nanomotion Sensors in Microbiology and Biology. Biosensors, 15(7), 455. https://doi.org/10.3390/bios15070455




