Recent Advances in Developing Cell-Free Protein Synthesis Biosensors for Medical Diagnostics and Environmental Monitoring
Abstract
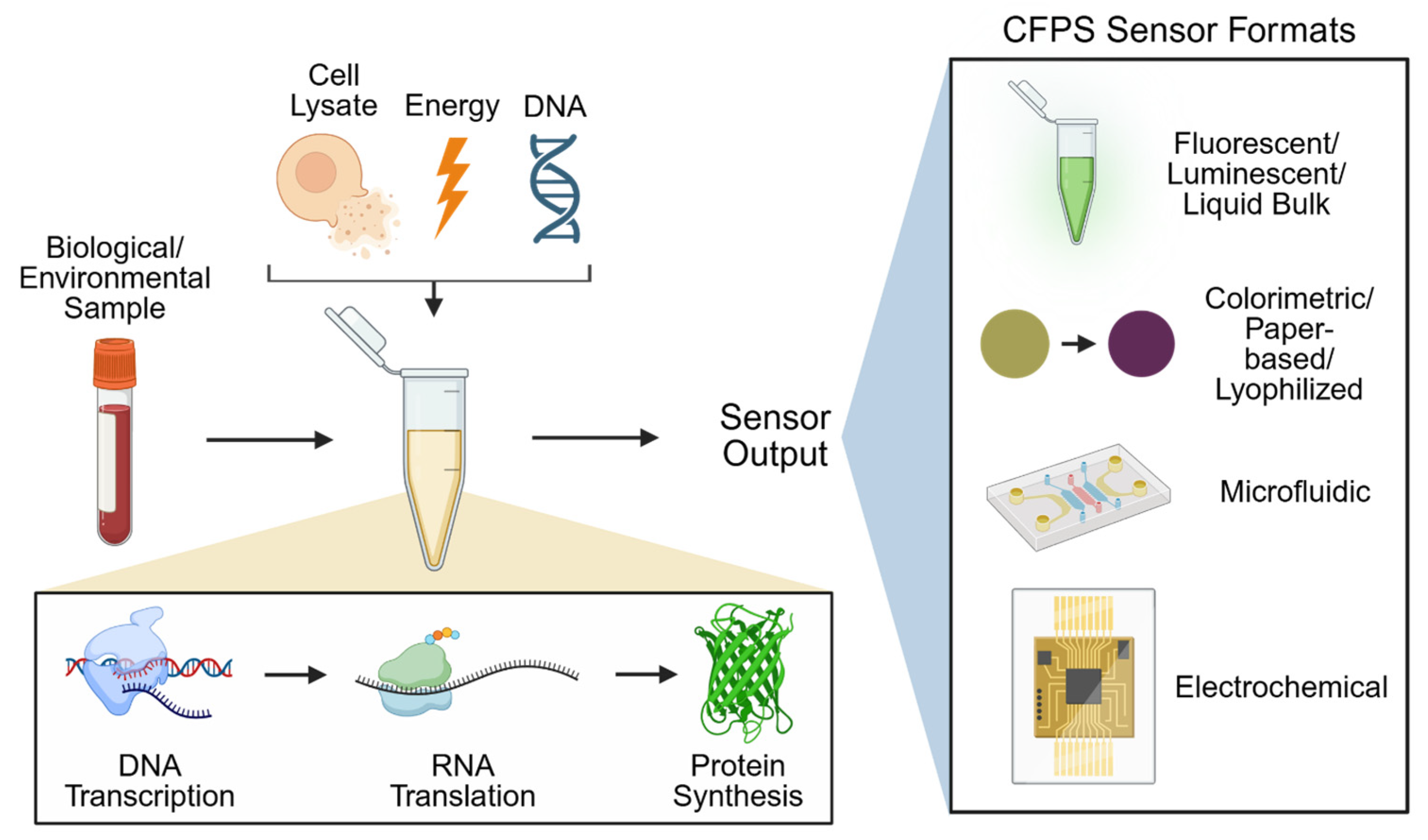
1. Introduction
2. Cell-Free Biosensors for Environmental and Medical Applications
2.1. Environmental Detection
2.2. Medical Diagnostics
3. Technological Advances and Optimization Strategies in Cell-Free Protein Synthesis for Biosensor Development
3.1. Optimization of Yield and Sensitivity
3.2. Preservation and Field Deployment Strategies
4. Integration of Synthetic Biology with Cell-Free Systems for Advanced Biosensor Applications
4.1. Complex Signal Processing and Multiplexed Systems
4.2. Novel Sensor Design Strategies
5. Emerging Materials and Future Directions
5.1. Advanced Materials for Cell-Free Biosensors
5.2. Future Challenges and Opportunities
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CFPS | Cell-free protein synthesis |
| LCMS | Liquid chromatography–mass spectrometry |
| aTF | Allosteric transcription factor |
| RAPID | Rapid adaptable portable in vitro detection |
| TLISA | T7 RNA polymerase-linked immunosensing assay |
| GABA | γ-aminobutyric acid |
| SLP | Supported lipid bilayer |
References
- Gui, Q.Y.; Lawson, T.; Shan, S.Y.; Yan, L.; Liu, Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef]
- Mihaela, G. A Short Review on Cell-Based Biosensing: Challenges and Breakthroughs in Biomedical Analysis. J. Biomed. Res. 2021, 35, 255–263. [Google Scholar] [CrossRef]
- Wang, T.; Lu, Y. Advances, Challenges and Future Trends of Cell-Free Transcription-Translation Biosensors. Biosensors 2022, 12, 318. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, D.M. Use of Cellular Synthetic Machinery for Biosensing Applications. Front. Pharmacol. 2019, 10, 1166. [Google Scholar] [CrossRef]
- Pardee, K. Perspective: Solidifying the Impact of Cell-Free Synthetic Biology Through Lyophilization. Biochem. Eng. J. 2018, 138, 91–97. [Google Scholar] [CrossRef]
- Jung, J.Y.; Rasor, B.J.; Rybnicky, G.A.; Silverman, A.D.; Standeven, J.; Kuhn, R.; Granito, T.; Ekas, H.M.; Wang, B.M.; Karim, A.S.; et al. At-Home, Cell-Free Synthetic Biology Education Modules for Transcriptional Regulation and Environmental Water Quality Monitoring. Acs Synth. Biol. 2023, 12, 2909–2921. [Google Scholar] [CrossRef]
- Hunt, J.P.; Barnett, R.J.; Robinson, H.; Soltani, M.; Nelson, J.A.D.; Bundy, B.C. Rapid Sensing of Clinically Relevant Glutamine Concentrations in Human Serum With Metabolically Engineered-Based Cell-Free Protein Synthesis. J. Biotechnol. 2021, 325, 389–394. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Guo, W.; Lu, Y. Advances in Cell-Free Biosensors: Principle, Mechanism, and Applications. Biotechnol. J. 2020, 15, 2000187. [Google Scholar] [CrossRef]
- Copeland, C.E.; Langlois, A.; Kim, J.; Kwon, Y.C. The Cell-Free System: A New Apparatus for Affordable, Sensitive, and Portable Healthcare. Biochem. Eng. J. 2021, 175, 108124. [Google Scholar] [CrossRef]
- Karig, D.K. Cell-Free Synthetic Biology for Environmental Sensing and Remediation. Curr. Opin. Biotech. 2017, 45, 69–75. [Google Scholar] [CrossRef]
- Dopp, J.L.; Tamiev, D.D.; Reuel, N.F. Cell-Free Supplement Mixtures: Elucidating the History and Biochemical Utility of Additives Used to Support Protein Synthesis in Extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef]
- Che, Y.B.; Xia, W.H.; Lu, F.P.; Chen, Z.; Liu, Y.H.; Cao, M.F.; He, N. Cell-Free Synthesis System: An Accessible Platform From Biosensing to Biomanufacturing. Microbiol. Res. 2025, 293, 128079. [Google Scholar] [CrossRef]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-Free Gene Expression: An Expanded Repertoire of Applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef]
- Gupta, S.; Sarkar, S.; Katranidis, A.; Bhattacharya, J. Development of a Cell-Free Optical Biosensor for Detection of a Broad Range of Mercury Contaminants in Water: A Plasmid DNA-Based Approach. Acs Omega 2019, 4, 9480–9487. [Google Scholar] [CrossRef]
- Hunt, J.P.; Zhao, E.L.; Free, T.J.; Soltani, M.; Warr, C.A.; Benedict, A.B.; Takahashi, M.K.; Griffitts, J.S.; Pitt, W.G.; Bundy, B.C. Towards Detection of SARS-CoV-2 RNA in Human Saliva: A Paper-Based Cell-Free Toehold Switch Biosensor With a Visual Bioluminescent Output. New Biotechnol. 2022, 66, 53–60. [Google Scholar] [CrossRef]
- Piorino, F.; Johnson, S.; Styczynski, M.P. A Cell-Free Biosensor for Assessment of Hyperhomocysteinemia. Acs Synth. Biol. 2023, 12, 2487–2492. [Google Scholar] [CrossRef]
- Free, T.J.; Tucker, R.W.; Simonson, K.M.; Smith, S.A.; Lindgren, C.M.; Pitt, W.G.; Bundy, B.C. Engineering At-Home Dilution and Filtration Methods to Enable Paper-Based Colorimetric Biosensing in Human Blood with Cell-Free Protein Synthesis. Biosensors 2023, 13, 104. [Google Scholar] [CrossRef]
- Free, T.J.; Talley, J.P.; Hyer, C.D.; Miller, C.J.; Griffitts, J.S.; Bundy, B.C. Engineering the Signal Resolution of a Paper-Based Cell-Free Glutamine Biosensor with Genetic Engineering, Metabolic Engineering, and Process Optimization. Sensors 2024, 24, 3073. [Google Scholar] [CrossRef]
- Tonooka, T. Microfluidic Device with an Integrated Freeze-Dried Cell-Free Protein Synthesis System for Small-Volume Biosensing. Micromachines 2021, 12, 27. [Google Scholar] [CrossRef]
- Wang, H.R.; Zhang, J.S.; Liu, Z.S.; Chen, M.M.; Ji, G.N.; Liu, L.Y.; Chang, Z.X.; Wang, Y.; Gao, Z.X.; Shi, H.M. CRISPR-Cas14a and Allosteric Transcription Factors Empowered Cell-Free Electrochemical Biosensor for Highly Sensitive and Stable Detection of Progesterone in Multiple Scenarios. Biosens. Bioelectron. 2025, 268, 116919. [Google Scholar] [CrossRef]
- Hunt, J.P.; Yang, S.O.; Wilding, K.M.; Bundy, B.C. The Growing Impact of Lyophilized Cell-Free Protein Expression Systems. Bioengineered 2017, 8, 325–330. [Google Scholar] [CrossRef]
- Gräwe, A.; Dreyer, A.; Vornholt, T.; Barteczko, U.; Buchholz, L.; Drews, G.; Ho, U.L.; Jackowski, M.E.; Kracht, M.; Lüders, J.; et al. A Paper-Based, Cell-Free Biosensor System for the Detection of Heavy Metals and Date Rape Drugs. PLoS ONE 2019, 14, e0210940. [Google Scholar] [CrossRef]
- Zhang, R.H.; Ruder, W.C. A New Environmental Biosensor for Cell Free Synthetic Biological Systems. Biophys. J. 2015, 108, 481a. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.K.; Zhao, C.; Bi, H.X.; Zhang, X.; Xue, B.; Li, C.Y.; Wang, S.; Yang, X.B.; Qiu, Z.G.; Wang, J.F.; et al. A Cell-Free Paper-Based Biosensor Dependent on Allosteric Transcription Factors (aTFs) for On-Site Detection of Harmful Metals Hg2+ and Pb2+ in Water. J. Hazard. Mater. 2022, 438, 129499. [Google Scholar] [CrossRef]
- Ekas, H.M.; Wang, B.; Silverman, A.D.; Lucks, J.B.; Karim, A.S.; Jewett, M.C. Engineering a PbrR-Based Biosensor for Cell-Free Detection of Lead at the Legal Limit. Acs Synth. Biol. 2024, 13, 3003–3012. [Google Scholar] [CrossRef]
- Beabout, K.; Bernhards, C.B.; Thakur, M.; Turner, K.B.; Cole, S.D.; Walper, S.A.; Chávez, J.L.; Lux, M.W. Optimization of Heavy Metal Sensors Based on Transcription Factors and Cell-Free Expression Systems. Acs Synth. Biol. 2021, 10, 3040–3054. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhu, K.L.; Chen, D.D.; Wang, J.; Wang, X.F.; Xu, A.; Wu, L.J.; Li, L.Z.; Chen, S.P. Monitoring Arsenic Using Genetically Encoded Biosensors: The Role of Evolved Regulatory Genes. Ecotox Environ. Safe 2021, 207, 111273. [Google Scholar] [CrossRef]
- Silverman, A.D.; Akova, U.; Alam, K.K.; Jewett, M.C.; Lucks, J.B. Design and Optimization of a Cell-Free Atrazine Biosensor. Acs Synth. Biol. 2020, 9, 671–677. [Google Scholar] [CrossRef]
- Liu, X.Y.; Silverman, A.D.; Alam, K.K.; Iverson, E.; Lucks, J.B.; Jewett, M.C.; Raman, S. Design of a Transcriptional Biosensor for the Portable, On-Demand Detection of Cyanuric Acid. Acs Synth. Biol. 2020, 9, 84–94. [Google Scholar] [CrossRef]
- Dong, X.Z.; Cheng, Q.Q.; Qi, S.; Qin, M.W.; Ding, N.; Sun, Y.H.; Xia, Y.; Zhang, Y.; Wang, Z.P. Broad-Spectrum Detection of Tetracyclines by Riboswitch-Based Cell-Free Expression Biosensing. J. Agr. Food Chem. 2023, 71, 9886–9895. [Google Scholar] [CrossRef]
- Duyen, T.T.M.; Matsuura, H.; Ujiie, K.; Muraoka, M.; Harada, K.; Hirata, K. Paper-Based Colorimetric Biosensor for Antibiotics Inhibiting Bacterial Protein Synthesis. J. Biosci. Bioeng. 2017, 123, 96–100. [Google Scholar] [CrossRef]
- Park, Y.J.; Choi, S.; Lee, K.W.; Park, S.Y.; Song, D.Y.; Yoo, T.H.; Kim, D.M. A Cell-Free Biosensor for Multiplexed and Sensitive Detection of Biological Warfare Agents. Biosens. Bioelectron. 2024, 257, 116331. [Google Scholar] [CrossRef]
- Voyvodic, P.L.; Pandi, A.; Koch, M.; Conejero, I.; Valjent, E.; Courtet, P.; Renard, E.; Faulon, J.L.; Bonnet, J. Plug-and-Play Metabolic Transducers Expand the Chemical Detection Space of Cell-Free Biosensors. Nat. Commun. 2019, 10, 1697. [Google Scholar] [CrossRef]
- Koch, M.; Pandi, A.; Borkowski, O.; Batista, A.C.; Faulon, J.L. Custom-Made Transcriptional Biosensors for Metabolic Engineering. Curr. Opin. Biotech. 2019, 59, 78–84. [Google Scholar] [CrossRef]
- Arce, A.; Chavez, F.G.; Gandini, C.; Puig, J.; Matute, T.; Haseloff, J.; Dalchau, N.; Molloy, J.; Pardee, K.; Federici, F. Decentralizing Cell-Free RNA Sensing with the Use of Low-Cost Cell Extracts. Front. Bioeng. Biotech. 2021, 9, 727584. [Google Scholar] [CrossRef]
- Lin, X.M.; Li, Y.T.; Li, Z.X.; Hua, R.; Xing, Y.Y.; Lu, Y. Portable Environment-Signal Detection Biosensors With Cell-Free Synthetic Biosystems. Rsc Adv. 2020, 10, 39261–39265. [Google Scholar] [CrossRef]
- Salehi, A.S.M.; Tang, M.J.S.; Smith, M.T.; Hunt, J.M.; Law, R.A.; Wood, D.W.; Bundy, B.C. Cell-Free Protein Synthesis Approach to Biosensing hTR(ss)-Specific Endocrine Disruptors. Anal. Chem. 2017, 89, 3395–3401. [Google Scholar] [CrossRef]
- Salehi, A.S.M.; Yang, S.O.; Earl, C.C.; Tang, M.J.S.; Hunt, J.P.; Smith, M.T.; Wood, D.W.; Bundy, B.C. Biosensing Estrogenic Endocrine Disruptors in Human Blood and Urine: A RAPID Cell-Free Protein Synthesis Approach. Toxicol. Appl. Pharm. 2018, 345, 19–25. [Google Scholar] [CrossRef]
- Hunt, J.P.; Galiardi, J.; Free, T.J.; Yang, S.O.; Poole, D.; Zhao, E.L.; Andersen, J.L.; Wood, D.W.; Bundy, B.C. Mechanistic Discoveries and Simulation-Guided Assay Optimization of Portable Hormone Biosensors With Cell-Free Protein Synthesis. Biotechnol. J. 2022, 17, e2100152. [Google Scholar] [CrossRef]
- Hunt, J.P.; Free, T.J.; Galiardi, J.; Watt, K.M.; Wood, D.W.; Bundy, B.C. Streamlining the Detection of Human Thyroid Receptor Ligand Interactions with XL1-Blue Cell-Free Protein Synthesis and Beta-Galactosidase Fusion Protein Biosensors. Life 2023, 13, 1972. [Google Scholar] [CrossRef]
- Wen, K.Y.; Cameron, L.; Chappell, J.; Jensen, K.; Bell, D.J.; Kelwick, R.; Kopniczky, M.; Davies, J.C.; Filloux, A.; Freemont, P.S. A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P.-Infected Respiratory Samples. Acs Synth. Biol. 2017, 6, 2293–2301. [Google Scholar] [CrossRef]
- Beabout, K.; Breedon, A.M.E.; Blum, S.M.; Miklos, A.E.; Lux, M.W.; Chávez, J.L.; Goodson, M.S. Detection of Bile Acids in Complex Matrices Using a Transcription Factor-Based Biosensor. Acs Biomater. Sci. Eng. 2022, 9, 5151–5162. [Google Scholar] [CrossRef]
- Talley, J.P.; Free, T.J.; Green, T.P.; Chipman, D.M.; Bundy, B.C. Eliminating Assay Background of a Low-Cost, Colorimetric Glutamine Biosensor by Engineering an Alternative Formulation of Cell-Free Protein Synthesis. Chemosensors 2025, 13, 206. [Google Scholar] [CrossRef]
- Hong, F.; Ma, D.; Wu, K.Y.; Mina, L.A.; Luiten, R.C.; Liu, Y.; Yan, H.; Green, A.A. Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators. Cell 2020, 180, 1018. [Google Scholar] [CrossRef]
- Chen, S.T.; Zhao, C.; Kang, X.D.; Zhang, X.; Xue, B.; Li, C.Y.; Wang, S.; Yang, X.B.; Li, C.; Qiu, Z.G.; et al. A Cell-Free Fluorescence Biosensor Based on Allosteric Transcription Factor NalC for Detection of Pentachlorophenol. Biotechnol. Lett. 2024, 46, 725–737. [Google Scholar] [CrossRef]
- Mcsweeney, M.A.; Patterson, A.T.; Loeffler, K.; de Larrea, R.C.L.; Mcnerney, M.P.; Kane, R.S.; Styczynski, M.P. A Modular Cell-Free Protein Biosensor Platform Using Split T7 RNA Polymerase. Sci. Adv. 2025, 11, eado6280. [Google Scholar] [CrossRef]
- Yenilmez, E.D.; Kökbas, U.; Alparslan, M.M.; Sanna, B.; Kartlasmis, K.; Kayrin, L.; Tuli, A. A New Biosensor for Fetal RHD Detection From Circulating Cell-Free Fetal DNA in Maternal Plasma. Febs J. 2016, 283, 237. [Google Scholar]
- Narahari, T.; Dahmer, J.; Sklavounos, A.; Kim, T.; Satkauskas, M.; Clotea, I.; Ho, M.; Lamanna, J.; Dixon, C.; Rackus, D.G.; et al. Portable Sample Processing for Molecular Assays: Application to Zika Virus Diagnostics. Lab. Chip 2022, 22, 1748–1763. [Google Scholar] [CrossRef]
- Chen, Y.B.; Xia, W.H.; Pan, Z.W.; Lu, F.P.; Liu, Y.H.; Cao, M.F.; He, N. Development of a Cell-Free, Toehold Switch-Based Biosensor for Rapid and Sensitive Zika Virus Detection. Anal. Chem. 2025, 97, 3486–3494. [Google Scholar] [CrossRef]
- Gaba, S.; Chauhan, N.; Chandra, R.; Jain, U. Future Advances of Artificial Biosensor Technology in Biomedical Applications. Talanta Open 2024, 9, 100301. [Google Scholar] [CrossRef]
- Cole, S.D.; Miklos, A.E.; Chiao, A.C.; Sun, Z.Z.; Lux, M.W. Methodologies for Preparation of Prokaryotic Extracts for Cell-Free Expression Systems. Syn. Syst. Biotechno 2020, 5, 252–267. [Google Scholar] [CrossRef]
- Dopp, J.L.; Jo, Y.R.; Reuel, N.F. Methods to Reduce Variability in E. coli-Based Cell-Free Protein Expression Experiments. Syn. Syst. Biotechno 2019, 4, 204–211. [Google Scholar] [CrossRef]
- Dopp, J.L.; Reuel, N.F. Process Optimization for Scalable Extract Preparation for Cell-Free Protein Synthesis. Biochem. Eng. J. 2018, 138, 21–28. [Google Scholar] [CrossRef]
- Banks, A.M.; Whitfield, C.J.; Brown, S.R.; Fulton, D.A.; Goodchild, S.A.; Grant, C.; Love, J.; Lendrem, D.W.; Fieldsend, J.E.; Howard, T.P. Key Reaction Components Affect the Kinetics and Performance Robustness of Cell-Free Protein Synthesis Reactions. Comput. Struct. Biotec 2022, 20, 218–229. [Google Scholar] [CrossRef]
- Spice, A.J.; Aw, R.; Bracewell, D.G.; Polizzi, K.M. Improving the Reaction Mix of a Cell-Free System Using a Design of Experiments Approach to Minimise Experimental Effort. Syn. Syst. Biotechno 2020, 5, 137–144. [Google Scholar] [CrossRef]
- Aw, R.; Polizzi, K.M. Biosensor-Assisted Engineering of a High-Yield Cell-Free Protein Synthesis Platform. Biotechnol. Bioeng. 2019, 116, 656–666. [Google Scholar] [CrossRef]
- Vezeau, G.E.; Salis, H.M. Tuning Cell-Free Composition Controls the Time Delay, Dynamics, and Productivity of TX-TL Expression. Acs Synth. Biol. 2021, 10, 2508–2519. [Google Scholar] [CrossRef]
- Chushak, Y.; Harbaugh, S.; Zimlich, K.; Alfred, B.; Chávez, J.; Kelley-Loughnane, N. Characterization of Synthetic Riboswitch in Cell-Free Protein Expression Systems. Rna Biol. 2021, 18, 1727–1738. [Google Scholar] [CrossRef]
- Copeland, C.E.; Kim, J.; Copeland, P.L.; Heitmeier, C.J.; Kwon, Y.C. Characterizing a New Fluorescent Protein for a Low Limit of Detection Sensing in the Cell-Free System. Acs Synth. Biol. 2022, 11, 2800–2810. [Google Scholar] [CrossRef]
- Li, W.X.; Xu, Y.M.; Zhang, Y.; Li, P.F.; Zhu, X.L.; Feng, C. Cell-Free Biosensing Genetic Circuit Coupled with Ribozyme Cleavage Reaction for Rapid and Sensitive Detection of Small Molecules. Acs Synth. Biol. 2023, 12, 1657–1666. [Google Scholar] [CrossRef]
- Ogawa, A. Suppressor tRNA-based Biosensors for Detecting Analytes. Anal. Sci. 2021, 37, 407–414. [Google Scholar] [CrossRef]
- Soltani, M.; Hunt, J.P.; Bundy, B.C. Rapid RNase Inhibitor Production to Enable Low-Cost, On-Demand Cell-Free Protein Synthesis Biosensor Use in Human Body Fluids. Biotechnol. Bioeng. 2021, 118, 3973–3983. [Google Scholar] [CrossRef]
- Soltani, M.; Bundy, B.C. Streamlining Cell-Free Protein Synthesis Biosensors for Use in Human Fluids: RNase Inhibitor Production During Extract Preparation. Biochem. Eng. J. 2022, 177, 108158. [Google Scholar] [CrossRef]
- Smith, S.A.; Lindgren, C.M.; Ebbert, L.E.; Free, T.J.; Nelson, J.A.D.; Simonson, K.M.; Hunt, J.P.; Bundy, B.C. “Just Add Small Molecules” Cell-Free Protein Synthesis: Combining DNA Template and Cell Extract Preparation Into a Single Fermentation. Biotechnol. Prog. 2023, 39, e3332. [Google Scholar] [CrossRef]
- Hunt, J.P.; Wilding, K.M.; Barnett, R.J.; Robinson, H.; Soltani, M.; Cho, J.E.; Bundy, B.C. Engineering Cell-Free Protein Synthesis for High-Yield Production and Human Serum Activity Assessment of Asparaginase: Toward On-Demand Treatment of Acute Lymphoblastic Leukemia. Biotechnol. J. 2020, 15, 1900294. [Google Scholar] [CrossRef]
- Blum, S.M.; Lee, M.S.; Mgboji, G.E.; Funk, V.L.; Beabout, K.; Harbaugh, S.V.; Roth, P.A.; Liem, A.T.; Miklos, A.E.; Emanuel, P.A.; et al. Impact of Porous Matrices and Concentration by Lyophilization on Cell-Free Expression. Acs Synth. Biol. 2021, 10, 1116–1131. [Google Scholar] [CrossRef]
- Zuppolini, S.; Salama, A.; Cruz-Maya, I.; Guarino, V.; Borriello, A. Cellulose Amphiphilic Materials: Chemistry, Process and Applications. Pharmaceutics 2022, 14, 386. [Google Scholar] [CrossRef]
- Nilsen, S.K.; Dahl, I.; Jorgensen, O.; Schneider, T. Micro-Fibre and Ultra-Micro-Fibre Cloths, Their Physical Characteristics, Cleaning Effect, Abrasion on Surfaces, Friction, and Wear Resistance. Build. Environ. 2002, 37, 1373–1378. [Google Scholar] [CrossRef]
- Niwa, T.; Sugimoto, R.; Watanabe, L.; Nakamura, S.; Ueda, T.; Taguchi, H. Large-Scale Analysis of Macromolecular Crowding Effects on Protein Aggregation Using a Reconstituted Cell-Free Translation System. Front. Microbiol. 2015, 6, 1113. [Google Scholar] [CrossRef]
- Carr, A.R.; Dopp, J.L.; Wu, K.Y.; Mousavi, P.S.; Jo, Y.R.; McNeley, C.E.; Lynch, Z.T.; Pardee, K.; Green, A.A.; Reuel, N.F. Toward Mail-in-Sensors for SARS-CoV-2 Detection: Interfacing Gel Switch Resonators with Cell-Free Toehold Switches. Acs Sensors 2022, 7, 806–815. [Google Scholar] [CrossRef]
- Dourbash, F.A.; Shestopalov, A.A.; Rothberg, L.J. Reduction of Non-Specific Adsorption in Label-Free Assays via Reversible Surface Blocking With Amphiphilic Sugars. Sensor Actuat B-Chem. 2022, 360, 131657. [Google Scholar] [CrossRef]
- Gervais, T.; Jensen, K.F. Mass Transport and Surface Reactions in Microfluidic Systems. Chem. Eng. Sci. 2006, 61, 1102–1121. [Google Scholar] [CrossRef]
- Nelis, J.; Elliott, C.; Campbell, K. “The Smartphone’s Guide to the Galaxy”: In Situ Analysis in Space. Biosensors 2018, 8, 96. [Google Scholar] [CrossRef]
- Reizabal, A.; Costa, C.M.; Pérez-Alvarez, L.; Vilas-Vilela, J.L.; Lanceros-Méndez, S. Silk Fibroin as Sustainable Advanced Material: Material Properties and Characteristics, Processing, and Applications. Adv. Funct. Mater. 2023, 33, 2210764. [Google Scholar] [CrossRef]
- Drachuk, I.; Harbaugh, S.; Chávez, J.L.; Kelley-Loughnane, N. Improving the Activity of DNA-Encoded Sensing Elements through Confinement in Silk Microcapsules. Acs Appl. Mater. Inter. 2020, 12, 48329–48339. [Google Scholar] [CrossRef]
- Li, Y.Y.; Lucci, T.; Dujovne, M.V.; Jung, J.K.; Capdevila, D.A.; Lucks, J.B. A Cell-Free Biosensor Signal Amplification Circuit With Polymerase Strand Recycling. Nat. Chem. Biol. 2025, 21, 949–958. [Google Scholar] [CrossRef]
- Mousavi, P.S.; Smith, S.J.; Chen, J.B.; Karlikow, M.; Tinafar, A.; Robinson, C.; Liu, W.H.; Ma, D.; Green, A.A.; Kelley, S.O.; et al. A Multiplexed, Electrochemical Interface for Gene-Circuit-Based Sensors. Nat. Chem. 2020, 12, 48–55. [Google Scholar] [CrossRef]
- Ma, D.; Li, Y.X.; Wu, K.Y.; Yan, Z.Q.; Tang, A.A.; Chaudhary, S.; Ticktin, Z.M.; Alcantar-Fernandez, J.; Moreno-Camacho, J.L.; Campos-Romero, A.; et al. Multi-Arm RNA Junctions Encoding Molecular Logic Unconstrained by Input Sequence for Versatile Cell-Free Diagnostics. Nat. Biomed. Eng. 2022, 6, 298. [Google Scholar] [CrossRef]
- Mathur, D.; Thakur, M.; Díaz, S.A.; Susumu, K.; Stewart, M.H.; Oh, E.; Walper, S.A.; Medintz, I.L. Hybrid Nucleic Acid-Quantum Dot Assemblies as Multiplexed Reporter Platforms for Cell-Free Transcription Translation-Based Biosensors. Acs Synth. Biol. 2022, 11, 4089–4102. [Google Scholar] [CrossRef]
- Peruzzi, J.A.; Galvez, N.R.; Kamat, N.P. Engineering Transmembrane Signal Transduction in Synthetic Membranes Using Two-Component Systems. P. Natl. Acad. Sci. USA 2023, 120, e2218610120. [Google Scholar] [CrossRef]
- Green, A.A.; Silver, P.A.; Collins, J.J.; Yin, P. Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell 2014, 159, 925–939. [Google Scholar] [CrossRef]
- Majumder, S.; Garamella, J.; Wang, Y.L.; DeNies, M.; Noireaux, V.; Liu, A.P. Cell-Sized Mechanosensitive and Biosensing Compartment Programmed With DNA. Chem. Commun. 2017, 53, 7349–7352. [Google Scholar] [CrossRef]
- Garamella, J.; Majumder, S.; Liu, A.P.; Noireaux, V. An Adaptive Synthetic Cell Based on Mechanosensing, Biosensing, and Inducible Gene Circuits. Acs Synth. Biol. 2019, 8, 1913–1920. [Google Scholar] [CrossRef]
- Harbaugh, S.V.; Silverman, A.D.; Chushak, Y.G.; Zimlich, K.; Wolfe, M.; Thavarajah, W.; Jewett, M.C.; Lucks, J.B.; Chavez, J.L. Engineering a Synthetic Dopamine-Responsive Riboswitch for Biosensing. Acs Synth. Biol. 2022, 11, 2275–2283. [Google Scholar] [CrossRef]
- Thavarajah, W.; Silverman, A.D.; Verosloff, M.S.; Kelley-Loughnane, N.; Jewett, M.C.; Lucks, J.B. Point-of-Use Detection of Environmental Fluoride a Cell-Free Riboswitch-Based Biosensor. Acs Synth. Biol. 2020, 9, 10–18. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Han, J.; Shin, J.; Park, K.S. Split T7 Switch-Mediated Cell-Free Protein Synthesis System for Detecting Target Nucleic Acids. Biosens. Bioelectron. 2024, 261, 116517. [Google Scholar] [CrossRef]
- Copeland, C.E.; Heitmeier, C.J.; Doan, K.D.; Lee, S.C.; Porche, K.B.; Kwon, Y.C. Expanding the Cell-Free Reporter Protein Toolbox by Employing a Split mNeonGreen System to Reduce Protein Synthesis Workload. Acs Synth. Biol. 2024, 13, 1663–1668. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lim, H.J.; Kim, D.M. Quantitative Analysis of γ-Aminobutyric Acid by Combined Cell-Free Protein Synthesis and Transamination Reactions. Acs Synth. Biol. 2022, 11, 1208–1212. [Google Scholar] [CrossRef]
- Sharpes, C.E.; McManus, J.B.; Blum, S.M.; Mgboji, G.E.; Lux, M.W. Assessment of Colorimetric Reporter Enzymes in the PURE System. Acs Synth. Biol. 2021, 10, 3205–3208. [Google Scholar] [CrossRef]
- Jayaram, A.K.; Pappa, A.M.; Ghosh, S.; Manzer, Z.A.; Traberg, W.C.; Knowles, T.P.J.; Daniel, S.; Owens, R.M. Biomembranes in Bioelectronic Sensing. Trends Biotechnol. 2022, 40, 107–123. [Google Scholar] [CrossRef]
- Manzer, Z.A.; Ghosh, S.; Roy, A.; Jacobs, M.L.; Carten, J.; Kamat, N.P.; Daniel, S. Cell-Free Synthesis Goes Electric: Dual Optical and Electronic Biosensor via Direct Channel Integration into a Supported Membrane Electrode. Acs Synth. Biol. 2023, 12, 502–510. [Google Scholar] [CrossRef]
- Benitez-Mateos, A.I.; Zeballos, N.; Comino, N.; de Redrojo, L.M.; Randelovic, T.; López-Gallego, F. Microcompartmentalized Cell-Free Protein Synthesis in Hydrogel μ-Channels. Acs Synth. Biol. 2020, 9, 2971–2978. [Google Scholar] [CrossRef]
- Boyd, M.A.; Thavarajah, W.; Lucks, J.B.; Kamat, N.P. Robust and Tunable Performance of a Cell-Free Biosensor Encapsulated in Lipid Vesicles. Sci. Adv. 2023, 9, eadd6605. [Google Scholar] [CrossRef]
- Cole, S.D.; Beabout, K.; Turner, K.B.; Smith, Z.K.; Funk, V.L.; Harbaugh, S.V.; Liem, A.T.; Roth, P.A.; Geier, B.A.; Emanuel, P.A.; et al. Quantification of Interlaboratory Cell-Free Protein Synthesis Variability. Acs Synth. Biol. 2019, 8, 2080–2091. [Google Scholar] [CrossRef]
- Kim, N.M.; Sinnott, R.W.; Sandoval, N.R. Transcription Factor-Based Biosensors and Inducible Systems in Non-Model Bacteria: Current Progress and Future Directions. Curr. Opin. Biotech. 2020, 64, 39–46. [Google Scholar] [CrossRef]


| Target Analyte | Detection Method/System | Limit of Detection | Selectivity/Specificity | Sample Matrix |
|---|---|---|---|---|
| Mercury [22] | Paper-based, dual- filter, smartphone readout | 6 μg/L | Selective for mercury (activation ratio >8–14 for Hg, <2 for others) | Water |
| Mercury [14] | merR gene, plasmid DNA, firefly luciferase/eGFP | 1 ppb | Selective for Hg2+; pH optimization and chelating agents enhance specificity | Water |
| Mercury [24] | Allosteric transcription factors (aTFs) | 0.5 nM | High selectivity for target metals; validated in real water samples with 91–123% recovery rates | Water |
| Lead [24] | aTFs | 0.1 nM | High selectivity for target metals; validated in real water samples with 91–123% recovery rates | Water |
| Lead [25] | Engineered PbrR mutants | 50 nM | Selective for lead | Water |
| Arsenic and mercury [26] | Optimized transcription factors | Arsenic ≤10 μg/L, Mercury ≤6 μg/L | Minimal response to nontoxic ions | Water |
| Tetracyclines [30] | Riboswitch-based, RNA aptamers | 0.4 μM | Broad-spectrum for tetracycline family | Milk samples |
| Biological warfare agents [32] | 16S rRNA targeting, retroreflective particles | 1 to 11 fM | Specific for B. anthracis, F. tularensis, Y. pestis, B. pseudomallei, B. abortus | Laboratory samples |
| Atrazine [28] | Metabolic pathway and cyanuric acid biosensor | 50 μM | Specific for atrazine via metabolic conversion | Laboratory samples |
| Target Analyte | Detection Method/System | Limit of Detection | Selectivity/Specificity | Sample Matrix |
|---|---|---|---|---|
| Thyroid receptor ligands [37] | Allosterically activated fusion protein | 48 to 75 nM | β-specific endocrine disruptors | Laboratory samples |
| Estrogenic compounds [38] | RAPID platform | 9 to 330 nM | selective for estrogenic activity | Human blood/urine |
| 3-oxo-C12-HSL [41] | Quorum sensing detection | 4.9 nM | Highly specific for 3-oxo-C12-HSL; results comparable to LC-MS | P. aeruginosa-infected sputum |
| Glutamine [7] | Metabolically engineered system | 10 μM | Inhibitor-based approach ensures glutamine specificity | Human serum |
| Bile acids [42] | Transcription factor-based | 0.61 μM | Selective for deoxycholic acid | Fecal water, wastewater, serum |
| Homocysteine [16] | Colorimetric detection | 1 μM | Selective detection at clinically relevant thresholds | Plasma |
| Progesterone [20] | CRISPR-Cas14a + aTFs | 67 pM to 0.33 μM | Single-nucleotide discrimination | 2 μL sample |
| Pentachlorophenol [45] | aTF NalC + NASBA | 0.002 μM | high specificity for pentachlorophenol; validated with 101–114% recovery | Environmental samples |
| Protein biomarkers [46] | Split T7 RNA polymerase (TLISA) | 50 to 200 nM | Various protein targets | Serum/saliva |
| SARS-CoV-2 RNA [15] | Paper-based toehold switches | 60 nM | High specificity for target RNA | Human saliva |
| System Type | Detection Method | Limit of Detection | Key Innovation | Sample Compatibility |
|---|---|---|---|---|
| Signal amplification [60] | Ribozyme cleavage circuits | 0.045 μM | Eliminates translation steps | Small molecules |
| Amplification circuit [76] | Polymerase strand recycling | 5 nm to 1 μM | T7 RNA polymerase recycling | Laboratory samples |
| Electrochemical multiplexing [77] | Gene-circuit-based sensors | 65 nM | Electrochemical readouts | Laboratory samples |
| Multiplexed pathogen detection [78] | Multi-arm RNA junctions | 20 aM | Molecular logic operations | Diagnostic samples |
| Quantum dot multiplexing [79] | Hybrid nucleic acid-QD assemblies | 1 enzyme unit 1 | Multiple enzymatic monitoring | Laboratory samples |
| CRISPR-based detection [20] | CRISPR-Cas systems integration | 4.2 pM | Signal processing enhancement | Complex biological matrices |
| Pathogen multiplexing [32] | 16S rRNA targeting | 2.4 nM | Simultaneous multi-pathogen ID | Laboratory samples |
| Metabolic transducers [33] | Plug-and-play cascades | 1 to 10 μM | Synthetic metabolic networks | Complex media/urine |
| System Type | Detection Method | Limit of Detection | Key Innovation | Sample Compatibility |
|---|---|---|---|---|
| Split reporter system [86] | Split T7 promoter | 10 pM | Three-way junction structure | Nucleic acids |
| Split protein system [87] | Split mNeonGreen | Not reported | Reduced synthesis workload | Laboratory samples |
| Metabolic biosensing [88] | GABA transaminase + CFPS | 47 μM | Enzymatic conversion approach | Laboratory samples |
| Dopamine detection [84] | Synthetic riboswitches | 0.48 μM | Engineered aptamer riboswitches | Human urine |
| Fluoride detection [85] | Natural riboswitches | 0.2 mM | B. cereus crcB riboswitch | Field conditions |
| Mechanosensitive systems [82] | MscL + biosensing | Not reported | Osmotic pressure integration | Synthetic liposomes |
| Adaptive synthetic cells [83] | Inducible genetic circuits | Not reported | Mechanosensitivity + gene expression | Synthetic systems |
| Colorimetric multiplexing [89] | Multi-enzyme systems | Not reported | Visual detection without instruments | Laboratory samples |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Green, T.P.; Talley, J.P.; Bundy, B.C. Recent Advances in Developing Cell-Free Protein Synthesis Biosensors for Medical Diagnostics and Environmental Monitoring. Biosensors 2025, 15, 499. https://doi.org/10.3390/bios15080499
Green TP, Talley JP, Bundy BC. Recent Advances in Developing Cell-Free Protein Synthesis Biosensors for Medical Diagnostics and Environmental Monitoring. Biosensors. 2025; 15(8):499. https://doi.org/10.3390/bios15080499
Chicago/Turabian StyleGreen, Tyler P., Joseph P. Talley, and Bradley C. Bundy. 2025. "Recent Advances in Developing Cell-Free Protein Synthesis Biosensors for Medical Diagnostics and Environmental Monitoring" Biosensors 15, no. 8: 499. https://doi.org/10.3390/bios15080499
APA StyleGreen, T. P., Talley, J. P., & Bundy, B. C. (2025). Recent Advances in Developing Cell-Free Protein Synthesis Biosensors for Medical Diagnostics and Environmental Monitoring. Biosensors, 15(8), 499. https://doi.org/10.3390/bios15080499





