Abstract
Aptamers have emerged as powerful molecular recognition elements for biosensing applications, offering high specificity, stability, and adaptability. This review explores key considerations in designing aptamer-based sensors (aptasensors), with a focus on biomarker selection, aptamer design, and detection and immobilization strategies. However, challenges such as biofluid stability and reversibility must be addressed to improve biosensor performance. In this study, the potential of aptamer-based platforms in diagnostics is explored, emphasizing their advantages and future applications. Looking ahead, advances in multifunctional aptamers, integration with nanomaterials, and computational optimization are highlighted as promising directions for enhancing their effectiveness in biosensing.
1. Introduction
Biosensing has become one of the most critical aspects of modern healthcare, enabling early detection and monitoring of a wide range of diseases. Biosensors can give a fast response using raw samples, which makes them excellent candidates for at-home tests and fast screening of diseases [1]. In recent years, biosensors have taken the spotlight in applications such as coronavirus disease (COVID-19) tests, which helped contain the pandemic’s spread. Every work location and every home performed a fast test to detect the presence of the virus using several biofluids, such as saliva, blood, and nasal mucosa. Biosensors also play an important role in diabetes management through the development of continuous glucose sensing. Moreover, emerging technologies such as robots, autonomous vehicles, and space exploration greatly benefit from the application of sensors and biosensors. The recent advancements in biosensing platforms can be related to the fast-paced development of artificial intelligence. The power of machine learning allowed big data analysis and the discovery of new correlations and predictions using biosensing information, particularly from wearable technology. The combination of molecular and physical data augments the information given to us to prevent and detect diseases and facilitate earlier predictions for effective intervention and improved patient care [1,2].
Today, what is limiting the applications of biosensors is the biosensing technology itself. Most sensing strategies rely on enzymatic detection, where antibodies labeled with enzymes enable affinity-based recognition and signal generation. There is an urgent need to develop novel sensing technologies able to detect an extended variety of biomolecules with fast response, accuracy, and reproducibility. Naturally occurring molecules such as antibodies and enzymes can only detect a limited range of targets. Aptamers expand the range of possible targets because, despite the challenges in the SELEX method, they can be selected and designed for, in principle, any target. Additionally, in medicine, accessibility is a vital factor with preventative care relying on the ability to track biomarkers in real-time rather than relying on costly and time-consuming laboratory tests. In this context, aptamers have been presented as ideal candidates to extend biosensing applications [3]. Aptamers have emerged as groundbreaking biosensing tools that can bridge the gap between traditional antibodies and small-molecule drugs [4]. They are also known as nucleic acid antibodies and are based on short, single-stranded DNA or RNA molecules that can fold into defined three-dimensional architectures, allowing them to bind to specific targets. The high tunable affinity of aptamers is made possible due to having an infinite number of possible base pairs, facilitating a wide range of bonding possibilities [5]. They can be tailored to have high affinity, selectivity, and specificity for their targets, allowing them to differentiate between nearly identical biomarkers, such as proteins that differ by a single amino acid [4].
An aptamer’s equilibrium of folded and unfolded states is fundamental to signal detection in biosensing applications, where the equilibrium of these two states can be shifted by the presence of a target biomarker [6]. An advantage of using aptamers is their intrinsic reversibility, enabling continuous and dynamic sensing where the surface can be refreshed (regenerated), returning to their original shape after each time a molecule is detected. While both antibodies and aptamers use non-covalent interactions, aptamers can be chemically optimized to control binding strength and stability [7,8]. Another benefit of using aptamers is their ability to target multiple biomarkers at once, known as multiplexed sensing or multiplexing. This is very beneficial in healthcare applications because measuring multiple biomarkers gives the user a clearer picture of what is possibly occurring in the body. For example, these multiplexed biosensors can detect multiple antibiotics, pathogens, and cancer biomarkers in a single analysis [9].
In order to develop new aptamers, a well-defined iterative procedure is employed, known as the Systematic Evolution of Ligands by Exponential Enrichment (SELEX). The SELEX method is a way of screening that allows for the iterative selection of highly specific aptamers for a defined target [10]. SELEX starts with a large library of base pairs, which increases the likelihood of identifying either DNA or RNA sequences that can bind to the desired target. These recurring DNA or RNA motifs can assemble and create complex structures for target recognition [4]. Aptamers exist in dynamic equilibrium between their low-energy and high-energy configurations (for example, folded and unfolded). The addition of the biomolecular target, to which the aptamer has a strong affinity, lowers the enthalpy of the higher energy state, thereby driving the aptamer to change shape dynamically. This change in configuration, driven by the presence of a biomolecule, can then be leveraged to generate output signals for sensing. The equilibrium dissociation constant KD represents the extent to which the complex dissociates into a free aptamer and a free target. It will mirror the binding affinity of an aptamer to its target molecule and provide a metric for evaluating aptasensor performance [11].
Aptamers are very versatile regarding their transduction mechanisms, which allows for a wide range of sensing applications. In aptamer-based biosensors, transduction refers to the process of converting the binding of a target molecule into a detectable signal. There are different ways to extract this signal, and the technique used depends on the application that is desired. A common approach is electrochemical aptasensors (E-AB), which use a three-electrode setup with an aptamer-modified working electrode. These sensors measure electrical signals corresponding to an electrochemical reaction, usually with a redox probe tethered to one end of the immobilized aptamer. When the target molecule binds to the immobilized aptamer, the aptamer will fold, causing the redox probe (attached to one end of the aptamer) to move closer or further away from the electrode surface, therefore causing a measurable change in the signal obtained. The signal in E-AB sensors is read through the change in current–voltage characteristics (e.g., cyclic voltammetry, differential pulse voltammetry, and impedance), and the compact size of aptamers is critical in reducing the distance between the electrode and probe, allowing for high sensitivity [12].
Another strategy that can be used is fluorescent detection, where an aptamer binds to its target and undergoes a conformational change that alters the lifetime of a tethered fluorescent probe [13]. Förster Resonance Energy Transfer (FRET) and quenching are two fluorescence-based techniques that can be used to enable the detection of targets. In quenching, a decrease in signal intensity is observed when a fluorescently labeled aptamer binds to a target molecule. FRET-based biosensors use fluorescence to detect biomolecules and changes in their microenvironment [14]. FRET is a non-radiative transfer of energy from an excited donor fluorophore molecule to a nearby acceptor molecule [9]. In these biosensors, the donors and acceptors are normally fluorescent molecules, proteins, or quantum dots. When a target biomolecule is present, it will alter the distance between the donor and acceptor, resulting in a change of relative fluorescence intensity. As the distances get closer together, FRET causes the donor’s fluorescence intensity to decrease while the acceptor’s increases. This change in relative intensity will be a measurable signal that allows for the detection and quantification of a biomolecule concentration.
However, making aptasensors is not trivial. The complete process of going from a health-related disease to having a reliable biosensor for disease detection can be a long and difficult path. Starting off, selecting the correct biofluid to target, such as blood, cerebrospinal fluid (CSF), urine, sweat, or tears, can be challenging and requires careful consideration. This choice is crucial because it will determine the sensor’s environment for operation and influence which aptamer is selected. After selecting a biofluid, the SELEX process is used to identify aptamers that can specifically bind to the target in the chosen biofluid environment. Once an aptamer is selected, it often requires further modification to enhance compatibility with the chosen transduction method to ensure an efficient signal. The aptamer then needs to be functionalized in order to obtain a signal or output. Finally, it can be integrated into a functional sensor and go through validation to ensure the device works as intended in real-world conditions. After all these steps, the device can be manufactured and deployed. This review will act as a guide to navigate the process of designing and implementing aptamer biosensors.
In this review, we highlight the incredible potential of aptamers in biosensing while demonstrating the steps from start to finish on how to begin using aptamers. There is an increased need for more research in the area of aptamers, and this review is proposed to invite readers to explore the possibilities aptamers can bring. Unlike several other excellent reviews on aptamers, the following sections are intended to bring readers practical knowledge on how to start aptamer-related research. We focus on the criteria that need to be considered to select the biomarker, biofluid, and aptamer, followed by a discussion on techniques that can be employed for aptamer-based biosensors and considerations for aptamer immobilization. Finally, we show groundbreaking examples of state-of-the-art work utilizing aptamers from the inside to the outside of the body.
2. How to Select a Biomarker
In designing aptamer-based sensing platforms, the choice of biomarker underlies any biosensor, and it should be carefully considered because it will influence the selection of the aptamer and the overall configuration of the device. According to the FDA, biomarkers could be defined as “characteristics that are objectively measured as indicators of health, disease, or a response to an exposure or intervention, including therapeutic interventions” [15].
When selecting a biomarker, the first question that arises is whether it is clinically relevant. This means that the biomarker of interest provides meaningful insights into a patient’s health status by acting as an indicator of disease presence, progression, or treatment response [16]. Specifically, biomarkers with FDA-approved applications offer a foundation for regulatory acceptance and integration into clinical workflows [17]. Secondly, it is important to consider the accessibility of the biofluid containing the biomarker, its concentration, possible interferents, and its compatibility with the sensing platform, since it directly impacts the feasibility of biomarker detection and biosensor design.
Biomarkers are commonly measured in biofluids such as serum, urine, saliva, blood, and cerebrospinal fluid (CSF), as well as exhaled air condensate and gastrointestinal fluids (Figure 1). Additionally, noninvasive sources such as sweat and interstitial fluid (ISF) are increasingly being explored for diagnostic purposes [18,19]. Each biofluid presents unique challenges; for example, blood and serum often contain high levels of proteins that can interfere with measurement. In contrast, sweat and interstitial fluid may require adjustments for ionic strength, pH, or composition variability [12]. Additionally, these biofluids offer the advantage of continuous monitoring with minimal discomfort, aligning with the growing demand for real-time diagnostic systems. However, real-time monitoring is not always necessary; it depends on the biomarker’s role in the disease. Biomarkers that undergo rapid changes, such as glucose or stress-related molecules, benefit significantly from dynamic monitoring [20,21,22]. Aptamer-based sensors are particularly suited for such applications due to their high specificity, fast response times, and adaptability to wearable technologies.
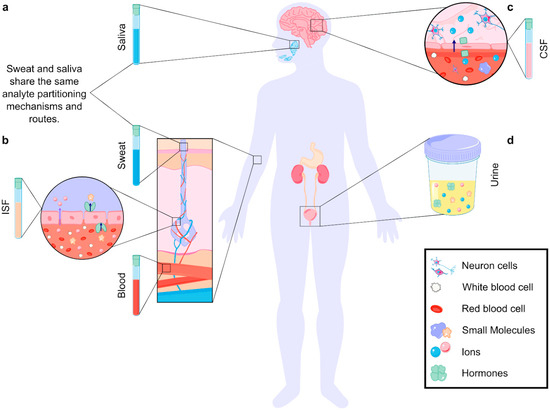
Figure 1.
Analyte partitioning in different biofluids. (a) In biofluids produced by glands (sweat, saliva), analytes travel from the blood, through ISF, and finally to the glands. Analytes are filtered in each step, making the analyte concentrations in blood differ from those in the biofluids [23]. (b) Analytes enter biofluids through paracellular (purple arrow) and transcellular (dark blue arrow, transporter-mediated) transport. The size of the analyte, cellular structure, and partitioning mechanism define the final concentration in the fluid [23]. (c) Analytes can travel from the blood to the cerebrospinal fluid (CSF) through the blood–brain barrier (BBB). Partitioning mechanisms include paracellular and transcellular transport (ion transport depicted) [24,25]. (d) Analytes can also be found in other noninvasive biofluids, such as urine.
Aptamers can be highly sensitive to biomarker concentrations, allowing for detection over a tunable range, from micromolar to picomolar levels [26]. This sensitivity makes them particularly useful for identifying biomarkers present at very low concentrations, which is often the case in the early stages of disease progression in cancer, neurodegenerative, or cardiovascular pathologies [27]. However, because aptamers typically exhibit a single, fixed binding affinity, they tend to have a narrow range of concentrations over which they provide accurate quantification, as they saturate quickly once the target concentration exceeds a certain threshold [28]. Unlike enzymes, aptamers do not catalyze reactions that regenerate the sensing interface, so their signal is directly tied to target occupancy, making saturation a key constraint. This fact makes them less suitable for detecting biomarkers whose concentrations vary widely under different physiological or pathological conditions. To overcome this problem, aptamers can be engineered through the addition of structural modifications and biosensor design strategies, expanding their working range [29,30].
One of the advantages of aptamers is that they can target a wide range of biomarkers with high specificity, including small molecules, ions, proteins, and even whole cells, forming non-covalent bonds via van der Waals, hydrogen bonding, and electrostatic interactions [31,32]. In contrast to aptamers, antibodies cannot target molecules that do not generate an immune response, as their production relies on the activation of the immune system, which typically responds only to foreign or immunogenic substances. Small molecules, certain lipids, and other compounds often fail to elicit such a response, making antibody generation challenging without conjugation to a larger carrier protein [33].
Overall, the biomarker selection should take into account the biofluid where the target will be detected. An example of the workflow is detailed here, employing glucose, an important biomarker that can be found in all biofluids, including blood, saliva, sweat, cerebrospinal fluid (CSF), and interstitial fluid (ISF). Suitable aptamers for glucose detection are flexible structures that are able to undergo conformational changes upon binding, allowing for a signal transduction mechanism that can be integrated into a sensing platform [34]. The selectivity of the aptamer relies on its ability to recognize glucose specifically while minimizing interactions with structurally similar molecules, such as fructose or galactose, which may be present in the biofluid. Once the main features of this aptamer are established by the properties of its target, the biofluid’s intrinsic composition needs to be considered. In sweat, glucose concentrations are significantly lower than in blood [35], requiring an aptamer with high sensitivity to detect minute fluctuations accurately and remain stable at variable ionic strengths and pH conditions. In CSF, where glucose is present at approximately 60% of blood glucose levels [36], the aptamer should be optimized for a narrower dynamic range, ensuring accurate quantification without saturation. Additionally, CSF is a relatively stable and low-protein environment [37], reducing interference but requiring aptamers with high specificity to avoid cross-reactivity with other metabolic components. Conversely, for gastrointestinal fluids, the highly variable pH (ranging from the acidic conditions in the stomach to the near-neutral environment of the intestines) [38] creates the need for an aptamer that maintains its structural integrity and binding functionality across a wide pH range, potentially through the incorporation of chemically modified nucleotides or protective surface coatings.
Continuing this rationale, C-reactive protein (CRP), an important inflammatory biomarker present in biofluids such as blood, saliva, sweat, cerebrospinal fluid (CSF), and interstitial fluid (ISF), will now be exemplified. Specific biofluid characteristics remain critical. Aptamers tailored for CRP detection must demonstrate high selectivity, effectively discriminating CRP from structurally or functionally related proteins like serum amyloid A (SAA), which may be present concurrently in these biofluids. Because CRP is a large protein (114 kDa), its concentration in sweat is lower compared to blood, requiring highly sensitive aptamers with robust behavior to ionic strength and pH variations. Likewise, the same CSF and gastrointestinal fluid characteristics explained before need to be considered when designing an aptasensor for CRP. CRP is a large protein that requires from small to moderately long aptamers to reach sufficient affinity (20–80 nucleotides) [39,40]. While longer aptamers effectively recognize and bind CRP, employing redox probes within electrochemical detection systems remains feasible but presents specific challenges. For example, the electron transfer distance increases, reducing signal strength and compromising detection sensitivity. Additionally, more complex folding dynamics can lead to slower response times and higher background noise due to nonspecific interactions. In this sense, other detection techniques can also be influenced by the response time, causing issues in fluorescent intensity, aggregation when using nanoparticles, etc. However, these drawbacks can be mitigated through strategic aptamer optimization, precise surface modification, and careful control of experimental conditions.
Aside from choosing a biomarker with specific relevance for the condition of interest, considering the physiological and biochemical differences among biofluids is essential for aptamer design for sensing purposes.
3. How to Select an Aptamer
The effectiveness of aptamer-based sensing platforms heavily depends on the ability of the aptamer to bind specifically and strongly to its target, which is determined by its secondary structure [41]. Whereas the primary structure of an aptamer is its linear sequence of nucleotides, its secondary structure refers to its three-dimensional configurations, such as stem loops, hairpins, and more complex G-quadruplex structures [42] (Figure 2). These configurations allow the aptamer to create the necessary architecture for high-affinity binding, essentially acting as a molecular “lock” that fits its target “key”. For that reason, one of the most important factors influencing the binding affinity of an aptamer is its overall shape [43]. The specific conformation of an aptamer determines how well it can fit into the binding pocket of its target, in contrast to antibodies with a rigid structure. Thus, the aforementioned non-covalent interactions, in addition to shape complementarity, contribute to the strength and specificity of the binding [44].
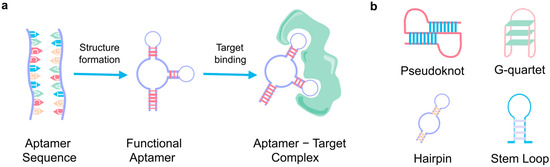
Figure 2.
Aptamer basics. (a) The primary aptamer sequence, folding into its three-dimensional structure and binding with its specific target. (b) The four main secondary structures of aptamers.
Particularly for small molecules, aptamers often start in a relatively unstructured or loosely folded state and undergo a significant conformational change upon binding [31]. This folding creates a precise binding pocket that tightly accommodates the small molecule, enhancing specificity. A unique feature of aptamers is their ability to distinguish between different chiral forms of a molecule, which is essential for targeting small molecules or peptides that may exist in multiple enantiomeric forms [45] and is critical across the pharmaceutical, agrochemical, food, and fragrance industries. Mirror-image molecules, despite having the same molecular formula, can exhibit markedly different biological activities, environmental behaviors, and sensory properties. This feature was explored by Challier et al. [46], who designed an electrochemical sensor capable of detecting one of the enantiomers of tyrosinamide, a small chiral molecule used as a model to exemplify the methodology. Their approach was based on the different diffusion rates between the molecule and the aptamer/molecule complex [46]. This selectivity is further enhanced by aptamers’ capacity to differentiate their target from structurally similar derivatives or analogs [47], making them extremely selective in complex biological environments, while antibodies and enzymes sometimes show cross-reactivity [48].
In the case of large molecules (i.e., proteins), aptamers generally retain more of their pre-folded secondary and tertiary structures even before binding. Rather than a complete structural reorganization, binding to a large target usually involves localized structural adjustments to optimize interactions across multiple binding sites [43,49]. Aptamers designed for proteins are key in detecting disease biomarkers, with their ability to recognize specific surface features. Due to the negatively charged phosphate backbone of aptamers, they tend to interact favorably with positively charged regions of proteins, forming stable and specific complexes [50]. Additionally, some aptamers are tailored to bind entire cells or cell surface proteins, enabling the detection of pathogens or abnormal cells [51]. For example, detecting circulating tumor cells (CTCs) in breast cancer is challenging due to their low concentration and phenotypic heterogeneity. Traditional methods, like antibody-based systems, rely on epithelial markers but may miss certain tumor cell subtypes [52]. Kolovskaya et al. explored RNA aptamers, which bind with high specificity to various breast cancer subtypes, employing magnetic separation, effectively isolating CTCs from blood samples. Due to their high affinity, structural versatility, and ability to target diverse CTCs, aptamers improved detection sensitivity in a minimally invasive manner [53]. Furthermore, Kolm et al. identified an aptamer that binds selectively to E. faecalis cells using a combined approach based on cell-SELEX and quantitative real-time PCR (polymerase chain reaction), providing a promising tool for the development of sensitive and specific detection of bacterial cells [54].
Once a target of interest is identified, selecting a suitable aptamer can follow one of three potential pathways [2]. First, a relevant aptamer may already be available in the literature, aligning well with the intended application. Therefore, the sequence of the aptamer can be ordered from a company or even synthesized in-house. Second, while an aptamer may be documented, its characteristics may make it unsuitable for the application’s specific requirements, such as sensitivity to nucleases, toxicity, and/or inappropriate size or shape. Lastly, if no aptamer is available or appropriate, a new sequence must be designed and selected through experimental processes. This aptamer selection could be performed in collaboration with researchers and specialized companies (Creative Biolabs, Novaptech, NeoVentures, AptamerLab, SomaLogic, Aptagen, Aptamer Group, Aptamer Sciences, NOXXON Pharma, etc.).
The next sections will include a description of the possibilities for aptamer selection, fabrication, and modification, as well as different strategies for their integration into sensing platforms.
3.1. Design, Fabrication, and Modification of Aptamers
The SELEX (Systematic Evolution of Ligands by Exponential Enrichment) process (Figure 3), the most widely used method for aptamer selection, is centered around the interaction between the aptamer and its target, which forms the foundation for successful downstream applications [55,56]. Aptamers function by binding to their targets with high specificity, and this interaction can vary depending on the relative sizes of the aptamer and its target. If the aptamer is larger than its target, it tends to incorporate the target into its structure. Conversely, when the target is larger, such as proteins, the aptamer may integrate into the structure or bind to the surface [57].
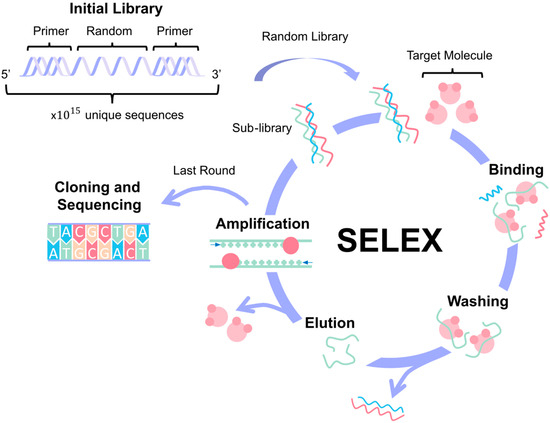
Figure 3.
Aptamer design. The Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process allows for aptamer design and selection based on the specific target molecule. It starts with the creation of an aptamer library where the sequences have a central random region flanked by constant primer regions. The process involves 1 to 16 cycles of binding, washing, elution, and amplification. In the last round, the high-affinity aptamers are identified for cloning and sequencing.
The SELEX process begins with generating a diverse library of single-stranded nucleic acids, which can be either DNA or RNA. This library consists of a central random sequence flanked by constant primer regions at both the 5′ and 3′ ends. The random sequence introduces vast variability, allowing for the potential discovery of aptamers with diverse binding properties. The length of this random region, which typically ranges from 20 to 80 nucleotides, and the choice of either natural or chemically modified nucleotides are critical factors that influence the library’s diversity and functionality [57]. The next question is how to choose a library. There are established libraries available to buy (Creative Biogene, Fusion BioLabs, Aptamer Group, etc.), and it is also possible to design a customized one. When creating the initial library, researchers should consider the target’s properties (charge, hydrophobicity, and size) and the intended use of the aptamer. It is also important to maximize sequence diversity and to include any specific sequences or structural features needed [58].
Initially, the library contains around 1015 unique sequences, plus the target, which is immobilized onto a solid support for the SELEX procedure. The process involves several iterative rounds of selection, typically between 1 and 16 cycles, though more may be necessary for highly specific targets. Each cycle consists of four main steps: binding, where the nucleic acids interact with the target molecule; washing/elution, to remove unbound or weakly bound sequences; amplification, usually via PCR or RT-PCR, to enrich the pool of bound sequences; and sequencing, to monitor the enrichment and identify the high-affinity aptamers [59]. The main factors that affect the SELEX process are the enrichment of high-affinity binders and the elimination of low-affinity or nonspecific sequences [2].
Traditional SELEX variants usually involve immobilizing the target molecule on a carrier matrix while the aptamers remain in solution (e.g., bead-based SELEX (Figure 4a) and microfluidic SELEX) [55,60]. However, the immobilization of the target can introduce biases or lead to structural alterations that may impact aptamer selection. To circumvent these limitations, alternative approaches such as microarray-based SELEX [61], capture SELEX [62], atomic force microscopy SELEX [63], and particle display SELEX [64] immobilize the aptamer library instead, keeping the target in a solution, as represented in Figure 4b. Another method, capillary electrophoresis (CE) SELEX [65], keeps both the target and the library in a solution to maintain a more natural interaction between them (Figure 4c). Moreover, Figure 4d shows specialized SELEX techniques such as cell-SELEX [53] and 3D cell-SELEX [66], which allow for the selection of aptamers that can specifically recognize certain cell types or conformations. In vivo SELEX (Figure 4e) represents an even more advanced approach, where an aptamer library is injected into a living organism, and aptamers that bind to target organs are harvested, offering a more physiologically relevant context for aptamer selection [67]. A different approach to SELEX has emerged, involving the use of computational bioinformatics methods like molecular docking and molecular dynamics (MD) simulations to design aptamers for various applications. This in silico strategy can also be integrated with SELEX to improve research efficiency [68].
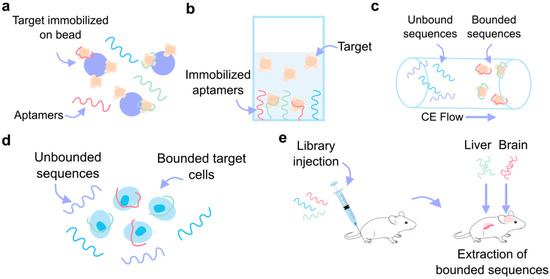
Figure 4.
SELEX variants. (a) Bead-based SELEX, where the target molecule is immobilized on the beads and the aptamer remains free in solution. (b) Target in solution SELEX, where the aptamers are immobilized to the substrate. (c) Capillary electrophoresis (CE) SELEX, where the unbounded and bounded sequences are separated based on their different flow rates over the electrophoresis gel. (d) Cell SELEX allows for the aptamer-based recognition of specific cells. (e) In vivo SELEX, where the aptamer library is injected into the animal model and the bound sequences are collected from organ tissue.
The structural diversity of the oligonucleotide library is a key factor that determines the success of SELEX. A library with greater diversity is more likely to contain aptamers capable of interacting effectively with a wide range of targets [2]. As mentioned before, both RNA and DNA aptamers can bind a specific target with high selectivity. Nevertheless, RNA oligonucleotides tend to offer higher structural complexity compared to DNA, which can enhance binding interactions. Conversely, DNA libraries provide certain advantages, such as increased chemical and biological stability, easier synthesis, and avoidance of the reverse transcription step necessary with RNA SELEX. Both RNA and DNA libraries have unique properties, and their structural diversity can be further increased by optimizing the ratio of different nucleotides during synthesis [2,69].
Despite advances in SELEX methodologies, several challenges remain, particularly when translating aptamer selection from controlled laboratory conditions to real-world biological environments [70]. Most aptamers are selected under highly defined buffer conditions, which often do not reflect the complexity of biological fluids. This discrepancy can lead to a limited number of aptamers performing well in actual biological contexts, such as in vivo conditions [42]. To overcome these limitations, further optimization and validation of aptamers in physiologically relevant environments are necessary, with the in vivo SELEX approach being an alternative [70].
An important aspect of this optimization process involves ensuring that the aptamer folds into its active conformation, which is essential for efficient binding. Several factors influence aptamer folding, including temperature, buffer composition, incubation time, and aptamer concentration [71]. Additionally, aptamer performance can be improved through sequence truncation, where nonessential nucleotides are removed to reduce synthesis costs and enhance binding efficiency [72]. The aptamer’s stability, binding affinity, or specificity can be increased by introducing modifications during the SELEX process or post-SELEX process [8,73], as shown in Table 1. Although these strategies could have important benefits over the aptamer performance, the cost and time investment in chemical synthesis, as well as the natural folding disruption of the aptamer, is something to consider at this stage [2].

Table 1.
Strategies for aptamer modifications.
Due to the extensive number of existing molecules, it is nearly impossible to have an aptamer for each of them. Thus, an interesting approach involves selecting aptamers for a specific feature of a family of molecules. These aptamers could be implemented in the separation, detection, or identification of similar molecules [93].
3.2. Getting Signal in Sensing Platforms Using Aptamers
The use of aptamers as biorecognition elements in biosensing, or aptasensors, is fundamentally based on the selective binding of the aptamer to its target molecule. When the target is present in a sample, it binds to the aptamer, and the binding event is transduced into a detectable signal. This signal can be generated through various physical mechanisms, depending on the biosensor format, and it is then quantified to determine the presence or concentration of the target molecule in the sample (Figure 5) [94].
Aptasensors can be configured in two primary assay formats: single-site binding and dual-site binding, depending on the size and nature of the target molecule. In single-site binding, the aptamer binds to a single target molecule, whereas dual-site binding involves aptamers interacting with two distinct sites on the target, which is particularly useful for larger biomolecules [94].
Typically, three main strategies are employed for target detection (capture, sandwich, or displacement strategies). In the capture strategy, an aptamer directly binds to the target analyte, generating a measurable signal due to a conformational change, making it suitable for simple and rapid detection [95]. It is important to consider that the aptamer needs to have a significant structure change, or it will not generate a measurable signal. The sandwich strategy employs two recognition elements: one immobilized to capture the target and another labeled to detect it, enhancing both specificity and signal amplification [96]. However, it involves a more complex assay design and requires additional reagents. In the displacement strategy, the target displaces a pre-bound molecule from the aptamer, disrupting the complex and triggering a detectable signal, which is particularly useful for competitive assays or small molecule detection, but may suffer from lower specificity if there are weak interactions between the pre-bound molecule and the aptamer, leading to nonspecific displacement. Each strategy offers distinct advantages and disadvantages depending on the target and desired sensitivity [97].
One of the unique features of aptamers is their ability to undergo conformational changes upon binding to their target molecules. These “aptamer switches” can alter their structure in response to binding, and this conformational shift can be exploited to transduce various signals (On and Off signals). For example, structural changes can generate fluorescence, colorimetric, or electrochemical signals, which can then be integrated into different biosensor platforms (Figure 5) [95]. However, converting an aptamer into a “structure-switching” biosensor poses significant challenges. The process often relies on trial and error, since established design principles for generating aptamer switches are still lacking. This makes the development of these biosensors a complex task, requiring empirical testing to optimize the aptamer’s ability to reliably produce a signal upon target binding [98].
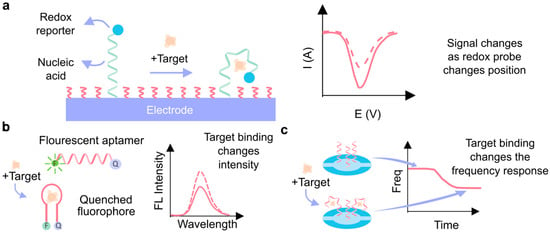
Figure 5.
Signal transduction techniques using aptamers. (a) Electrochemical aptasensor. In the presence of the target molecule, the aptamer undergoes a conformational change, which is transduced by the redox reporter and measured by electrochemical techniques. (b) Fluorescent aptasensor. Binding with the target molecule produces a change in the optical tag (fluorophore). (c) Quartz crystal microbalance aptasensor, which correlates with the mass accumulated on the quartz crystal resonator with the change in its frequency [99].
While aptamer switches can convert binding events into measurable signals through conformational changes, their inherent signal strength is sometimes limited, which is why enzyme-assisted target recycling is employed to amplify the signal through repeated cycles of target release and rebinding. Enzyme-assisted target recycling in aptasensors leverages the catalytic activity of enzymes to repeatedly release and reuse target molecules after they bind to an aptamer. In this strategy, once the target binds to the aptamer and initiates a signal, a nuclease enzyme dissociates the target from the aptamer, allowing it to interact with another aptamer molecule. This process enhances signal amplification by enabling multiple rounds of binding events with the same target molecule, improving the sensitivity and detection limits of the aptasensor [100]. Zhan et al. [101] employed this method to develop a fluorescence aptasensor capable of detecting low levels of Aflatoxin B1, a harmful mycotoxin in agricultural products, thanks to exonuclease-assisted target recycling amplification. This method allows low detection limits (0.36 ng/mL), ensuring reliable monitoring even in trace amounts [101].
Another possible approach to prepare an aptasensor involves the design of split aptamers. Here, a specific aptamer is divided into two or more separate, non-functional fragments that can selectively reassemble in the presence of the target. However, designing split aptamers is challenging due to the need to preserve the 3D structure of the original aptamer, which can be significantly disrupted if the division occurs at an incorrect site [102]. An example of this strategy is proposed by Chang et al. [103], where a colorimetric method using split aptamers and gold nanoparticles allowed them to detect estradiol, a key estrogen hormone that, in high levels, may cause breast cancer, liver damage, hormonal imbalances, etc. They found that depending on where they were splitting the aptamer, the dynamic range and sensitivity changed, showing the importance of choosing the appropriate split fragment [103].
Regarding the signals obtained from the sensing platform, four primary types are commonly used: optical, electrical, mass-sensitive, and micromechanical (Figure 5). Table 2 provides examples of each signal type based on the employed detection strategy.

Table 2.
Types of signals in aptasensor design, detection strategy, and its application in laboratory settings.
3.3. Immobilization Strategies and Considerations
The detection strategy used in aptasensing largely determines how the aptamer must function in the sample. For strategies that do not require the signal generated by the aptamer to be localized close to the sensing platform, such as in most fluorescence, colorimetric, and chemiluminescence techniques, aptamers are most commonly employed free in solution. However, in situations involving flow and sensing techniques that benefit from having the aptamer-generated signal close to the sensing platform, particularly for enhancing signal readouts (e.g., electrochemical, mass-sensitive, and micromechanical techniques), aptamers are typically immobilized on the sensing platforms. Because aptamer proximity is key in these approaches, aptamer length must be taken into consideration. Although shorter aptamers increase signal proximity to the sensing platform, longer aptamers may be required for larger bioanalytes to allow for proper aptamer folding. Nanoparticles are also beneficial in immobilization contexts, as they can impart aptamers with biochemical capabilities that enhance detection by the sensing platform and increase the surface area for aptamer immobilization.
The specific method of aptamer immobilization significantly impacts their sensitivity, stability, and accessibility [133,134]. Consequently, various immobilization techniques have been developed to achieve optimal results, with careful consideration given to the target analyte and biofluid when selecting an aptamer immobilization strategy. Below, we review different approaches to this process (Figure 6).
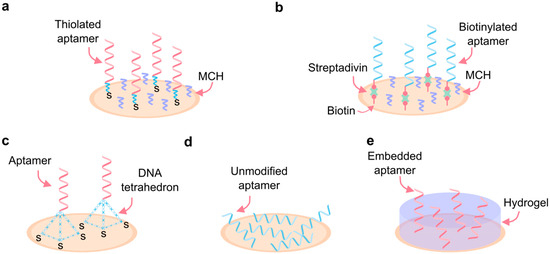
Figure 6.
Immobilization of aptamers. (a) High-density covalent binding to the surface by thiol-metal interaction. (b) High-density affinity-based binding by streptavidin/avidin-biotin interactions. (c) Low-density DNA tetrahedron aptamer surface binding. (d) Adsorption binding of unmodified aptamers. (e) Immobilization by embedding aptamers in a hydrogel matrix. MCH: 6-Mercapto-1-hexanol.
For analytes present at high concentrations in a target biofluid, a common immobilization method involves the covalent or affinity-based binding of aptamers to the surface of interest (e.g., an electrode or nanoparticle). Although covalent and affinity-based bonds can make aptamers more rigid and less flexible, high analyte concentrations in biofluids can help counteract the resulting sensitivity reduction. The relative concentration of an analyte depends on its physiological concentration range and the biosensor’s detection limits. Analytes, such as glucose, which are present in high quantities under physiological conditions and can easily fall within a biosensor’s detection range, may be considered high-concentration analytes.
One widely used technique relies on thiol–metal interactions, where aptamers are either directly labeled with a thiol group or attached via a thiol-containing alkyl chain or poly(thymine) linker. The thiol groups interact with the metal, usually gold, on the electrode surface, forming a covalent bond [138,139]. Streptavidin/avidin–biotin interactions, a common affinity-based binding technique, involve tethering biotin-labeled aptamers to avidin-modified surfaces and provide similar aptamer properties to covalent bonding techniques [140]. Specifically, these methods offer strong, stable aptamer attachment to surfaces.
One limitation of this technique is the nonspecific adsorption of aptamers onto the surface due to electrostatic or hydrophobic interactions, which reduces sensor sensitivity by altering aptamer conformation. Backfilling, which involves introducing mercaptohexanol (MCH) either after or alongside the aptamers, helps reduce nonspecific adsorption by blocking unoccupied spaces on the electrode surface [141,142].
For analytes present in low concentrations in biofluids, detection may benefit from aptamer immobilization via adsorption. Adsorption utilizes the negatively charged DNA backbone to adhere aptamers to positively charged surfaces. This technique does not require aptamer modification, making it the simplest of all methods. Maintaining aptamers in their native state also makes them particularly desirable for low-concentration analytes, as their ability to fold easily makes them highly sensitive [142]. However, this method may not be ideal in scenarios involving fluid flow, as aptamers can easily become displaced by moving fluid. In such cases, covalent or affinity-based immobilization techniques may be more appropriate [143].
For large bioanalytes, the use of DNA nanostructures can be advantageous. In this approach, aptamers are tethered in controlled quantities to DNA-based tetrahedrons, which can be fabricated in various sizes depending on the analyte’s dimensions. These structures are then placed on the surface, allowing for precise spacing of aptamers [144]. In addition to providing adequate spacing for the binding of large bioanalytes, the precise control offered by DNA tetrahedrons can reduce steric hindrance and electrostatic repulsion, which can occur when aptamers are too closely spaced [145].
In biofluids with harsh conditions, such as extreme pH or high salt concentrations, embedding aptamers in a matrix can enhance their stability and maintain performance. In this technique, matrices such as hydrogels and polymeric films create ideal conditions for aptamers. Although this approach limits the degrees of freedom of the aptamers, it enhances their stability under harsh conditions while preserving their performance [146].
An important consideration in biofluids is the presence of other biological molecules, such as proteins, amino acids, and nucleic acids. These molecules can adhere to the electrode surface and cause “biofouling,” which impedes the binding of the target analyte. While few techniques exist to address this challenge, some methods used for aptamers include zwitterionic peptides, aromatic thiols, and antifouling polymeric coatings [147,148].
In all these techniques, various factors during the aptamer immobilization process can be adjusted to optimize their function and immobilization. Key parameters include the ionic strength of the buffer used; pH; and aptamer incubation time, concentration, and conformation (open or closed) [149].
4. Challenges and Opportunities in the Use of Aptamers in Sensors
Aptamers, as nucleic acid molecules with high specificity and affinity for their targets, offer significant potential in sensing applications. Their advantages include ease of generation, low manufacturing costs, minimal batch-to-batch variation, reversible folding properties, and low immunogenicity when compared to monoclonal antibodies [31,150].
Despite these benefits, the development and application of aptamers face significant challenges. The SELEX process is often labor-intensive and inefficient. This method requires multiple cycles of selection and amplification to enrich high-affinity aptamers, which can introduce biases, such as the loss of rare binding sequences and unintended amplification of non-target sequences. These issues can compromise the quality and binding affinity of the selected aptamers [151]. Researchers are making efforts to refine the selection protocols, optimize library design, and incorporate new analytical and computational techniques, thereby overcoming SELEX’s inherent challenges while enhancing the specificity and efficiency of aptamer generation [152].
Another critical limitation is the performance disparity between in vitro and in vivo conditions. Aptamers selected in vitro may fail to function effectively in biological systems due to their intrinsic physicochemical properties, such as ionic strength, pH, and the presence of competing molecules [151]. Additionally, they may lose their structure and binding capability under extreme temperatures, limiting their usability in diverse sensing environments [153]. Therefore, in vivo applications depend on animal models to generate aptamers compatible with complex biological environments, along with chemical modifications to improve resistance to nuclease degradation and prolong action times [31,70]. All these facts, while improving stability and functionality, increase the complexity and cost of aptamer development. Regarding the reversibility of aptasensors, incomplete release of target molecules during regeneration leaves binding sites partially occupied. This residual occupancy reduces the sensor’s sensitivity and performance over repeated use cycles [154]. Efforts are underway to develop smart regeneration strategies using electrical or chemical cues to reset the aptamer conformation after target binding.
Despite these challenges, aptamers also present significant opportunities in biosensor development. Their synthetic nature allows for high batch-to-batch reproducibility and cost-effective production [155]. Their ability to undergo structural modifications enables fine-tuning for improved affinity and specificity [150]. Moreover, aptamers can be integrated into different sensing platforms, expanding their versatility. The potential for real-time point-of-care, wearable, and implantable applications further highlights their growing relevance in diagnostics and prognostication of diseases.
5. Diagnostic Applications of Aptamers
Aptamers have revolutionized biosensing by enabling the rapid and real-time detection of biomarkers for a wide range of diagnostic applications. Their reversibility, tunable concentration range, versatility in selecting targets, and stability have shown they are well suited for being applied in the real world. An aptamer’s ability to respond dynamically to changes in biomarker concentrations allows for continuous monitoring, a key aspect in pharmacokinetics, drug delivery, in vitro metabolite detection, and therapeutic drug monitoring. This is also important for neurological applications, where going through the blood–brain barrier (BBB) is a major challenge. The BBB is extremely selective and restricts the entry of many molecules, making delivering pharmaceuticals a huge challenge. Aptamers have become an increasingly useful tool for being able to target very specific molecules and diffuse passively into the BBB due to their small size. Additionally, the use of aptamer-based biosensors in point-of-care (POC) diagnostics and wearable technologies enables noninvasive, high-sensitivity detection of biomarkers, paving the way for improved disease monitoring and personalized medicine.
5.1. Integrated and Point-of-Care Aptasensors
One of the many ways aptamer-based biosensors are being explored is for the in vitro monitoring of metabolites. Metabolites are usually measured using techniques like mass spectroscopy, nuclear magnetic resonance, and different electrochemical and optical methods. These techniques, however, require the sample to be extracted, prepared, and analyzed, making real-time measurement not possible. The traditional metabolite monitoring techniques, like those mentioned, often lack time resolution and are limited to single-time-point and single-parameter measurements, making it difficult to track dynamic metabolic changes. Using aptamers allows for the use of multiplexing and reversible sensing as an advantage over current techniques. Yiling Yang et al. [156] developed a dual-purpose electrochemical aptamer-based biosensor designed for tracking pH and metabolite concentrations in cell culture media over a period of time (Figure 7a). This biosensor was able to continuously monitor and track the pH through the shift in redox probe potential to see changes in the cell culture environment and phenylalanine levels to track metabolic activity. Silver ion cytotoxicity is a concern for many continuous cell culture monitoring systems. Ag/AgCl reference electrodes can cause silver ions to go into the medium and be toxic to cells, disrupting activity. There are some ways to help this issue, such as encapsulation to mitigate this toxicity (which can help slow down the diffusion of silver ions, using alternative reference electrodes that are non-toxic), or constant medium changes (which can be timely and costly). To address this issue, the researchers developed a leak-free electrode to mitigate silver ion cytotoxicity, ensuring long-term stability and accuracy. Using these sensors, the pH and phenylalanine levels were successfully monitored for 72 h in multiple cell lines, showing the potential for real-time and continuous metabolic tracking [156].
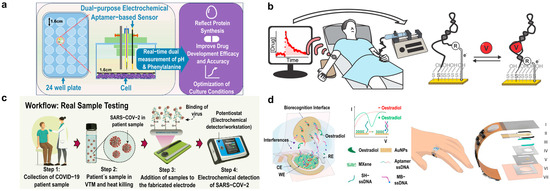
Figure 7.
Diagnostic applications of aptamers. (a) Electrochemical aptamer-based sensor for pH and metabolite monitoring in cell culture media for up to 72 h in multiple cell lines. Adapted from [155]. (b) Therapeutic drug monitoring with electrochemical aptamer-based sensor for vancomycin detection. Adapted from [157]. (c) Point-of-care electrochemical aptamer-based sensor for COVID-19 detection. Adapted from [158]. (d) Skin-interfaced electrochemical aptamer-based sensor for picomolar detection of oestradiol in sweat. Adapted from [126].
E-AB sensors are being developed for therapeutic drug monitoring (TDM) and high-precision controlled drug delivery. Philippe Dauphin-Ducharme et al. [157] presented an E-AB sensor designed for vancomycin, a drug requiring tight dosing control (Figure 7b). Vancomycin is used to treat bacterial infections; however, it has a very narrow therapeutic window, meaning that if someone is in the wrong range, it can be toxic to the kidneys and cause hearing damage. Current TDM methods are often utilized through blood sampling in a lab, which can have slow results and lack real-time measurement. This device was able to demonstrate the measurement of vancomycin in finger-prick samples, which is quicker and still very precise. The results demonstrated the potential for personalized, feedback-controlled drug administration directly in blood and in vivo, as they were able to enable the quick single-step detection of specific molecules [157].
Aptamers also have shown promise for point-of-care (POC) and at-home diagnostics. A notable example is their use in COVID-19 detection, where aptamers have gained increasing attention as an alternative to antibodies. Narlawar Shrikrishna et al. [158] developed a screen-printed carbon electrode-based electrochemical aptasensor that demonstrated reliable detection of the virus in various environments, offering a fast and effective diagnostic tool for widespread use (Figure 7c). Using SELEX, the receptor binding domain of SARS-CoV-2 was selected as the target, making this a highly specific aptamer. This aptasensor offers a rapid and cost-effective device while highlighting the potential of aptamers for POC applications [158].
Aptamer-based biosensors are also being explored for diabetes management technologies, such as for glucose and insulin detection. There are many different devices on the market for diabetes management, but this aptamer sensing strategy shows a way to capture important biomarkers, such as insulin resistance, for a quicker diagnosis process. Jihong Wang et al. [159] presented a surface-enhanced Raman scattering aptasensor that uses gold bipyramidal nanoparticles to enhance the sensitivity of the signal. This sensor can detect multiple biomarkers related to diabetes, like insulin, which can also be used to differentiate type 1 from type 2 diabetes. The highly selective sensor can detect the changes in molecules with the presence of the aptamer, and these structural changes can indicate glucose or insulin binding. The researchers were also able to validate their testing with saliva samples and showed that it can be a noninvasive technology. This precise simultaneous monitoring of both glucose and insulin is important and has become a widespread tool for diagnosis [159].
5.2. Implantable Aptasensors
Another area of focus has been the development of implantable sensors for continuous pharmacokinetic analysis. Pharmacokinetic analysis is vital for seeing how drugs move in the body in order to ensure correct dosing is given to avoid toxicity. A common issue for many pharmacokinetic models is the lack of real-time measurements, which often rely on approximations. Matthew McDonough et al. [160] showed the ability of high-resolution temporal datasets to model dynamic pharmacokinetic profiles with strong statistical reliability. They achieved this by using E-AB sensors to monitor plasma concentrations of tobramycin in rats and statistically compared this to traditional one- and two-compartment pharmacokinetic models, therefore also understanding the impact of kidney function. Using these sensors allowed for rapid results and the ability to monitor hundreds of data points at once, and after monitoring the drug elimination rates, it was found that kidney function significantly affected the rate of tobramycin elimination [160].
Similarly, monitoring drug levels in real-time is crucial for the diagnosis and management of drug intoxication and overdose, where traditional methods rely on blood draws and do not provide immediate results. Obtin Alkhamis et al. [161] developed two cocaine detection sensors: an in vitro fluorescent sensor and an electrochemical aptamer-based sensor designed for real-time measurements in vivo. The first sensor was a fluorescent sensor that detected cocaine in vitro, and the second was an E-AB sensor that allows for real-time and high-frequency cocaine monitoring in the circulating blood streams of live animals. They also developed new aptamers through SELEX that could specifically bind to cocaine with stronger binding than previous aptamers, allowing for improved cocaine detection for both in vivo and in vitro environments, offering potential applications for clinical diagnostics and pharmacology research [161].
Aptamers have also been increasingly used for neuropharmacology, particularly for tracking drug transport across the BBB and monitoring neurotransmitter levels. Their specificity and high affinity are a huge advantage when it comes to working with such complicated systems as the brain, where highly specific targeting is required. Additionally, the small size of aptamers makes them ideal for drug transport through the BBB. Julian Gerson et al. utilized E-AB sensors to measure vancomycin transport across the BBB in real-time, achieving temporal resolution on the order of seconds. They focused on tracking the movement of the drug from the injection site in the veins, measuring drug concentrations in both locations, allowing for spatial resolution [162].
Neurotransmitter monitoring is essential for understanding neurological disorders as well as natural brain function. Chuanzhen Zhao et al. [163] developed an implantable aptamer-field-effect transistor (FET) neuroprobe for in vivo neurotransmitter monitoring of serotonin. These neuroprobes were fabricated using microelectromechanical system technology and tested in simulated tissue matrices like brain tissue and under electrical stimulation to mimic the release of serotonin. The FET neuroprobes allow for a high spatial resolution since they can bind directly and do so in seconds, allowing them to monitor neurotransmitter changes in real time. The sensors demonstrated functionality across different testing environments, such as stimulated brain tissue and electrical stimulation, which shows their potential for continuous neurotransmitter monitoring in neural research and for medical diagnostics [163].
5.3. Wearable Aptasensors
Aptamers have emerged as a game-changing tool for wearable biosensors, expanding the range of detectable analytes and improving the feasibility of continuous monitoring. Unlike antibodies, which have been widely used in biosensing, aptamers offer key advantages for wearable applications due to their stability, ease of modification, and ability to undergo reversible binding, making them ideal for continuous, real-time monitoring. Enzyme-based biosensors have been the cornerstone of wearable sensing, but the number of analytes detectable by enzymes is limited to a few small metabolites. The introduction of aptamers has introduced groundbreaking possibilities, allowing for the sensing of a broader range of biomarkers, including hormones, cytokines involved in immune response, stress, and metabolic health. Aptamers have shown great promise in monitoring analytes in noninvasive biofluids for wearable applications [164].
Ye, Cui et al. [126] reported a significant advance in the field by developing a wearable aptamer nanobiosensor for noninvasive monitoring of female hormones from sweat (Figure 7d). This study’s innovation lies in its ability to measure hormones like estrogen with high sensitivity and specificity, which were previously difficult to monitor continuously and noninvasively. By addressing the challenge of wearable biosensors for hormone detection, this sensor enables real-time reproductive health tracking, providing valuable insights for fertility management and hormone-related disorders [126].
Aptamers have also allowed for the study of other hormones in noninvasive biofluids such as cortisol. Cortisol regulates metabolism and most of the stress and immune response. Continuous tracking of cortisol in noninvasive biofluids, such as sweat and interstitial fluid, is crucial for understanding and managing stress-related health conditions. Unlike traditional blood-based tests, which provide only a snapshot of cortisol levels at a single point in time, continuous monitoring allows for real-time tracking of diurnal patterns and acute stress responses. This is particularly important for detecting chronic stress, adrenal disorders, and mental health conditions such as depression and anxiety.
Recent studies have demonstrated the unique capabilities of aptamer-based wearable cortisol sensors, showcasing both the versatility and the technical advantages of aptamer technology. Singh et al. [165] developed a noninvasive, wearable electrochemical sensor for real-time cortisol monitoring using a pseudoknot-assisted aptamer. A pseudoknot is a complex secondary structure formed by folding the aptamer into interlocking loops, which enhances the binding stability and reduces background noise, improving signal clarity. The sensor integrated a flexible microfluidic system, which allowed for the separation of old and new sweat, ensuring that the cortisol readings were accurate and reflective of real-time physiological changes. This work would not have been possible without aptamers, as antibody-based systems lack the ability to regenerate signals and maintain specificity over prolonged periods in complex biological fluids like sweat [165].
Wang et al. [166] introduced an innovative field-effect transistor (FET) sensor platform for wearable cortisol monitoring, where the binding of cortisol to an aptamer immobilized on a thin film of indium oxide (In2O3) produced a measurable electrical signal. FET sensors work by detecting changes in the electrical field at the sensor surface, which alters the conductivity of the material. With this approach, electrochemical detection is possible without the need to use a redox probe. The sensor was incorporated into a smartwatch, enabling continuous and wireless monitoring of cortisol levels. The FET configuration allowed the sensor to be highly sensitive and low-power, making it ideal for long-term use in wearable devices [166].
Aptamers have also made a great impact in interstitial fluid analysis. Interstitial fluid (ISF) has emerged as a highly valuable biofluid for wearable biosensors due to its rich composition of biomarkers that reflect physiological and pathological states. ISF surrounds the cells in tissues and serves as a transport medium for nutrients, waste products, hormones, electrolytes, and signaling molecules. ISF can be accessed noninvasively through microneedle technology, offering a promising alternative for continuous health monitoring without the discomfort and complexity of blood sampling. Wearable biosensors that sample ISF can provide real-time insights into metabolic status, drug pharmacokinetics, immune responses, and electrolyte balance, opening the door to personalized medicine and early disease detection. However, the dynamic and complex nature of ISF presents significant technical challenges for biosensing, including low biomarker concentrations, signal interference from complex biological matrices, and sensor stability over time. Aptamers have addressed many of these challenges, making continuous ISF microneedle-based analysis feasible for the first time. Microneedles provide a minimally invasive route for accessing biomarkers in the dermal layer while maintaining high user comfort and compliance.
Wu et al. [167] developed a microneedle-based aptamer sensor for continuous therapeutic drug monitoring in ISF. Therapeutic drug monitoring (TDM) is critical for optimizing drug dosing, reducing side effects, and improving treatment outcomes. The sensor used aptamers specific to the target drug molecules, allowing for real-time pharmacokinetic tracking with high sensitivity and specificity. The ability to track drug concentration in real time represents a significant advancement for precision medicine, allowing clinicians to adjust dosing dynamically based on individual patient responses [167].
Friedel et al. [168] presented a microneedle-based electrochemical aptamer sensor used for real-time molecular monitoring of phenylalanine, a biomarker clinically important for managing phenylketonuria (PKU)—a metabolic disorder where elevated phenylalanine levels can cause neurological damage if untreated. The use of aptamers provided high specificity and reversible binding, allowing for continuous, real-time monitoring without sensor degradation. The sensor achieved consistent signal detection over 90 days of dry storage and a clinically relevant lag time (~20 min), reinforcing the potential of aptamers for pharmacokinetics and chronic disease management [168].
The use of aptamer-based microneedles opened several opportunities for tracking molecules in ISF. The study by Zheng et al. [169] presented an innovative hydrogel-based microneedle patch for the extraction and analysis of interstitial fluid (ISF) biomarkers. The hydrogel microneedles swell upon skin insertion, facilitating efficient ISF collection for ex vivo analysis. The biosensor demonstrated the ability to detect four key biomarkers using aptamers: glucose (for diabetes monitoring), ATP (for cellular energy balance), L-tyrosinamide (a metabolic and cancer biomarker), and thrombin (for blood coagulation). The aptamer-based strand displacement strategy allowed for reagentless detection, where target binding induced fluorescence by separating the quencher from the fluorophore. This approach achieved high sensitivity and specificity, thanks to the strong thermodynamic stability of aptamer-target binding. The use of aptamers allowed for reversible, specific binding and signal generation without post-processing steps, making the system highly suitable for continuous health monitoring. The combination of hydrogel microneedles and aptamer-based detection represents a significant advance in noninvasive biomarker monitoring, particularly for metabolic and hematologic health tracking [169].
The opportunities unlocked by aptamers for wearable biosensors are substantial. Their ability to target a wide range of biomarkers means that future wearables could monitor not only metabolic health but also immune response, mental health, and even early-stage cancer markers. Aptamers’ small size and ease of synthesis allow for multiplexed sensing platforms, where a single wearable could monitor dozens of biomarkers simultaneously. This multiplexing capability could lead to a new generation of personalized health devices, providing continuous, real-time insights into an individual’s physiological state [170]. The development of flexible, low-power electronics and improved signal processing algorithms will further enhance the utility and accuracy of aptamer-based wearables. By overcoming current challenges, aptamers have the potential to transform wearable biosensing from single-analyte monitoring to comprehensive, dynamic health tracking, unlocking new possibilities for early disease detection, chronic disease management, and personalized medicine.
6. Outlook and Future Directions
As aptamer-based technologies continue to advance, efforts are focused on addressing key challenges to improve their efficiency, stability, and versatility in sensing applications. The current limitations of the SELEX process, including its high cost and low yield, are driving the development of more efficient fabrication strategies. Future research is likely to benefit from computational tools that predict aptamer-target interactions, reducing the reliance on time-consuming trial and error and large combinatorial libraries. In parallel, integrating aptamers with other biomolecules such as enzymes and antibodies could create hybrid systems that harness the strengths of each component, potentially leading to enhanced specificity and multifunctional sensing platforms. Another critical area of development is improving aptamer stability in complex biofluids, where degradation and structural changes can compromise performance. Using aptamers as building blocks for two-dimensional and three-dimensional nanostructures, in addition to their integration with smart, multifunctional nanomaterials such as metal–organic frameworks or flexible polymers, offers a promising approach to preserving their functional conformation and improving signal transduction. A final important advancement in aptamer research is the development of multifunctional aptamers, offering greater versatility, binding multiple targets simultaneously, or switching between binding states in response to stimuli such as light or pH.
Beyond medical applications, aptamers hold great potential in environmental monitoring, where highly selective biosensors could detect pollutants, toxins, and pathogens in water, soil, and air. These advancements will drive the next generation of aptamer-based technologies, making them more robust, scalable, and adaptable to real-world challenges.
Funding
This research was funded by Rice University startup grant (grant number F10000370).
Acknowledgments
B.M., J.N.G., K.V., and J.R.S. acknowledge the support from the startup grant at Rice University.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| E-ABs | Electrochemical aptasensors |
| FRET | Förster Resonance Energy Transfer |
| CF | Cerebrospinal fluid |
| FDA | Food and Drug Administration |
| ISF | Interstitial fluid |
| CTCs | Circulating tumor cells |
| PCR | Polymerase chain reaction |
| MDs | Molecular dynamics |
| CEA | Carcinoembryonic antigen |
| PSA | Prostate-specific antigen |
| LDL | Low-density lipoprotein |
| rPfLDH | Plasmodium falciparum lactate dehydrogenase |
| PD-L1 | Programmed death ligand-1 |
| EVs | Extracellular vesicles |
| CRP | C-reactive protein |
| cTn-I | Cardiac troponin I |
| MCH | Mercaptohexanol |
| BBB | Blood–brain barrier |
| POC | Point-of-care |
| TDM | Therapeutic drug monitoring |
| FET | Field-effect transistor |
| PKU | Phenylketonuria |
| ATP | Adenosine triphosphate |
References
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Currás, N.; Dauphin-Ducharme, P.; Scida, K.; Chávez, J.L. From the Beaker to the Body: Translational Challenges for Electrochemical, Aptamer-Based Sensors. Anal. Methods 2020, 12, 1288–1310. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Yang, L.F.; Ling, M.; Kacherovsky, N.; Pun, S.H. Aptamers 101: Aptamer Discovery and in Vitro Applications in Biosensors and Separations. Chem. Sci. 2023, 14, 4961–4978. [Google Scholar] [CrossRef] [PubMed]
- Gelinas, A.D.; Davies, D.R.; Janjic, N. Embracing Proteins: Structural Themes in Aptamer–Protein Complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef]
- Requena, M.D.; Gray, B.P.; Sullenger, B.A. Protocol for Purification of Cells in Their Native State Using Reversible Aptamer-Antidote Pairs. STAR Protoc. 2023, 4, 102348. [Google Scholar] [CrossRef]
- Gray, B.P.; Requena, M.D.; Nichols, M.D.; Sullenger, B.A. Aptamers as Reversible Sorting Ligands for Preparation of Cells in Their Native State. Cell Chem. Biol. 2020, 27, 232–244.e7. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, H.; Gubu, A.; Yu, S.; Zhang, H.; Dai, H.; Zhang, Y.; Zhang, B.; Ma, Y.; Lu, A.; et al. Chemically Modified Aptamers for Improving Binding Affinity to the Target Proteins via Enhanced Non-Covalent Bonding. Front. Cell Dev. Biol. 2023, 11, 1091809. [Google Scholar] [CrossRef]
- Grabowska, I.; Zapotoczny, S.; Chlopicki, S. Multiplex Electrochemical Aptasensors for Detection of Endothelial Dysfunction: Ready for Prime Time? Trends Anal. Chem. 2023, 169, 117372. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three Decades of Nucleic Acid Aptamer Technologies: Lessons Learned, Progress and Opportunities on Aptamer Development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Yuhan, J.; Zhu, L.; Zhu, L.; Huang, K.; He, X.; Xu, W. Cell-Specific Aptamers as Potential Drugs in Therapeutic Applications: A Review of Current Progress. J. Control. Release 2022, 346, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Lukas, H.; Wang, M.; Lee, Y.; Gao, W. Nucleic Acid-Based Wearable and Implantable Electrochemical Sensors. Chem. Soc. Rev. 2024, 53, 7960–7982. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Zhang, Y.; Cai, J.; Cai, W.; Gao, T. Aptamer-Based Fluorescent Biosensors. Curr. Med. Chem. 2011, 18, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Noumani, A.; Yadav, A.K.; Solanki, P.R. FRET Based Biosensor: Principle Applications Recent Advances and Challenges. Diagnostics 2023, 13, 1375. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Focus Area: Biomarkers. Available online: https://www.fda.gov/science-research/focus-areas-regulatory-science-report/focus-area-biomarkers (accessed on 6 February 2025).
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and Valuable Tools towards Diagnosis, Prognosis and Treatment of COVID-19 and Other Diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Biomarkers at FDA. Available online: https://www.fda.gov/science-research/about-science-research-fda/biomarkers-fda (accessed on 6 February 2025).
- Chen, L.; Yang, G.; Qu, F. Aptamer-Based Sensors for Fluid Biopsies of Protein Disease Markers. Talanta 2024, 276, 126246. [Google Scholar] [CrossRef]
- Salama, R.; Arshavsky-Graham, S.; Sella-Tavor, O.; Massad-Ivanir, N.; Segal, E. Design Considerations of Aptasensors for Continuous Monitoring of Biomarkers in Digestive Tract Fluids. Talanta 2022, 239, 123124. [Google Scholar] [CrossRef]
- Lu, J.K.; Wang, W.; Goh, J.; Maier, A.B. A Practical Guide for Selecting Continuous Monitoring Wearable Devices for Community-Dwelling Adults. Heliyon 2024, 10, e33488. [Google Scholar] [CrossRef]
- Ellison, J.M.; Stegmann, J.M.; Colner, S.L.; Michael, R.H.; Sharma, M.K.; Ervin, K.R.; Horwitz, D.L. Rapid Changes in Postprandial Blood Glucose Produce Concentration Differences at Finger, Forearm, and Thigh Sampling Sites. Diabetes Care 2002, 25, 961–964. [Google Scholar] [CrossRef]
- Caplin, A.; Chen, F.S.; Beauchamp, M.R.; Puterman, E. The Effects of Exercise Intensity on the Cortisol Response to a Subsequent Acute Psychosocial Stressor. Psychoneuroendocrinology 2021, 131, 105336. [Google Scholar] [CrossRef]
- Heikenfeld, J.; Jajack, A.; Feldman, B.; Granger, S.W.; Gaitonde, S.; Begtrup, G.; Katchman, B.A. Accessing Analytes in Biofluids for Peripheral Biochemical Monitoring. Nat. Biotechnol. 2019, 37, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Ye, M.; Levy, A.; Rothstein, J.; Bergles, D.; Searson, P.C. The Blood-Brain Barrier: An Engineering Perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The Blood–Brain Barrier: Structure, Regulation and Drug Delivery. Sig. Transduct. Target. Ther. 2023, 8, 217. [Google Scholar] [CrossRef]
- Ștefan, G.; Hosu, O.; De Wael, K.; Lobo-Castañón, M.J.; Cristea, C. Aptamers in Biomedicine: Selection Strategies and Recent Advances. Electrochim. Acta 2021, 376, 137994. [Google Scholar] [CrossRef]
- Li, Y.; TAM, W.W.; Yu, Y.; Zhuo, Z.; Xue, Z.; Tsang, C.; Qiao, X.; Wang, X.; Wang, W.; Li, Y.; et al. The Application of Aptamer in Biomarker Discovery. Biomark. Res. 2023, 11, 70. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, Y.; Di, Y.; Xiu, C.; He, L.; Liao, S.; Li, D.; Huang, B. DNA Aptamers from Whole-Serum SELEX as New Diagnostic Agents against Gastric Cancer. RSC Adv. 2019, 9, 950–957. [Google Scholar] [CrossRef]
- Zhou, L.; Ou, L.-J.; Chu, X.; Shen, G.-L.; Yu, R.-Q. Aptamer-Based Rolling Circle Amplification: A Platform for Electrochemical Detection of Protein. Anal. Chem. 2007, 79, 7492–7500. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lai, R.Y. A Reagentless and Reusable Electrochemical Aptamer-Based Sensor for Rapid Detection of Ampicillin in Complex Samples. Talanta 2018, 176, 619–624. [Google Scholar] [CrossRef]
- McKeague, M.; DeRosa, M.C. Challenges and Opportunities for Small Molecule Aptamer Development. J. Nucleic Acids 2012, 2012, 748913. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ennifar, E.; Nakamura, Y. Thermodynamic Study of Aptamers Binding to Their Target Proteins. Biochimie 2018, 145, 91–97. [Google Scholar] [CrossRef]
- Mjaaland, S.; Fossum, S. Modulation of Immune Responses with Monoclonal Antibodies. I. Effects on Regional Lymph Node Morphology and on Anti-Hapten Responses to Haptenized Monoclonal Antibodies. Eur. J. Immunol. 1990, 20, 1457–1461. [Google Scholar] [CrossRef]
- Huang, P.-J.J.; Liu, J. Simultaneous Detection of L-Lactate and D-Glucose Using DNA Aptamers in Human Blood Serum. Angew. Chem. Int. Ed. 2023, 62, e202212879. [Google Scholar] [CrossRef] [PubMed]
- Karpova, E.V.; Shcherbacheva, E.V.; Galushin, A.A.; Vokhmyanina, D.V.; Karyakina, E.E.; Karyakin, A.A. Noninvasive Diabetes Monitoring through Continuous Analysis of Sweat Using Flow-Through Glucose Biosensor. Anal. Chem. 2019, 91, 3778–3783. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.C.; Xing, X.W.; Zhang, J.T.; He, M.W.; Ma, Y.B.; Wu, L.; Wang, X.; Wang, H.F.; Yu, S.Y. Correlation between Blood Glucose and Cerebrospinal Fluid Glucose Levels in Patients with Differences in Glucose Metabolism. Front. Neurol. 2023, 14, 1103026. [Google Scholar] [CrossRef]
- Schilde, L.M.; Kösters, S.; Steinbach, S.; Schork, K.; Eisenacher, M.; Galozzi, S.; Turewicz, M.; Barkovits, K.; Mollenhauer, B.; Marcus, K.; et al. Protein Variability in Cerebrospinal Fluid and Its Possible Implications for Neurological Protein Biomarker Research. PLoS ONE 2018, 13, e0206478. [Google Scholar] [CrossRef]
- Khadke, S.; MacDougall, F.; Islam, T. Chapter Five—Novel Formulation Approaches for Gastrointestinal Targeting: Characterization and Animal Model Considerations. In Oral Delivery of Therapeutic Peptides and Proteins; Tyagi, P., Subramony, J.A., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 167–198. ISBN 978-0-12-821061-1. [Google Scholar]
- Hwang, J.; Seo, Y.; Jo, Y.; Son, J.; Choi, J. Aptamer-Conjugated Live Human Immune Cell Based Biosensors for the Accurate Detection of C-Reactive Protein. Sci. Rep. 2016, 6, 34778. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-H.; Wen, C.-Y.; Hong, C.-Y.; Lai, J.-C. Evaluation of Aptamer Specificity with or without Primers Using Clinical Samples for C-Reactive Protein by Magnetic-Assisted Rapid Aptamer Selection. RSC Adv. 2017, 7, 42856–42865. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Wang, Z.; Newbigging, A.M.; Reid, M.S.; Li, X.-F.; Le, X.C. Aptamers Facilitating Amplified Detection of Biomolecules. Anal. Chem. 2015, 87, 274–292. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Heuer, C.; Jiang, X.; Segal, E. Aptasensors versus Immunosensors—Which Will Prevail? Eng. Life Sci. 2022, 22, 319–333. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. Adaptive Recognition by Nucleic Acid Aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef]
- Davies, D.R.; Gelinas, A.D.; Zhang, C.; Rohloff, J.C.; Carter, J.D.; O’Connell, D.; Waugh, S.M.; Wolk, S.K.; Mayfield, W.S.; Burgin, A.B.; et al. Unique Motifs and Hydrophobic Interactions Shape the Binding of Modified DNA Ligands to Protein Targets. Proc. Natl. Acad. Sci. USA 2012, 109, 19971–19976. [Google Scholar] [CrossRef] [PubMed]
- Domsicova, M.; Korcekova, J.; Poturnayova, A.; Breier, A. New Insights into Aptamers: An Alternative to Antibodies in the Detection of Molecular Biomarkers. Int. J. Mol. Sci. 2024, 25, 6833. [Google Scholar] [CrossRef]
- Challier, L.; Mavré, F.; Moreau, J.; Fave, C.; Schöllhorn, B.; Marchal, D.; Peyrin, E.; Noël, V.; Limoges, B. Simple and Highly Enantioselective Electrochemical Aptamer-Based Binding Assay for Trace Detection of Chiral Compounds. Anal. Chem. 2012, 84, 5415–5420. [Google Scholar] [CrossRef]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-Resolution Molecular Discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Gaster, R.S.; Hall, D.A.; Wang, S.X. Autoassembly Protein Arrays for Analyzing Antibody Cross-Reactivity. Nano Lett. 2011, 11, 2579–2583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, Z.; Liu, D.; Jiang, H.; Zhang, Z.-K.; Lu, A.; Zhang, B.-T.; Yu, Y.; Zhang, G. Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int. J. Mol. Sci. 2021, 22, 4093. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-Type-Specific, Aptamer-Functionalized Agents for Targeted Disease Therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating Tumor Cells: Biology and Clinical Significance. Sig. Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef]
- Kolovskaya, O.S.; Zyuzyukina, A.V.; Dassie, J.P.; Zamay, G.S.; Zamay, T.N.; Boyakova, N.V.; Khorzhevskii, V.A.; Kirichenko, D.A.; Lapin, I.N.; Shchugoreva, I.A.; et al. Monitoring of Breast Cancer Progression via Aptamer-Based Detection of Circulating Tumor Cells in Clinical Blood Samples. Front. Mol. Biosci. 2023, 10, 1184285. [Google Scholar] [CrossRef]
- Kolm, C.; Cervenka, I.; Aschl, U.J.; Baumann, N.; Jakwerth, S.; Krska, R.; Mach, R.L.; Sommer, R.; DeRosa, M.C.; Kirschner, A.K.T.; et al. DNA Aptamers against Bacterial Cells Can Be Efficiently Selected by a SELEX Process Using State-of-the Art qPCR and Ultra-Deep Sequencing. Sci. Rep. 2020, 10, 20917. [Google Scholar] [CrossRef] [PubMed]
- Nagar, D.N.; Yathirajarao, T.; Kumar, P.; Kushwaha, P.; Suman, P. Bead-Based SELEX for Aptamers Selection and Their Application in Detection of Diverse Antigens. In Immunodiagnostic Technologies from Laboratory to Point-Of-Care Testing; Suman, P., Chandra, P., Eds.; Springer: Singapore, 2021; pp. 125–139. ISBN 9789811558238. [Google Scholar]
- Kohlberger, M.; Gadermaier, G. SELEX: Critical Factors and Optimization Strategies for Successful Aptamer Selection. Biotechnol. Appl. Biochem. 2022, 69, 1771–1792. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhu, J.; Shen, G.; Deng, Y.; Geng, X.; Wang, L. Improving Aptamer Performance: Key Factors and Strategies. Microchim. Acta 2023, 190, 255. [Google Scholar] [CrossRef]
- Vorobyeva, M.A.; Davydova, A.S.; Vorobjev, P.E.; Pyshnyi, D.V.; Venyaminova, A.G. Key Aspects of Nucleic Acid Library Design for in Vitro Selection. Int. J. Mol. Sci. 2018, 19, 470. [Google Scholar] [CrossRef]
- Gopinath, S.C.B. Methods Developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef]
- Olsen, T.; Zhu, J.; Kim, J.; Pei, R.; Stojanovic, M.N.; Lin, Q. An Integrated Microfluidic SELEX Approach Using Combined Electrokinetic and Hydrodynamic Manipulation. SLAS Technol. 2017, 22, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, H.; Sun, W.; Xu, D.; He, F. Rapid Selection of Aptamers Based on Protein Microarray. RSC Adv. 2019, 9, 9762–9768. [Google Scholar] [CrossRef]
- Niu, C.; Zhang, C.; Liu, J. Capture-SELEX of DNA Aptamers for Estradiol Specifically and Estrogenic Compounds Collectively. Environ. Sci. Technol. 2022, 56, 17702–17711. [Google Scholar] [CrossRef]
- Takenaka, M.; Okumura, Y.; Amino, T.; Miyachi, Y.; Ogino, C.; Kondo, A. DNA-Duplex Linker for AFM-SELEX of DNA Aptamer against Human Serum Albumin. Bioorganic Med. Chem. Lett. 2017, 27, 954–957. [Google Scholar] [CrossRef]
- Gordon, C.K.L.; Wu, D.; Pusuluri, A.; Feagin, T.A.; Csordas, A.T.; Eisenstein, M.S.; Hawker, C.J.; Niu, J.; Soh, H.T. Click-Particle Display for Base-Modified Aptamer Discovery. ACS Chem. Biol. 2019, 14, 2652–2662. [Google Scholar] [CrossRef]
- Li, L.; Zhou, J.; Wang, K.; Chen, X.; Fu, L.; Wang, Y. Screening and Identification of Specific Aptamers for Shellfish Allergen Tropomyosin with Capillary Electrophoresis-SELEX. Food Anal. Methods 2022, 15, 1535–1544. [Google Scholar] [CrossRef]
- Nelissen, F.H.T.; Peeters, W.J.M.; Roelofs, T.P.; Nagelkerke, A.; Span, P.N.; Heus, H.A. Improving Breast Cancer Treatment Specificity Using Aptamers Obtained by 3D Cell-SELEX. Pharmaceuticals 2021, 14, 349. [Google Scholar] [CrossRef] [PubMed]
- Civit, L.; Theodorou, I.; Frey, F.; Weber, H.; Lingnau, A.; Gröber, C.; Blank, M.; Dambrune, C.; Stunden, J.; Beyer, M.; et al. Targeting Hormone Refractory Prostate Cancer by in Vivo Selected DNA Libraries in an Orthotopic Xenograft Mouse Model. Sci. Rep. 2019, 9, 4976. [Google Scholar] [CrossRef]
- Buglak, A.A.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. Methods and Applications of In Silico Aptamer Design and Modeling. Int. J. Mol. Sci. 2020, 21, 8420. [Google Scholar] [CrossRef]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic Acid Aptamers: Clinical Applications and Promising New Horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef]
- Sola, M.; Menon, A.P.; Moreno, B.; Meraviglia-Crivelli, D.; Soldevilla, M.M.; Cartón-García, F.; Pastor, F. Aptamers Against Live Targets: Is In Vivo SELEX Finally Coming to the Edge? Mol. Ther. Nucleic Acids 2020, 21, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Dickey, D.D.; Giangrande, P.H.; Thiel, W.H. 744. Optimizing Conditions for Aptamer Folding Using a High-Throughput Aptamer Fluorescence Binding and Internalization (AFBI) Assay. Mol. Ther. 2016, 24, S293. [Google Scholar] [CrossRef]
- Ma, P.; Ye, H.; Guo, H.; Ma, X.; Yue, L.; Wang, Z. Aptamer Truncation Strategy Assisted by Molecular Docking and Sensitive Detection of T-2 Toxin Using SYBR Green I as a Signal Amplifier. Food Chem. 2022, 381, 132171. [Google Scholar] [CrossRef]
- Ji, D.; Feng, H.; Liew, S.W.; Kwok, C.K. Modified Nucleic Acid Aptamers: Development, Characterization, and Biological Applications. Trends Biotechnol. 2023, 41, 1360–1384. [Google Scholar] [CrossRef]
- Shum, K.T.; Tanner, J.A. Differential Inhibitory Activities and Stabilisation of DNA Aptamers against the SARS Coronavirus Helicase. ChemBioChem 2008, 9, 3037–3045. [Google Scholar] [CrossRef]
- Haruta, K.; Otaki, N.; Nagamine, M.; Kayo, T.; Sasaki, A.; Hiramoto, S.; Takahashi, M.; Hota, K.; Sato, H.; Yamazaki, H. A Novel PEGylation Method for Improving the Pharmacokinetic Properties of Anti-Interleukin-17A RNA Aptamers. Nucleic Acid Ther. 2017, 27, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Amaya-González, S.; López-López, L.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Affinity of Aptamers Binding 33-Mer Gliadin Peptide and Gluten Proteins: Influence of Immobilization and Labeling Tags. Anal. Chim. Acta 2015, 873, 63–70. [Google Scholar] [CrossRef]
- Alves Ferreira-Bravo, I.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-Deoxy-2′-Fluoroarabinonucleotide (FANA) Aptamers That Bind HIV-1 Reverse Transcriptase with Picomolar Affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef]
- Kasahara, Y.; Irisawa, Y.; Fujita, H.; Yahara, A.; Ozaki, H.; Obika, S.; Kuwahara, M. Capillary Electrophoresis–Systematic Evolution of Ligands by Exponential Enrichment Selection of Base- and Sugar-Modified DNA Aptamers: Target Binding Dominated by 2′-O,4′-C-Methylene-Bridged/Locked Nucleic Acid Primer. Anal. Chem. 2013, 85, 4961–4967. [Google Scholar] [CrossRef]
- Abeydeera, N.D.; Egli, M.; Cox, N.; Mercier, K.; Conde, J.N.; Pallan, P.S.; Mizurini, D.M.; Sierant, M.; Hibti, F.-E.; Hassell, T.; et al. Evoking Picomolar Binding in RNA by a Single Phosphorodithioate Linkage. Nucleic Acids Res. 2016, 44, 8052–8064. [Google Scholar] [CrossRef] [PubMed]
- Kohn, E.M.; Konovalov, K.; Gomez, C.A.; Hoover, G.N.; Yik, A.K.; Huang, X.; Martell, J.D. Terminal Alkyne-Modified DNA Aptamers with Enhanced Protein Binding Affinities. ACS Chem. Biol. 2023, 18, 1976–1984. [Google Scholar] [CrossRef]
- Matsunaga, K.-I.; Kimoto, M.; Lim, V.W.; Tan, H.P.; Wong, Y.Q.; Sun, W.; Vasoo, S.; Leo, Y.S.; Hirao, I. High-Affinity Five/Six-Letter DNA Aptamers with Superior Specificity Enabling the Detection of Dengue NS1 Protein Variants beyond the Serotype Identification. Nucleic Acids Res. 2021, 49, 11407–11424. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, H.; Kataoka, Y.; Kuwahara, M.; Horii, K.; Shiratori, I.; Waga, I. A High Affinity Modified DNA Aptamer Containing Base-Appended Bases for Human β-Defensin. Anal. Biochem. 2020, 594, 113627. [Google Scholar] [CrossRef]
- Morihiro, K.; Hasegawa, O.; Kasahara, Y.; Mori, S.; Kasai, T.; Kuwahara, M.; Obika, S. Azobenzene-Modified DNA Aptamers Evolved by Capillary Electrophoresis (CE)-SELEX Method. Bioorganic Med. Chem. Lett. 2021, 31, 127607. [Google Scholar] [CrossRef]
- Kulkarni, O.; Pawar, R.D.; Purschke, W.; Eulberg, D.; Selve, N.; Buchner, K.; Ninichuk, V.; Segerer, S.; Vielhauer, V.; Klussmann, S.; et al. Spiegelmer Inhibition of CCL2/MCP-1 Ameliorates Lupus Nephritis in MRL-(Fas)Lpr Mice. J. Am. Soc. Nephrol. 2007, 18, 2350–2358. [Google Scholar] [CrossRef]
- Oberthür, D.; Achenbach, J.; Gabdulkhakov, A.; Buchner, K.; Maasch, C.; Falke, S.; Rehders, D.; Klussmann, S.; Betzel, C. Crystal Structure of a Mirror-Image L-RNA Aptamer (Spiegelmer) in Complex with the Natural L-Protein Target CCL2. Nat. Commun. 2015, 6, 6923. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Cho, S.; Lin, R.; Gedi, V.; Park, S.; Ahn, C.W.; Lee, D.-K.; Lee, M.-H.; Lee, S.; Kim, S. Nonbiodegradable Spiegelmer-Driven Colorimetric Biosensor for Bisphenol A Detection. Biosensors 2022, 12, 864. [Google Scholar] [CrossRef]
- Litke, J.L.; Jaffrey, S.R. Designing and Expressing Circular RNA Aptamers to Regulate Mammalian Cell Biology. Methods Mol. Biol. 2023, 2570, 223–234. [Google Scholar] [CrossRef]
- Liang, H.; Yan, Z.; Tong, Y.; Chen, S.; Li, J.; Chen, L.; Yang, H. Circular Bivalent Aptamers Enhance the Activation of the Regenerative Signaling Pathway for Repairing Liver Injury In Vivo. Chem. Commun. 2023, 59, 1621–1624. [Google Scholar] [CrossRef]
- Ding, C.; Zhang, C.; Cheng, S.; Xian, Y. Multivalent Aptamer Functionalized Ag2S Nanodots/Hybrid Cell Membrane-Coated Magnetic Nanobioprobe for the Ultrasensitive Isolation and Detection of Circulating Tumor Cells. Adv. Funct. Mater. 2020, 30, 1909781. [Google Scholar] [CrossRef]
- Zhang, Z.; Pandey, R.; Li, J.; Gu, J.; White, D.; Stacey, H.D.; Ang, J.C.; Steinberg, C.-J.; Capretta, A.; Filipe, C.D.M.; et al. High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva. Angew. Chem. Int. Ed. 2021, 60, 24266–24274. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Yuan, L.; Jin, K.; Han, X.; Tian, Y.; Zhou, N. Electrochemical Detection of Tobramycin Based on Enzymes-Assisted Dual Signal Amplification by Using a Novel Truncated Aptamer with High Affinity. Biosens. Bioelectron. 2018, 122, 254–262. [Google Scholar] [CrossRef]
- Dhiman, A.; Anand, A.; Malhotra, A.; Khan, E.; Santra, V.; Kumar, A.; Sharma, T.K. Rational Truncation of Aptamer for Cross-Species Application to Detect Krait Envenomation. Sci. Rep. 2018, 8, 17795. [Google Scholar] [CrossRef]
- Mei, H.; Bing, T.; Yang, X.; Qi, C.; Chang, T.; Liu, X.; Cao, Z.; Shangguan, D. Functional-Group Specific Aptamers Indirectly Recognizing Compounds with Alkyl Amino Group. Anal. Chem. 2012, 84, 7323–7329. [Google Scholar] [CrossRef]
- Sequeira-Antunes, B.; Ferreira, H.A. Nucleic Acid Aptamer-Based Biosensors: A Review. Biomedicines 2023, 11, 3201. [Google Scholar] [CrossRef]
- Wolfe, M.; Cramer, A.; Webb, S.; Goorskey, E.; Chushak, Y.; Mirau, P.; Arroyo-Currás, N.; Chávez, J.L. Rational Approach to Optimizing Conformation-Switching Aptamers for Biosensing Applications. ACS Sens. 2024, 9, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.B.; Gu, M.B. Aptamer-Based Sandwich-Type Biosensors. J. Biol. Eng. 2017, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Feagin, T.A.; Maganzini, N.; Soh, H.T. Strategies for Creating Structure-Switching Aptamers. ACS Sens. 2018, 3, 1611–1615. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Özgür, E.; Denizli, A. Quartz Crystal Microbalance-Based Aptasensors for Medical Diagnosis. Micromachines 2022, 13, 1441. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, J.; Lu, F. Aptamers Used for Biosensors and Targeted Therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef]
- Zhan, H.; Yang, S.; Li, C.; Liu, R.; Chen, W.; Wang, X.; Zhao, Y.; Xu, K. A Highly Sensitive Competitive Aptasensor for AFB1 Detection Based on an Exonuclease-Assisted Target Recycling Amplification Strategy. Anal. Methods 2022, 15, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Debiais, M.; Lelievre, A.; Smietana, M.; Müller, S. Splitting Aptamers and Nucleic Acid Enzymes for the Development of Advanced Biosensors. Nucleic Acids Res. 2020, 48, 3400–3422. [Google Scholar] [CrossRef]
- Chang, C.-C.; Yeh, C.-Y. Using Simple-Structured Split Aptamer for Gold Nanoparticle-Based Colorimetric Detection of Estradiol. Anal. Sci. 2021, 37, 479–483. [Google Scholar] [CrossRef]
- Chen, C.-H.; Jong, Y.-J.; Chao, Y.-Y.; Wang, C.-C.; Chen, Y.-L. Fluorescent Aptasensor Based on Conformational Switch–Induced Hybridization for Facile Detection of β-Amyloid Oligomers. Anal. Bioanal. Chem. 2022, 414, 8155–8165. [Google Scholar] [CrossRef]
- Pei, X.; Wu, X.; Xiong, J.; Wang, G.; Tao, G.; Ma, Y.; Li, N. Competitive Aptasensor for the Ultrasensitive Multiplexed Detection of Cancer Biomarkers by Fluorescent Nanoparticle Counting. Analyst 2020, 145, 3612–3619. [Google Scholar] [CrossRef] [PubMed]
- Serebrennikova, K.V.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. A Fluorescence Resonance Energy Transfer Aptasensor for Aflatoxin B1 Based on Ligand-Induced ssDNA Displacement. Molecules 2023, 28, 7889. [Google Scholar] [CrossRef]
- Zhang, B.; Wei, C. An Aptasensor for the Label-Free Detection of Thrombin Based on Turn-on Fluorescent DNA-Templated Cu/Ag Nanoclusters. RSC Adv. 2020, 10, 35374–35380. [Google Scholar] [CrossRef]
- Zhang, S.; Song, G.; Yang, Z.; Kang, K.; Liu, X. A Label-Free Fluorescence Aptamer Sensor for Point-of-Care Serotonin Detection. Talanta 2024, 277, 126363. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Huang, R.; Xiong, L.; Chen, F.; Sun, W.; Yu, L. A Colorimetric Aptasensor for Ochratoxin A Detection Based on Tetramethylrhodamine Charge Effect-Assisted Silver Enhancement. Biosensors 2023, 13, 468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Q.; Ye, L. Colorimetric Aptasensing of Microcystin-LR Using DNA-Conjugated Polydiacetylene. Anal. Bioanal. Chem. 2024, 416, 7131–7140. [Google Scholar] [CrossRef]
- Li, G.; Yu, T.; Li, H.; Wan, B.; Tan, X.; Zhou, X.; Liang, J.; Zhou, Z. Colorimetric Aptasensors for Sensitive Low-Density Lipoprotein Detection Based on Reduced Oxide Graphene@molybdenum Disulfide-Ferrocene Nanosheets with Peroxidase-like Activity. Anal. Methods 2024, 17, 136–144. [Google Scholar] [CrossRef]
- Ogunmolasuyi, A.M.; Fogel, R.; Hoppe, H.; Goldring, D.; Limson, J. A Microfluidic Paper Analytical Device Using Capture Aptamers for the Detection of PfLDH in Blood Matrices. Malar. J. 2022, 21, 174. [Google Scholar] [CrossRef]
- Lv, X.; Frahat Foda, M.; He, J.; Zhou, J.; Cai, J. Robust and Facile Label-Free Colorimetric Aptasensor for Ochratoxin A Detection Using Aptamer-Enhanced Oxidase-like Activity of MnO2 Nanoflowers. Food Chem. 2023, 401, 134144. [Google Scholar] [CrossRef]
- Khan, A.; Di, K.; Khan, H.; He, N.; Li, Z. Rapid Capturing and Chemiluminescent Sensing of Programmed Death Ligand-1 Expressing Extracellular Vesicles. Biosensors 2022, 12, 281. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Sun, Y.; Luo, C. A Novel Chemiluminescence Sensor for Alpha-Fetoprotein Detection Based on an Aptamer–Luminol Modified Magnetic Graphene Oxide and Copper-Based MOF Composite. Anal. Methods 2024, 16, 5723–5732. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Sun, Y.; Dai, Y.; Gao, D.; Wang, X.; Luo, C. A Chemiluminescence Aptasensor for Sensitive Detection of Carcinoembryonic Antigen Based on Dual Aptamer-Conjugates Biorecognition. Sens. Actuators B Chem. 2021, 326, 128833. [Google Scholar] [CrossRef]
- Yan, X.; Ji, Y.; Xiao, Y.; Xue, X.; Liu, J.; Li, S.; Ai, F.; Zheng, X. One-Pot Label-Free Dual-Aptasensor as a Chemiluminescent Tool Kit Simultaneously Detect Adenosine Triphosphate and Chloramphenicol in Foods. Talanta 2021, 229, 122226. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ma, J.; Neng, J.; Yang, B.; Wang, Y.; Xing, F. A Novel and Label-Free Chemiluminescence Detection of Zearalenone Based on a Truncated Aptamer Conjugated with a G-Quadruplex DNAzyme. Biosensors 2023, 13, 118. [Google Scholar] [CrossRef]
- Han, X.; Qin, M.; Lu, X.; Wang, Q.; Zhang, Y.; Wang, Z. A Highly Sensitive SERS Aptasensor Using a Novel Truncated Aptamer for the Detection of Deoxynivalenol. Sens. Actuators B Chem. 2025, 424, 136908. [Google Scholar] [CrossRef]
- Chen, F.; Huang, Y.; Liu, Y.; Zhuang, Y.; Cao, X.; Qin, X. SERS Analysis Platform Based on Aptamer Recognition-Release Strategy for Efficient and Sensitive Diagnosis of Colorectal Precancerous Lesions. Int. J. Nanomed. 2024, 19, 10009–10021. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Park, J.; Kim, W.; Jo, S.; Kim, M.; Kim, C.; Park, H.; Bang, D.; Lee, W.; Park, J. Bio-Inspired Ag Nanovilli-Based Sandwich-Type SERS Aptasensor for Ultrasensitive and Selective Detection of 25-Hydroxy Vitamin D3. Biosens. Bioelectron. 2021, 188, 113341. [Google Scholar] [CrossRef]
- Park, K.S.; Choi, A.; Kim, H.J.; Park, I.; Eom, M.-S.; Yeo, S.-G.; Son, R.G.; Park, T.-I.; Lee, G.; Soh, H.T.; et al. Ultra-Sensitive Label-Free SERS Biosensor with High-Throughput Screened DNA Aptamer for Universal Detection of SARS-CoV-2 Variants from Clinical Samples. Biosens. Bioelectron. 2023, 228, 115202. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Wang, R.; Li, D. A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers. Micromachines 2022, 13, 1036. [Google Scholar] [CrossRef]
- Villalonga, A.; Parrado, C.; Díaz, R.; Sánchez, A.; Mayol, B.; Martínez-Ruíz, P.; Vilela, D.; Villalonga, R. Supramolecular Enzymatic Labeling for Aptamer Switch-Based Electrochemical Biosensor. Biosensors 2022, 12, 514. [Google Scholar] [CrossRef]
- Villalonga, A.; Díaz, R.; Ojeda, I.; Sánchez, A.; Mayol, B.; Martínez-Ruiz, P.; Villalonga, R.; Vilela, D. Sandwich-Type Electrochemical Aptasensor with Supramolecular Architecture for Prostate-Specific Antigen. Molecules 2024, 29, 4714. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Wang, M.; Min, J.; Tay, R.Y.; Lukas, H.; Sempionatto, J.R.; Li, J.; Xu, C.; Gao, W. A Wearable Aptamer Nanobiosensor for Non-Invasive Female Hormone Monitoring. Nat. Nanotechnol. 2024, 19, 330–337. [Google Scholar] [CrossRef]
- Alharbi, M.A.; Rhouati, A.; Zourob, M. Development of a Label-Free Electrochemical Aptasensor for Rift Valley Fever Virus Detection. Sci. Rep. 2024, 14, 23892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Karimi-Maleh, H. In Situ Synthesis of Label-Free Electrochemical Aptasensor-Based Sandwich-like AuNPs/PPy/Ti3C2Tx for Ultrasensitive Detection of Lead Ions as Hazardous Pollutants in Environmental Fluids. Chemosphere 2023, 324, 138302. [Google Scholar] [CrossRef]
- Cennamo, N.; Pasquardini, L.; Arcadio, F.; Lunelli, L.; Vanzetti, L.; Carafa, V.; Altucci, L.; Zeni, L. SARS-CoV-2 Spike Protein Detection through a Plasmonic D-Shaped Plastic Optical Fiber Aptasensor. Talanta 2021, 233, 122532. [Google Scholar] [CrossRef] [PubMed]
- Vu, C.-A.; Yang, W.-W.; Chan, H.W.-H.; Chen, W.-Y. An Aptamer Sandwich Assay for Protein Biomarker Detection by Solution-Gated Silicon Nanowire Field-Effect Transistor Systems. Microchem. J. 2024, 207, 112245. [Google Scholar] [CrossRef]
- Senturk, H.; Eksin, E.; Işık, Ö.; İlaslan, Z.; Mısırlı, F.; Erdem, A. Impedimetric Aptasensor for Lysozyme Detection Based on Carbon Nanofibres Enriched Screen-Printed Electrodes. Electrochim. Acta 2021, 377, 138078. [Google Scholar] [CrossRef]
- Park, K. Impedance Technique-Based Label-Free Electrochemical Aptasensor for Thrombin Using Single-Walled Carbon Nanotubes-Casted Screen-Printed Carbon Electrode. Sensors 2022, 22, 2699. [Google Scholar] [CrossRef]
- Yuan, M.; Song, Z.; Fei, J.; Wang, X.; Xu, F.; Cao, H.; Yu, J. Aptasensor for Lead(II) Based on the Use of a Quartz Crystal Microbalance Modified with Gold Nanoparticles. Microchim. Acta 2017, 184, 1397–1403. [Google Scholar] [CrossRef]
- Dong, Z.-M.; Zhao, G.-C. Quartz Crystal Microbalance Aptasensor for Sensitive Detection of Mercury(II) Based on Signal Amplification with Gold Nanoparticles. Sensors 2012, 12, 7080–7094. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, S.; Zhang, X.; Du, C.; Si, S.; Chen, J. A High-Frequency QCM Biosensing Platform for Label-Free Detection of the SARS-CoV-2 Spike Receptor-Binding Domain: An Aptasensor and an Immunosensor. Analyst 2023, 148, 719–723. [Google Scholar] [CrossRef]
- Ji, J.; Pang, Y.; Li, D.; Huang, Z.; Zhang, Z.; Xue, N.; Xu, Y.; Mu, X. An Aptamer-Based Shear Horizontal Surface Acoustic Wave Biosensor with a CVD-Grown Single-Layered Graphene Film for High-Sensitivity Detection of a Label-Free Endotoxin. Microsyst. Nanoeng. 2020, 6, 4. [Google Scholar] [CrossRef]
- Gao, F.; Li, Z.Q.; Li, J.Y.; Li, Z.Y.; Wei, J.; Ding, X.; Wang, L.; Zhai, L.Y.; Nan, J.C.; Wang, C. Aptamer-Based Microcantilever Biosensors for Label-Free PSA Detection. IEEE Sens. J. 2024, 25, 266–273. [Google Scholar] [CrossRef]
- Vishnubhotla, R.; Montgomery, C.B.; Steffens, K.L.; Semancik, S. Conformational Changes of Immobilized Polythymine Due to External Stressors Studied with Temperature-Controlled Electrochemical Microdevices. Langmuir 2021, 37, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Jarczewska, M.; Ziółkowski, R.; Górski, Ł.; Malinowska, E. Application of RNA Aptamers as Recognition Layers for the Electrochemical Analysis of C-Reactive Protein. Electroanalysis 2018, 30, 658–664. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Electrochemical Aptasensor for Human Osteopontin Detection Using a DNA Aptamer Selected by SELEX. Anal. Chim. Acta 2017, 987, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Levicky, R.; Herne, T.M.; Tarlov, M.J.; Satija, S.K. Using Self-Assembly To Control the Structure of DNA Monolayers on Gold: A Neutron Reflectivity Study. J. Am. Chem. Soc. 1998, 120, 9787–9792. [Google Scholar] [CrossRef]
- Meng, X.; O’Hare, D.; Ladame, S. Surface Immobilization Strategies for the Development of Electrochemical Nucleic Acid Sensors. Biosens. Bioelectron. 2023, 237, 115440. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Kamath, R.; Xiong, X.; Tan, W.; Fan, Z.H. Aptamer-Enabled Efficient Isolation of Cancer Cells from Whole Blood Using a Microfluidic Device. Anal. Chem. 2012, 84, 4199–4206. [Google Scholar] [CrossRef]
- Pei, H.; Lu, N.; Wen, Y.; Song, S.; Liu, Y.; Yan, H.; Fan, C. A DNA Nanostructure-Based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef]
- Aliakbarinodehi, N.; Jolly, P.; Bhalla, N.; Miodek, A.; De Micheli, G.; Estrela, P.; Carrara, S. Aptamer-Based Field-Effect Biosensor for Tenofovir Detection. Sci. Rep. 2017, 7, 44409. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Oligonucleotide-Functionalized Hydrogels as Stimuli Responsive Materials and Biosensors. Soft Matter 2011, 7, 6757–6767. [Google Scholar] [CrossRef]
- Oberhaus, F.V.; Frense, D.; Beckmann, D. Immobilization Techniques for Aptamers on Gold Electrodes for the Electrochemical Detection of Proteins: A Review. Biosensors 2020, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wang, G.; Hein, R.; Liu, N.; Luo, X.; Davis, J.J. Antifouling Strategies for Selective In Vitro and In Vivo Sensing. Chem. Rev. 2020, 120, 3852–3889. [Google Scholar] [CrossRef]
- Liu, Y.; Canoura, J.; Alkhamis, O.; Xiao, Y. Immobilization Strategies for Enhancing Sensitivity of Electrochemical Aptamer-Based Sensors. ACS Appl. Mater. Interfaces 2021, 13, 9491–9499. [Google Scholar] [CrossRef]
- Fallah, A.; Imani Fooladi, A.A.; Havaei, S.A.; Mahboobi, M.; Sedighian, H. Recent Advances in Aptamer Discovery, Modification and Improving Performance. Biochem. Biophys. Rep. 2024, 40, 101852. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. 2021, 60, 16800–16823. [Google Scholar] [CrossRef]
- Komarova, N.; Kuznetsov, A. Inside the Black Box: What Makes SELEX Better? Molecules 2019, 24, 3598. [Google Scholar] [CrossRef]
- Oliveira, R.; Pinho, E.; Barros, M.M.; Azevedo, N.F.; Almeida, C. In Vitro Selection of DNA Aptamers against Staphylococcal Enterotoxin A. Sci. Rep. 2024, 14, 11345. [Google Scholar] [CrossRef]
- Jia, Y.; Chen, S.; Wang, Q.; Li, J. Recent Progress in Biosensor Regeneration Techniques. Nanoscale 2024, 16, 2834–2846. [Google Scholar] [CrossRef]
- Thiviyanathan, V.; Gorenstein, D.G. Aptamers and the Next Generation of Diagnostic Reagents. Proteom. Clin. Appl. 2012, 6, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gao, X.; Widdicombe, B.; Zhang, X.; Zielinski, J.L.; Cheng, T.; Gunatilaka, A.; Leung, K.K.; Plaxco, K.W.; Rajasekharan Unnithan, R.; et al. Dual-Purpose Aptamer-Based Sensors for Real-Time, Multiplexable Monitoring of Metabolites in Cell Culture Media. ACS Nano 2024, 18, 26127–26139. [Google Scholar] [CrossRef] [PubMed]
- Dauphin-Ducharme, P.; Yang, K.; Arroyo-Currás, N.; Ploense, K.L.; Zhang, Y.; Gerson, J.; Kurnik, M.; Kippin, T.E.; Stojanovic, M.N.; Plaxco, K.W. Electrochemical Aptamer-Based Sensors for Improved Therapeutic Drug Monitoring and High-Precision, Feedback-Controlled Drug Delivery. ACS Sens. 2019, 4, 2832–2837. [Google Scholar] [CrossRef]
- Shrikrishna, N.S.; Halder, S.; Kesarwani, V.; Nagamani, K.; Gandhi, S. Unveiling the Potential: High-Affinity Aptamers for Point of Care Detection of SARS-CoV-2 RBD Protein and It’s Validation in Clinical Samples. Chem. Eng. J. 2024, 493, 152841. [Google Scholar] [CrossRef]
- Wang, J.; Guo, S.; Park, E.; Lee, S.; Park, Y.; Han, X.X.; Zhao, B.; Jung, Y.M. SERS-Based Aptamer Sensing Strategy for Diabetes Biomarker Detection. Anal. Chem. 2024, 96, 20082–20089. [Google Scholar] [CrossRef]
- McDonough, M.H.; Stocker, S.L.; Kippin, T.E.; Meiring, W.; Plaxco, K.W. Using Seconds-resolved Pharmacokinetic Datasets to Assess Pharmacokinetic Models Encompassing Time-varying Physiology. Brit. J. Clin. Pharma. 2023, 89, 2798–2812. [Google Scholar] [CrossRef] [PubMed]
- Alkhamis, O.; Canoura, J.; Wu, Y.; Emmons, N.A.; Wang, Y.; Honeywell, K.M.; Plaxco, K.W.; Kippin, T.E.; Xiao, Y. High-Affinity Aptamers for In Vitro and In Vivo Cocaine Sensing. J. Am. Chem. Soc. 2024, 146, 3230–3240. [Google Scholar] [CrossRef]
- Gerson, J.; Erdal, M.K.; Dauphin-Ducharme, P.; Idili, A.; Hespanha, J.P.; Plaxco, K.W.; Kippin, T.E. A High-precision View of Intercompartmental Drug Transport via Simultaneous, Seconds-resolved, in Situ Measurements in the Vein and Brain. Br. J Pharmacol. 2024, 181, 3869–3885. [Google Scholar] [CrossRef]
- Zhao, C.; Cheung, K.M.; Huang, I.-W.; Yang, H.; Nakatsuka, N.; Liu, W.; Cao, Y.; Man, T.; Weiss, P.S.; Monbouquette, H.G.; et al. Implantable Aptamer–Field-Effect Transistor Neuroprobes for in Vivo Neurotransmitter Monitoring. Sci. Adv. 2021, 7, eabj7422. [Google Scholar] [CrossRef]
- Rabiee, N.; Rabiee, M. Wearable Aptasensors. Anal. Chem. 2024, 96, 19160–19182. [Google Scholar] [CrossRef]
- Singh, N.K.; Chung, S.; Chang, A.-Y.; Wang, J.; Hall, D.A. A Non-Invasive Wearable Stress Patch for Real-Time Cortisol Monitoring Using a Pseudoknot-Assisted Aptamer. Biosens. Bioelectron. 2023, 227, 115097. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, C.; Wang, Z.; Yang, K.-A.; Cheng, X.; Liu, W.; Yu, W.; Lin, S.; Zhao, Y.; Cheung, K.M.; et al. Wearable Aptamer-Field-Effect Transistor Sensing System for Noninvasive Cortisol Monitoring. Sci. Adv. 2022, 8, eabk0967. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Tehrani, F.; Teymourian, H.; Mack, J.; Shaver, A.; Reynoso, M.; Kavner, J.; Huang, N.; Furmidge, A.; Duvvuri, A.; et al. Microneedle Aptamer-Based Sensors for Continuous, Real-Time Therapeutic Drug Monitoring. Anal. Chem. 2022, 94, 8335–8345. [Google Scholar] [CrossRef] [PubMed]
- Friedel, M.; Werbovetz, B.; Drexelius, A.; Watkins, Z.; Bali, A.; Plaxco, K.W.; Heikenfeld, J. Continuous Molecular Monitoring of Human Dermal Interstitial Fluid with Microneedle-Enabled Electrochemical Aptamer Sensors. Lab A Chip 2023, 23, 3289–3299. [Google Scholar] [CrossRef]
- Zheng, H.; GhavamiNejad, A.; GhavamiNejad, P.; Samarikhalaj, M.; Giacca, A.; Poudineh, M. Hydrogel Microneedle-Assisted Assay Integrating Aptamer Probes and Fluorescence Detection for Reagentless Biomarker Quantification. ACS Sens. 2022, 7, 2387–2399. [Google Scholar] [CrossRef]
- Childs, A.; Mayol, B.; Lasalde-Ramírez, J.A.; Song, Y.; Sempionatto, J.R.; Gao, W. Diving into Sweat: Advances, Challenges, and Future Directions in Wearable Sweat Sensing. ACS Nano 2024, 18, 24605–24616. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).