Probing the Influence of Specular Reflection and Overexposure on Backscattering Mueller Matrix Polarimetry for Tissue Imaging and Sensing
Abstract
1. Introduction
2. Materials and Methods
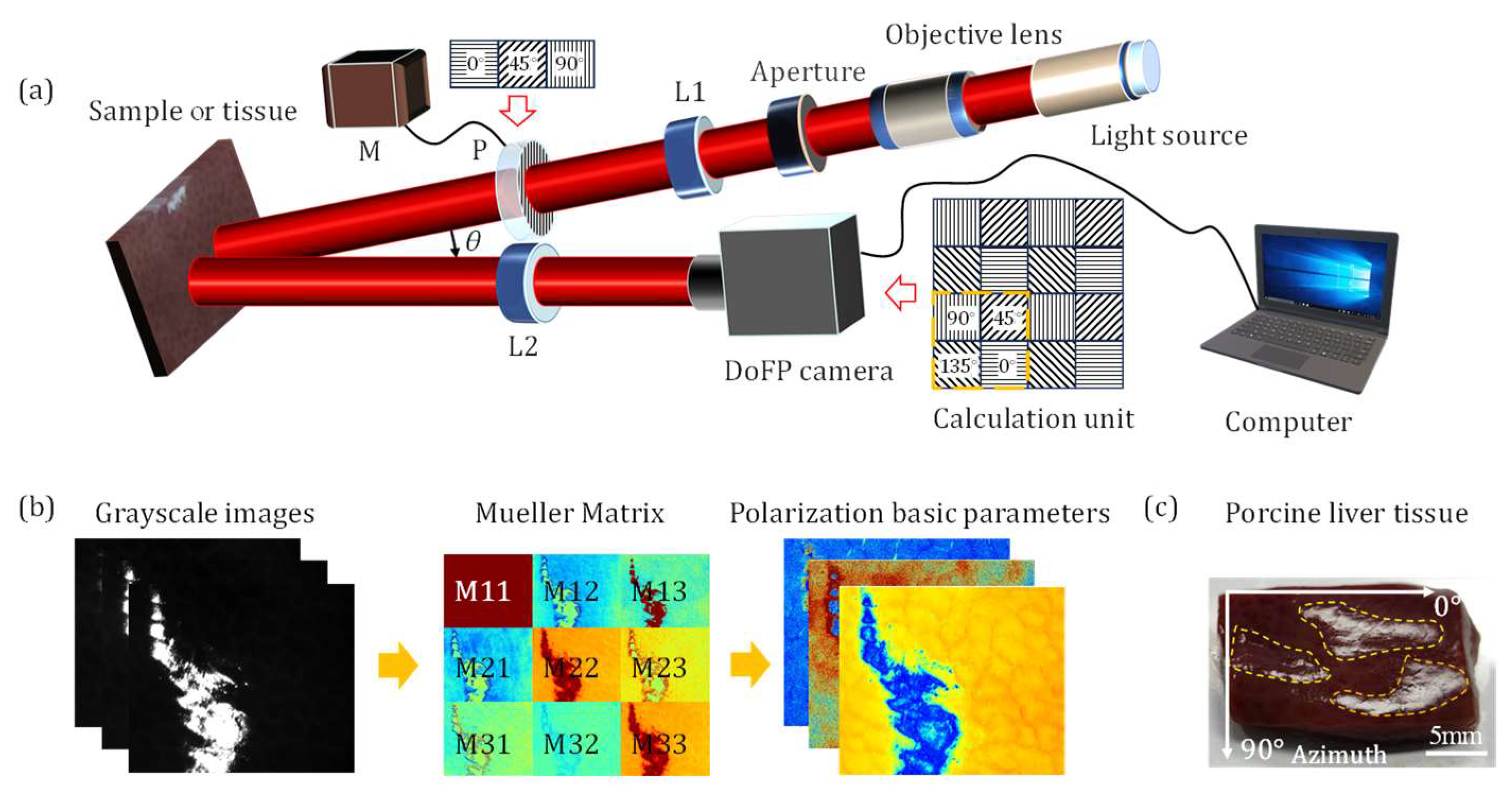
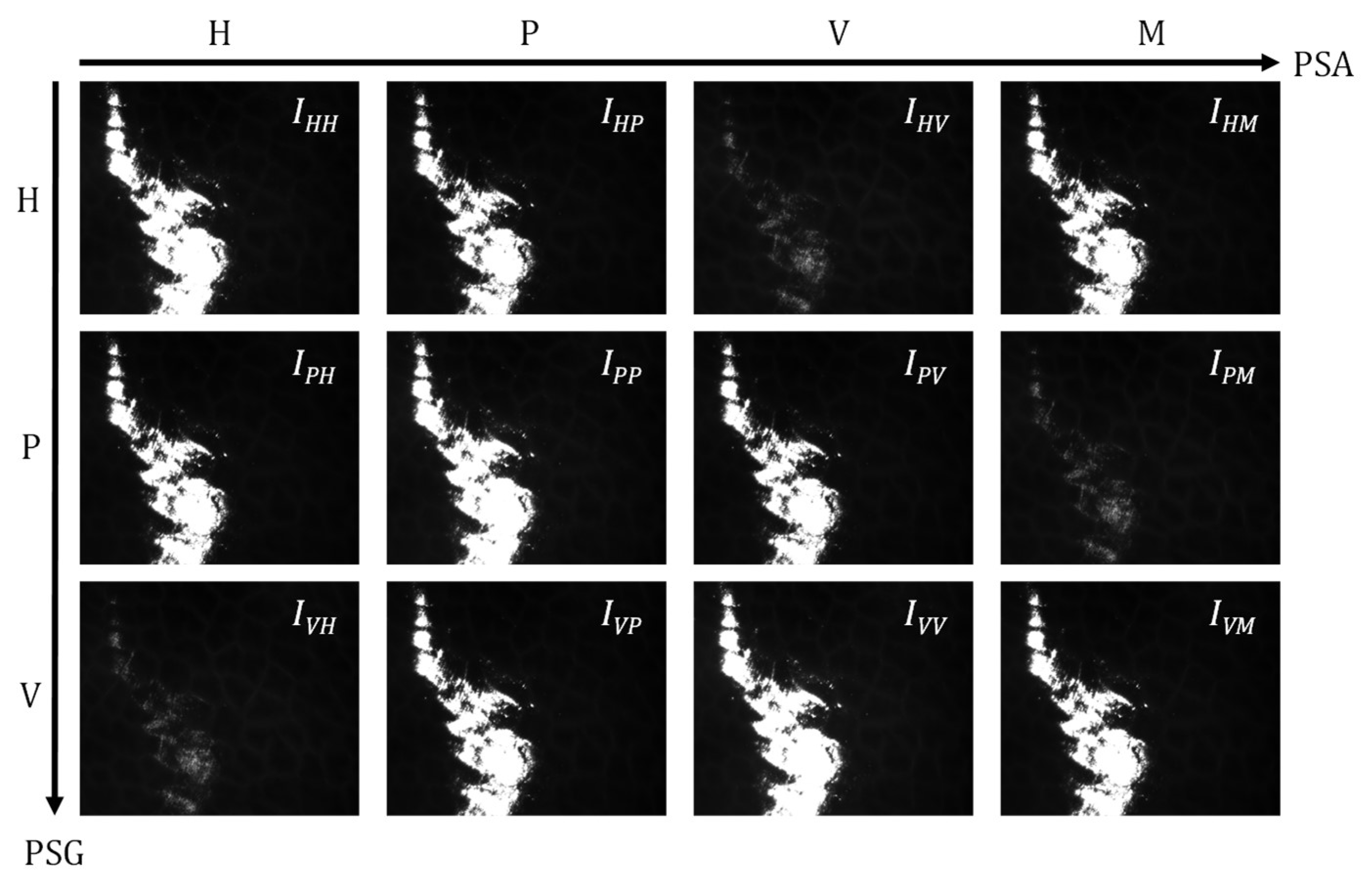
2.1. Sample and Experimental Setup
2.2. Mueller Matrix Transformation (MMT) and Mueller Matrix Polar Decomposition (MMPD) Parameters
2.3. Analysis Methods
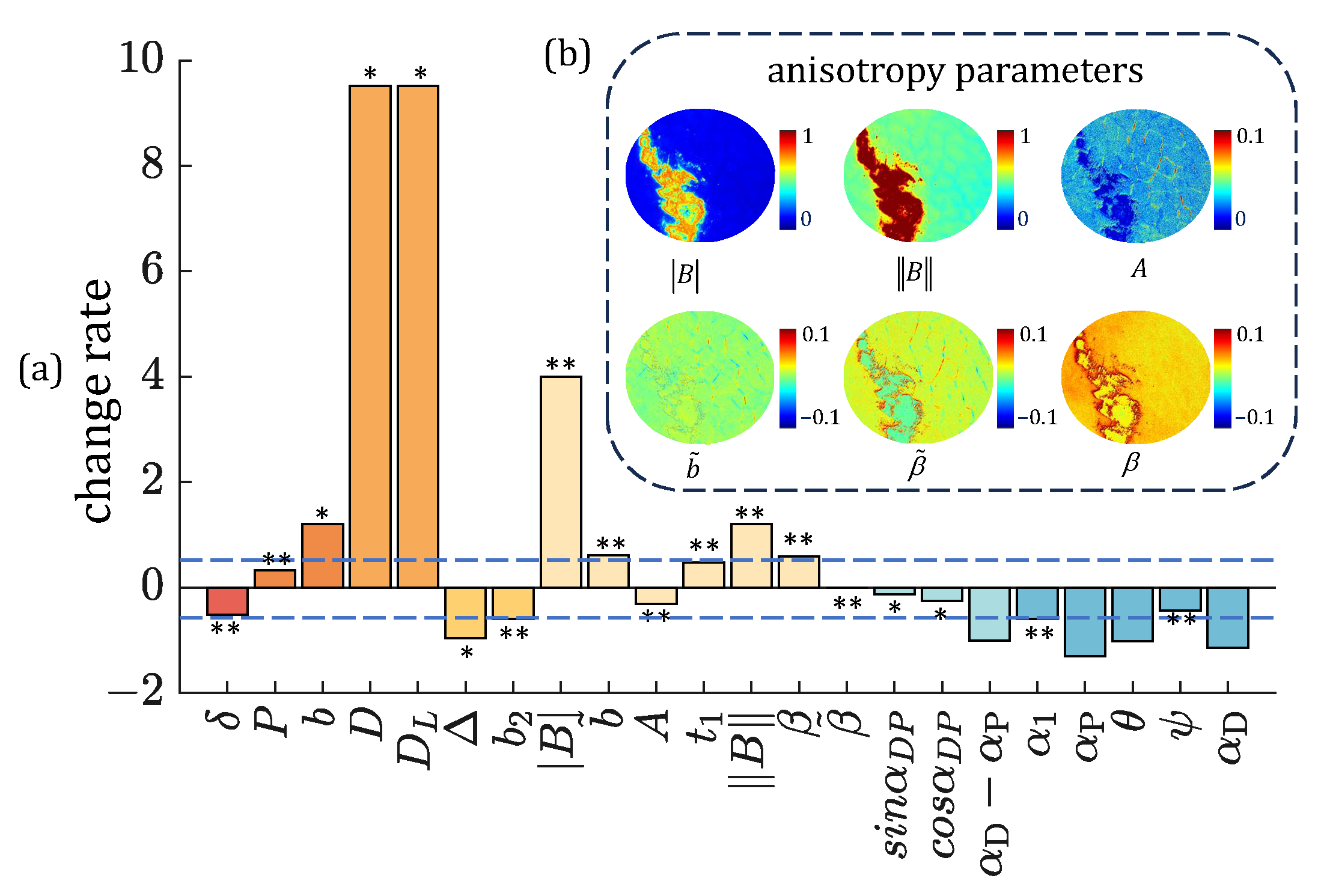
3. Results
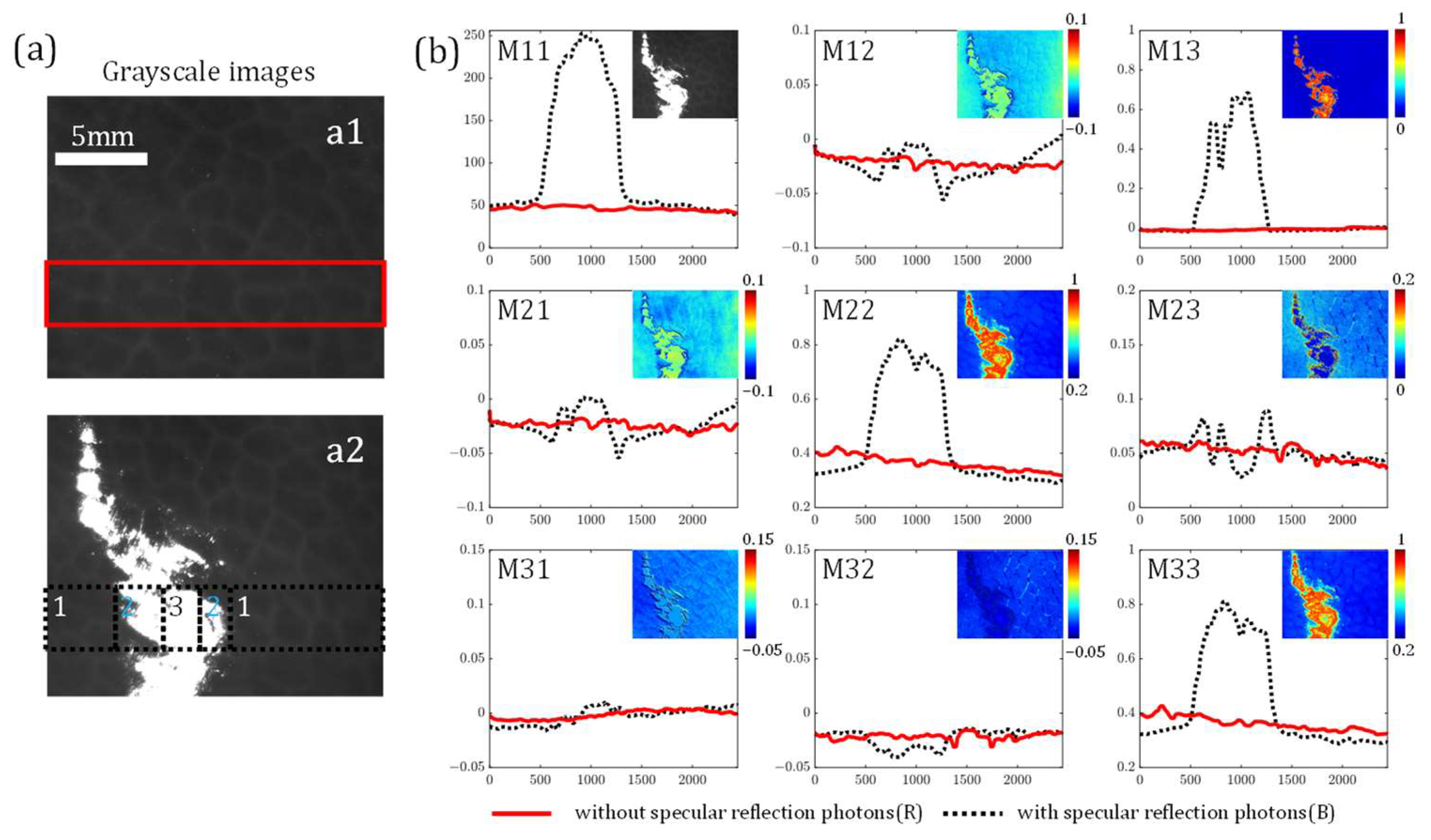
3.1. Influence of Specular Reflection on MM Elements
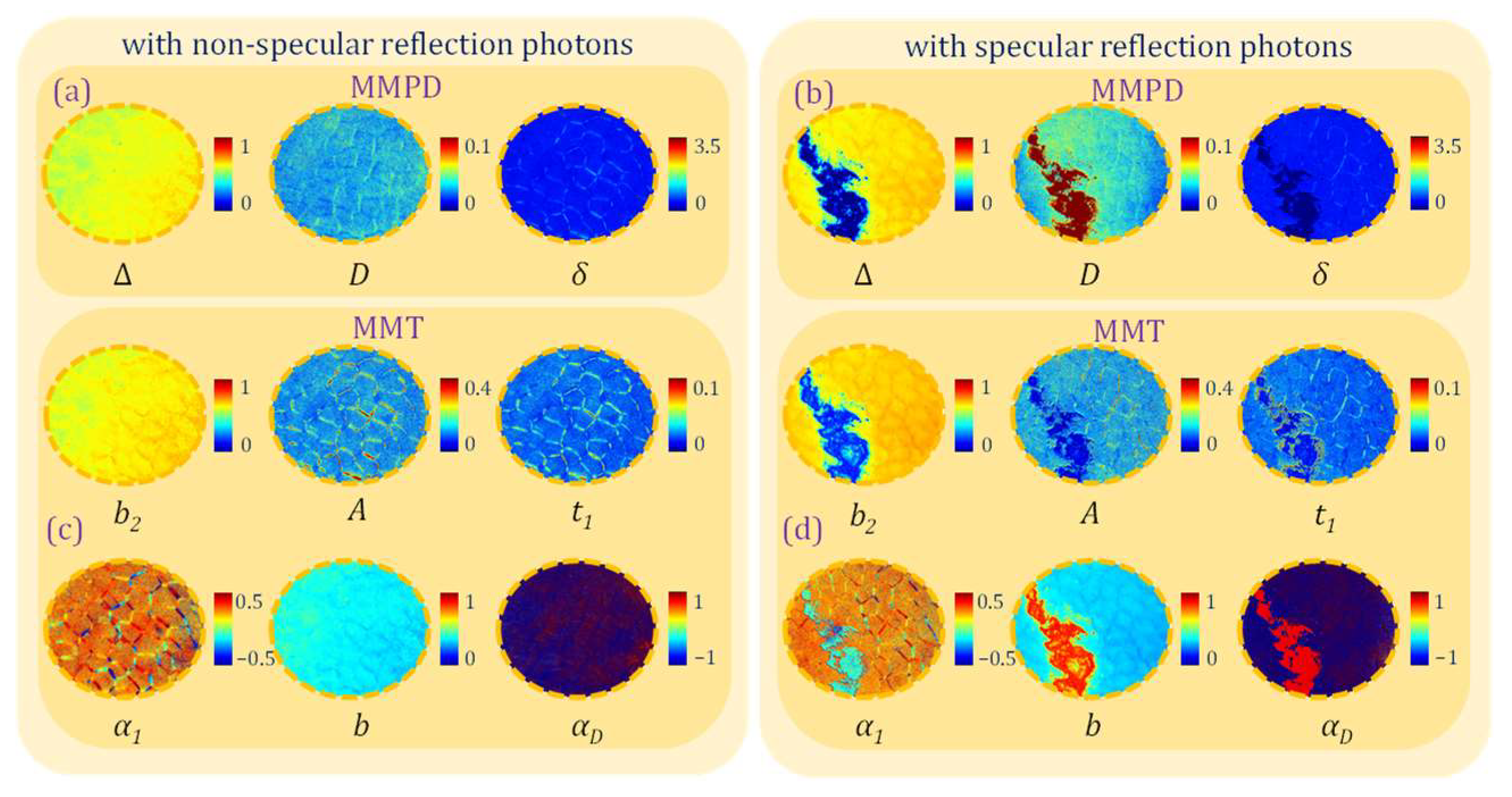
3.2. Polarization Measurement Errors Under Overexposure Induced by Specular Reflection
3.3. Polarimetric Parameters Selection Strategies Under Overexposure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramella-Roman, J.C.; Saytashev, I.; Piccini, M. A review of polarization-based imaging technologies for clinical and preclinical applications. J. Opt. 2020, 22, 123001. [Google Scholar] [CrossRef]
- Tukimin, S.N.; Karman, S.B.; Ahmad, M.Y.; Zaman, W.S.W.K. Polarized light-based cancer cell detection techniques: A review. IEEE Sens. J. 2019, 19, 9010–9025. [Google Scholar] [CrossRef]
- Ghosh, N.; Vitkin, I.A. Tissue polarimetry: Concepts, challenges, applications, and outlook. J. Biomed. Opt. 2011, 16, 110801. [Google Scholar] [CrossRef] [PubMed]
- Tuchin, V.V. Polarized light interaction with tissues. J. Biomed. Opt. 2016, 21, 071114. [Google Scholar] [CrossRef]
- He, C.; He, H.; Chang, J.; Chen, B.; Ma, H.; Booth, M.J. Polarisation optics for biomedical and clinical applications: A review. Light. Sci. Appl. 2021, 10, 194. [Google Scholar] [CrossRef]
- Chue-Sang, J.; Gonzalez, M.; Pierre, A.; Laughrey, M.; Saytashev, I.; Novikova, T.; Ramella-Roman, J.C. Optical phantoms for biomedical polarimetry: A review. J. Biomed. Opt. 2019, 24, 030901. [Google Scholar] [CrossRef]
- Qi, J.; Elson, D.S. Mueller polarimetric imaging for surgical and diagnostic applications: A review. J. Biophotonics 2017, 10, 950–982. [Google Scholar] [CrossRef]
- Ignatenko, D.N.; Shkirin, A.V.; Lobachevsky, Y.P.; Gudkov, S.V. Applications of Mueller matrix polarimetry to biological and agricultural diagnostics: A review. Appl. Sci. 2022, 12, 5258. [Google Scholar] [CrossRef]
- He, H.; Zeng, N.; Du, E.; Guo, Y.; Li, D.; Liao, R.; Ma, H. A possible quantitative Mueller matrix transformation technique for anisotropic scattering media. Photonics Lasers Med. 2013, 2, 129–137. [Google Scholar] [CrossRef]
- Lu, S.Y.; Chipman, R.A. Interpretation of Mueller matrices based on polar decomposition. J. Opt. Soc. Am. A 1996, 13, 1106–1113. [Google Scholar] [CrossRef]
- Cloude, S.R. Group theory and polarisation algebra. Optik 1986, 75, 26–36. [Google Scholar]
- Ortega-Quijano, N.; Arce-Diego, J.L. Mueller matrix differential decomposition. Opt. Lett. 2011, 36, 1942–1944. [Google Scholar] [CrossRef]
- Jiao, W.; Zhang, Z.; Zeng, N.; Hao, R.; He, H.; He, C.; Ma, H. Complex spatial illumination scheme optimization of backscattering Mueller matrix polarimetry for tissue imaging and biosensing. Biosensors 2024, 14, 208. [Google Scholar] [CrossRef]
- Kumar, N.; Kumar Nayak, J.; Pradhan, A.; Ghosh, N. Mueller matrix-based characterization of cervical tissue sections: A quantitative comparison of polar and differential decomposition methods. J. Biomed. Opt. 2024, 29, 052916. [Google Scholar] [CrossRef] [PubMed]
- Bargo, P.R.; Kollias, N. Measurement of skin texture through polarization imaging. Br. J. Dermatol. 2010, 162, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical technologies for the improvement of skin cancer diagnosis: A review. Sensors 2021, 21, 252. [Google Scholar] [CrossRef]
- Manhas, S.; Vizet, J.; Deby, S.; Vanel, J.C.; Boito, P.; Verdier, M.; De Martino, A.; Pagnoux, D. Demonstration of full 4 × 4 Mueller polarimetry through an optical fiber for endoscopic applications. Opt. Express 2015, 23, 3047–3054. [Google Scholar] [CrossRef]
- Qi, J.; Tatla, T.; Nissanka-Jayasuriya, E.; Yuan, A.Y.; Stoyanov, D.; Elson, D.S. Surgical polarimetric endoscopy for the detection of laryngeal cancer. Nat. Biomed. Eng. 2023, 7, 971–985. [Google Scholar] [CrossRef]
- Trout, R.M.; Gnanatheepam, E.; Gado, A.; Reik, C.; Ramella-Roman, J.C.; Hunter, M.; Schnelldorfer, T.; Georgakoudi, I. Polarization enhanced laparoscope for improved visualization of tissue structural changes associated with peritoneal cancer metastasis. Biomed. Opt. Express 2022, 13, 571–589. [Google Scholar] [CrossRef]
- Horiguchi, N.; Tahara, T.; Kawamura, T.; Okubo, M.; Tahara, S.; Nagasaka, M.; Nakagawa, Y.; Shibata, T.; Ohmiya, N. A Comparative Study of White Light Endoscopy, Chromoendoscopy and Magnifying Endoscopy with Narrow Band Imaging in the Diagnosis of Early Gastric Cancer after Helicobacter pylori Eradication. J. Gastrointestin. Liver Dis. 2017, 26, 357–362. [Google Scholar] [CrossRef]
- Lier, M.C.; Vlek, S.L.; Ankersmit, M.; van de Ven, P.M.; Dekker, J.J.; Bleeker, M.C.; Mijatovic, V.; Tuynman, J.B. Comparison of enhanced laparoscopic imaging techniques in endometriosis surgery: A diagnostic accuracy study. Surg. Endosc. 2020, 34, 96–104. [Google Scholar] [CrossRef]
- Alali, S.; Vitkin, A. Polarized light imaging in biomedicine: Emerging Mueller matrix methodologies for bulk tissue assessment. J. Biomed. Opt. 2015, 20, 061104. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Elson, D.S. A high definition Mueller polarimetric endoscope for tissue characterisation. Sci. Rep. 2016, 6, 25953. [Google Scholar] [CrossRef]
- Garcia, M.; Gruev, V. Optical characterization of rigid endoscopes and polarization calibration methods. Opt. Express 2017, 25, 15713–15728. [Google Scholar] [CrossRef]
- Iannucci, L.E.; Gruev, V.; Lake, S.P. Influence of signal-to-noise ratio on DoLP and AoP measurements during reflectance-mode division-of-focal plane Stokes polarimetry of biological tissues. Biomed. Opt. Express 2024, 15, 2798–2810. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Zeng, N.; Jiao, W.; He, H.; He, C.; Ma, H. Cartesian coordinates transformation for backscattering computational polarimetry. Opt. Express 2024, 32, 32294–32308. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Le Brun, G.; Le Jeune, B. Mueller matrix decomposition for biological tissue analysis. Opt. Commun. 2013, 293, 4–9. [Google Scholar] [CrossRef]
- Földesy, P.; Siket, M.; Nagy, A.; Jánoki, I. Correction of overexposure in laser speckle contrast imaging. Opt. Express 2022, 30, 21523–21534. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, D.; Chen, X.; Zhu, L.; Chen, X.; Xie, Z.; Huang, G.; Gao, J.; Shi, W.; Cui, G. Visible near-infrared hyperspectral imaging and supervised classification for the detection of small intestinal necrosis tissue in vivo. Biomed. Opt. Express 2022, 13, 6061–6080. [Google Scholar] [CrossRef]
- Qi, J.; Ye, M.; Singh, M.; Clancy, N.T.; Elson, D.S. Narrow band 3 × 3 Mueller polarimetric endoscopy. Biomed. Opt. Express 2013, 4, 2433–2449. [Google Scholar] [CrossRef]
- Zhang, Z.; Hao, R.; Shao, C.; Mi, C.; He, H.; He, C.; Du, E.; Liu, S.; Wu, J.; Ma, H. Analysis and optimization of aberration induced by oblique incidence for in-vivo tissue polarimetry. Opt. Lett. 2023, 48, 6136–6139. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Yao, Y.; Pei, H.; Hu, Z.; Zhang, F.; Wang, J.; Yu, G.; Huang, C.; Liu, H.; Tao, L.; et al. Mueller matrix imaging of pathological slides with plastic coverslips. Opt. Express 2023, 31, 15682–15696. [Google Scholar] [CrossRef] [PubMed]
- Swami, M.; Manhas, S.; Buddhiwant, P.; Ghosh, N.; Uppal, A.; Gupta, P. Polar decomposition of 3 × 3 Mueller matrix: A tool for quantitative tissue polarimetry. Opt. Express 2006, 14, 9324–9337. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.; Quach, T.; Vo, Q. Analysis of polarization features of human breast cancer tissue by Mueller matrix visualization. J. Biomed. Opt. 2024, 29, 052917. [Google Scholar] [CrossRef]
- Li, P.; Dong, Y.; Wan, J.; He, H.; Aziz, T.; Ma, H. Polaromics: Deriving polarization parameters from a Mueller matrix for quantitative characterization of biomedical specimen. J. Phys. D Appl. Phys. 2021, 55, 034002. [Google Scholar] [CrossRef]
- Khaliq, A.; Ashraf, S.; Gul, B.; Ahmad, I. Comparative study of 3 × 3 Mueller matrix transformation and polar decomposition. Opt. Commun. 2021, 485, 126756. [Google Scholar] [CrossRef]
- Ghosh, N.; Wood, M.F.; Vitkin, I.A. Mueller matrix decomposition for extraction of individual polarization parameters from complex turbid media exhibiting multiple scattering, optical activity, and linear birefringence. J. Biomed. Opt. 2008, 13, 044036. [Google Scholar] [CrossRef]
- Chen, J.L.; Lee, H.J. An efficient algorithm for form structure extraction using strip projection. Pattern Recognit. 1998, 31, 1353–1368. [Google Scholar] [CrossRef]
| 3 × 3 Mueller matrix transformation (MMT) parameters | |
| 3 × 3 Mueller matrix polar decomposition (MMPD) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, W.; Zeng, N.; Hao, R.; Ma, H.; He, C.; He, H. Probing the Influence of Specular Reflection and Overexposure on Backscattering Mueller Matrix Polarimetry for Tissue Imaging and Sensing. Biosensors 2025, 15, 333. https://doi.org/10.3390/bios15050333
Jiao W, Zeng N, Hao R, Ma H, He C, He H. Probing the Influence of Specular Reflection and Overexposure on Backscattering Mueller Matrix Polarimetry for Tissue Imaging and Sensing. Biosensors. 2025; 15(5):333. https://doi.org/10.3390/bios15050333
Chicago/Turabian StyleJiao, Wei, Nan Zeng, Rui Hao, Hui Ma, Chao He, and Honghui He. 2025. "Probing the Influence of Specular Reflection and Overexposure on Backscattering Mueller Matrix Polarimetry for Tissue Imaging and Sensing" Biosensors 15, no. 5: 333. https://doi.org/10.3390/bios15050333
APA StyleJiao, W., Zeng, N., Hao, R., Ma, H., He, C., & He, H. (2025). Probing the Influence of Specular Reflection and Overexposure on Backscattering Mueller Matrix Polarimetry for Tissue Imaging and Sensing. Biosensors, 15(5), 333. https://doi.org/10.3390/bios15050333






