Recent Electrochemical Advancements for Liquid-Biopsy Nucleic Acid Detection for Point-of-Care Prostate Cancer Diagnostics and Prognostics
Abstract
1. Introduction
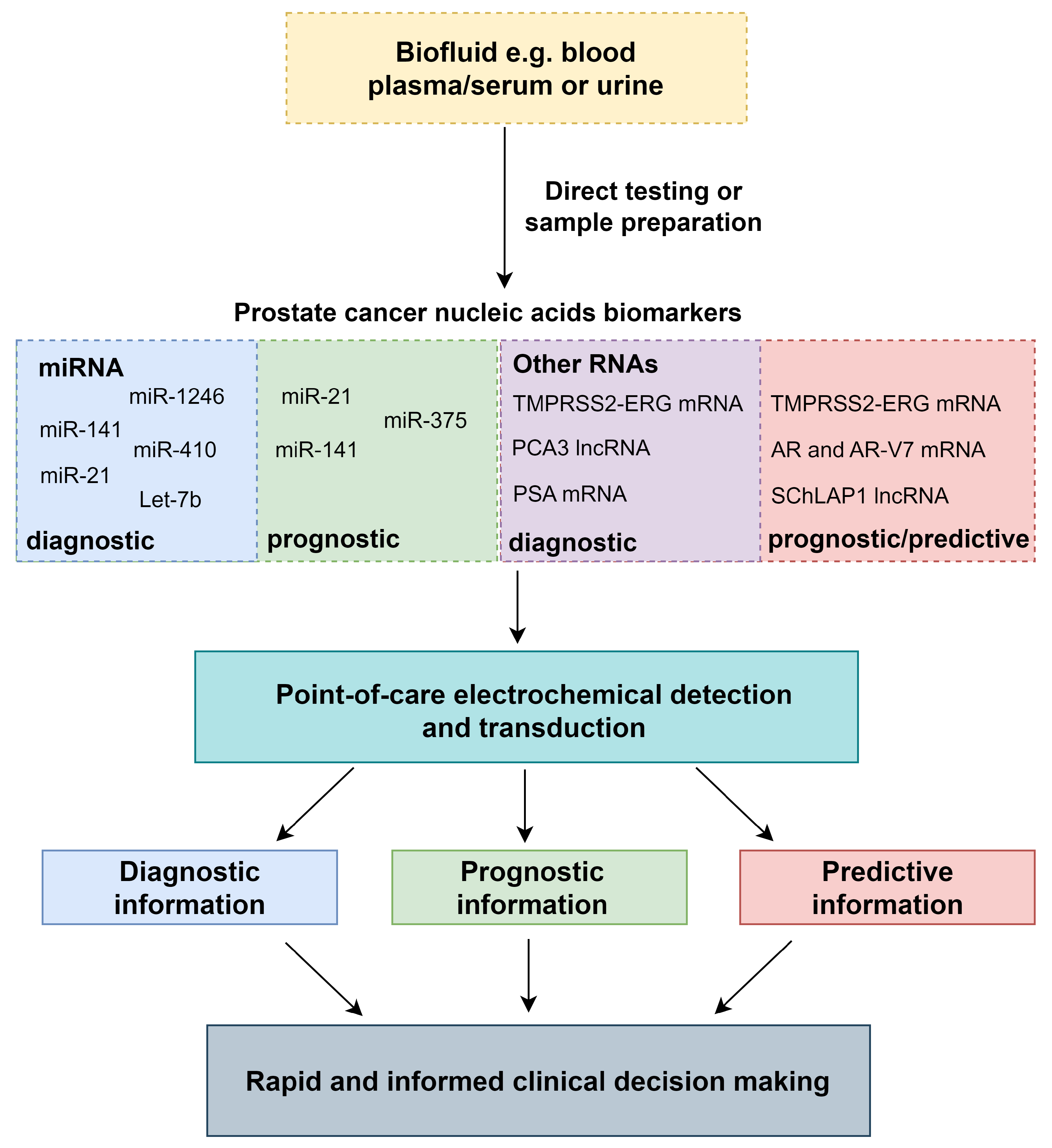
2. Circulating and Exosomal Nucleic Acid Biomarkers for PCa Diagnosis and Prognosis
2.1. MicroRNAs
2.2. mRNAs and lncRNAs
2.3. Biofluid Considerations and Sample Preparation
3. Point-of-Care Electrochemical Techniques
3.1. Electrochemical Impedance Spectroscopy
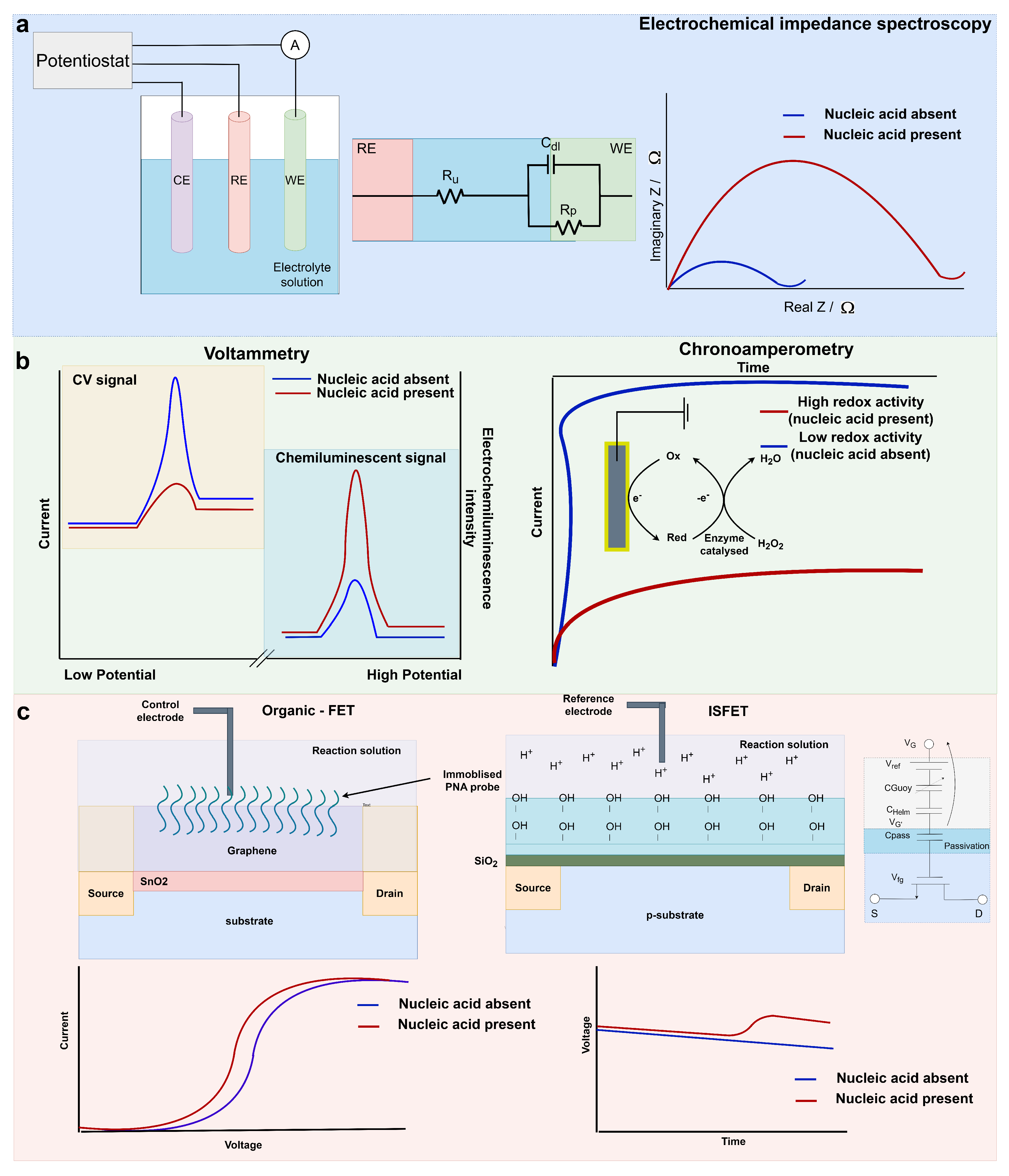
3.2. Voltammetry and Chronoamperometry
3.2.1. Voltammetry
3.2.2. Chronoamperometry
| Bio-Electrical Detection Method | Bio-Recognition Element | Nucleic Acid Target | Limit of Detection | Quantitative Range | Endogenous Detection | References |
|---|---|---|---|---|---|---|
| EIS | ssDNA probe on chitosan and carbon nanotubes | PCA3 lncRNA | 0.128 nM | N/A | cell line | [73] |
| EIS | printed carbon electrode, chondroitin sulfate stabilised AuNPs and ssDNA probe | PCA3 lncRNA | 83 pM | N/A | N/A | [77] |
| EIS | SPCE, AuNPs and aptamer | PCA3 lncRNA | 1 fM | 0.1 pM to 10 nM | spiked artificial urine | [43] |
| EIS | AuNPs, peptide nanotubes and ssDNA probe | miR-410 | 3.9 fM | 10 fM to 300 pM | spiked serum | [37] |
| chronoamperometry | framework nucleic acid electrode and ssDNA probe | miR-21, miR-141 and Let-7a | 10 fM (miR-21) and 1 aM (miR-141) | 10 aM to 1 pM (miR-141) | cell line | [96] |
| chronoamperometry | RPA and peroxidase-mimicking nanozymes | TMPRSS2-ERG, PCA3, SChLAP1 and KLK2 nucleic acids | 50 copies | N/A | urine and serum samples | [95] |
| chronoamperometry | screen-printed carbon electrode and biotinylated ssDNA probe | exosomal miR-451 and miR-21 | 10 pM | 10 pM to 100 nM | extracted exosomal RNA from urine samples | [83] |
| chronoamperometry | gold nanoparticles and sandwich assay | PCA3 and PSA mRNA | 4.4 and 1.5 pM | 25 pM to 10 nM (PCA3), 25 pM to 1 nM (PSA) | extracted RNA from urine samples | [42] |
| chronoamperometry | RT-LAMP, magnetic beads and SPCE | PCA3 lncRNA and PSA mRNA | N/A | N/A | extracted RNA from urine samples | [41] |
| chemoluminescence and CV | AuNPs, Ru complexes and DNA probes | miR-21 and miR-141 | 6.3 and 8.6 fM | 0.02 pM to 150 pM (miR-21), 0.03 pM to 150 pM (miR-141) | N/A | [82] |
| DPV | SWCNT dendritic Au nanostructure and peptide nucleic acid probe | miR-21 | 0.01 fM | 0.01 fM to 1 M | spiked serum | [65] |
| SWV and EIS | MoS2/AuNPs/AgNW and signal amplification | miR-21 and miR-141 | 0.1 fM | 1 fM to 1 nM | spiked serum | [19] |
| SWV | redox labelled DNA hairpins on Au electrode and recycling signal amplification | miR-21 and miR-141 | 4.2 and 3.0 fM | 5 fM to 50 pM | cell lines | [92] |
| SWV | ssDNA probe and gold working electrode | miR-375 | 11.7 aM | 10 aM to 1 nM | cell lines and spiked serum | [23] |
| graphene FET | peptide nucleic acids immobilised on graphene oxide nanosheet | miR-21, miR-1246 and Let-7b | 10 fM | 10 fM to 10 nM | urine samples | [21] |
| solution-gated graphene FET | ssDNA probe immobolised on Au gate | miR-21 | 0.01 aM | 0.01 aM to 1 pM | blood serum patient samples | [20] |
| ISFET | target-specific RT-LAMP and pH-sensing passivation layer | AR-V7, TMPRSS2-ERG, YAP1 and AR-FL mRNA | 5–8 aM | 5–8 aM to 5–8 pM | cell lines, spiked serum and plasma | [84,97] |
3.3. Potentiometric Sensing Using Field-Effect Transistors
4. REASSURED Criteria and Future Directions for PCa PoC Devices
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PoC | point of care |
| PCa | prostate cancer |
| PSA | prostate-specific antigen |
| ADT | androgen-deprivation therapies |
| AR | androgen receptor |
| PCA3 | prostate cancer antigen 3 |
| ssDNA | single-stranded DNA |
| EIS | electrochemical impedance spectroscopy |
| AuNP | gold nanoparticle |
| SPCE | screen-printed carbon electrode |
| MWCNT | multi-walled carbon nanotubes |
| CV | cyclic voltammetry |
| SWV | square-wave voltammetry |
| DPV | differential-pulse voltammetry |
| FET | field-effect transistors |
| ISFET | ion-sensitive FET |
| PTEN | phosphatase and tensin homolog |
| AKT | protein kinase B |
| mTOR | mammalian target of rapamycin |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Schroder, F.H.; Hugosson, J.; Roobol, M.J.; Tammela, T.L.J.; Zappa, M.; Kwiatkowski, M.; Lujan, M.; Maattanen, L.; Lilja, H.; Denis, L.J.; et al. The European Randomized Study of Screening for Prostate Cancer— Prostate Cancer Mortality at 13 Years of Follow-up. Lancet 2014, 384, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Donovan, J.L.; Turner, E.L.; Metcalfe, C.; Young, G.J.; Walsh, E.I.; Lane, J.A.; Noble, S.; Oliver, S.E.; Evans, S.; et al. Effect of a low-intensity PSA-based screening intervention on prostate cancer mortality: The CAP randomized clinical trial. JAMA-J. Am. Med. Assoc. 2018, 319, 883–895. [Google Scholar] [CrossRef]
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers that differentiate benign prostatic hyperplasia from prostate cancer: A literature review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef]
- Canter, D.J.; Branch, C.; Shelnutt, J.; Foreman, A.J.; Lehman, A.M.; Sama, V.; Edwards, D.K.; Abran, J. The 17-Gene Genomic Prostate Score Assay Is Prognostic for Biochemical Failure in Men with Localized Prostate Cancer after Radiation Therapy at a Community Cancer Center. Adv. Radiat. Oncol. 2023, 8, 101193. [Google Scholar] [CrossRef]
- Fine, N.D.; LaPolla, F.; Epstein, M.; Loeb, S.; Dani, H. Genomic classifiers for treatment selection in newly diagnosed prostate cancer. BJU Int. 2019, 124, 578–586. [Google Scholar] [CrossRef]
- Dal Pra, A.; Ghadjar, P.; Hayoz, S.; Liu, V.Y.; Spratt, D.E.; Thompson, D.J.; Davicioni, E.; Huang, H.C.; Zhao, X.; Liu, Y.; et al. Validation of the Decipher genomic classifier in patients receiving salvage radiotherapy without hormone therapy after radical prostatectomy—An ancillary study of the SAKK 09/10 randomized clinical trial. Ann. Oncol. 2022, 33, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Oh-hohenhorst, S.J.; Lange, T. Role of Metastasis-Related microRNAs in Prostate Cancer. Cancers 2021, 13, 4492. [Google Scholar] [CrossRef]
- Rana, S.; Valbuena, G.N.; Curry, E.; Bevan, C.L.; Keun, H.C. MicroRNAs as biomarkers for prostate cancer prognosis: A systematic review and a systematic reanalysis of public data. Br. J. Cancer 2022, 126, 502–513. [Google Scholar] [CrossRef]
- Gittelman, M.C.; Hertzman, B.; Bailen, J.; Williams, T.; Koziol, I.; Henderson, R.J.; Efros, M.; Bidair, M.; Ward, J.F. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: A prospective multicenter clinical study. J. Urol. 2013, 190, 64–69. [Google Scholar] [CrossRef]
- Nicholson, A.; Mahon, J.; Boland, A.; Beale, S.; Dwan, K.; Fleeman, N.; Hockenhull, J.; Dundar, Y. The clinical effectiveness and cost-effectiveness of the PROGENSA® prostate cancer antigen 3 assay and the prostate health index in the diagnosis of prostate cancer: A systematic review and economic evaluation. Health Technol. Assess. 2015, 19, 1–191. [Google Scholar] [CrossRef] [PubMed]
- Tward, J.D.; Schlomm, T.; Bardot, S.; Canter, D.J.; Scroggins, T.; Freedland, S.J.; Lenz, L.; Flake, D.D.; Cohen, T.; Brawer, M.K.; et al. Personalizing Localized Prostate Cancer: Validation of a Combined Clinical Cell-cycle Risk (CCR) Score Threshold for Prognosticating Benefit From Multimodality Therapy. Clin. Genitourin. Cancer 2021, 19, 296–304. [Google Scholar] [CrossRef]
- Brooks, M.A.; Thomas, L.; Magi-Galluzzi, C.; Li, J.; Crager, M.R.; Lu, R.; Abran, J.; Aboushwareb, T.; Klein, E.A. GPS Assay Association with Long-Term Cancer Outcomes: Twenty-Year Risk of Distant Metastasis and Prostate Cancer–Specific Mortality. JCO Precis. Oncol. 2021, 5, 442–449. [Google Scholar] [CrossRef]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; Von Stetten, F. Loop-mediated isothermal amplification (LAMP)-review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Tétreault, N.; De Guire, V. MiRNAs: Their discovery, biogenesis and mechanism of action. Clin. Biochem. 2013, 46, 842–845. [Google Scholar] [CrossRef]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomarkers Med. 2013, 7, 623–631. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Tian, R.; Li, Y.; Bai, J. Hierarchical assembled nanomaterial paper based analytical devices for simultaneously electrochemical detection of microRNAs. Anal. Chim. Acta 2019, 1058, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Ren, Z.; Zhang, H.; Li, Z.; Xue, C.; Wang, J.; Zhang, D.; Yang, H.; Wang, X.; Li, J. Unamplified and Real-Time Label-Free miRNA-21 Detection Using Solution-Gated Graphene Transistors in Prostate Cancer Diagnosis. Adv. Sci. 2023, 10, e2205886. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.; Cho, Y.S.; Kim, Y.; Tae, J.H.; No, T.I.; Shim, J.S.; Jeong, Y.; Kang, S.H.; Lee, K.H. Electrical Cartridge Sensor Enables Reliable and Direct Identification of MicroRNAs in Urine of Patients. ACS Sens. 2021, 6, 833–841. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, J.; Li, B.; Liu, J.; Xu, J.J.; Chen, H.Y. Dual-Mode SERS and Electrochemical Detection of miRNA Based on Popcorn-like Gold Nanofilms and Toehold-Mediated Strand Displacement Amplification Reaction. Anal. Chem. 2021, 93, 6120–6127. [Google Scholar] [CrossRef] [PubMed]
- Jeong, B.; Kim, Y.J.; Jeong, J.Y.; Kim, Y.J. Label-free electrochemical quantification of microRNA-375 in prostate cancer cells. J. Electroanal. Chem. 2019, 846, 113127. [Google Scholar] [CrossRef]
- Ghorbanmehr, N.; Gharbi, S.; Korsching, E.; Tavallaei, M.; Einollahi, B.; Mowla, S.J. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine—promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019, 79, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Porzycki, P.; Ciszkowicz, E.; Semik, M.; Tyrka, M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int. Urol. Nephrol. 2018, 50, 1619–1626. [Google Scholar] [CrossRef]
- Arisan, E.D.; Rencuzogullari, O.; Freitas, I.L.; Radzali, S.; Keskin, B.; Kothari, A.; Warford, A.; Uysal-Onganer, P. Upregulated wnt-11 and mir-21 expression trigger epithelial mesenchymal transition in aggressive prostate cancer cells. Biology 2020, 9, 52. [Google Scholar] [CrossRef]
- Stafford, M.Y.; Willoughby, C.E.; Walsh, C.P.; McKenna, D.J. Prognostic value of miR-21 for prostate cancer: A systematic review and meta-analysis. Biosci. Rep. 2022, 42, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Dong, Y.; Wang, K.J.; Deng, Z.; Zhang, W.; Shen, H.F. Plasma exosomal miR-125a-5p and miR-141-5p as non-invasive biomarkers for prostate cancer. Neoplasma 2021, 67, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Qin, X.J.; Cao, D.L.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Ye, D.W. An elevated serum miR-141 level in patients with bone-metastatic prostate cancer is correlated with more bone lesions. Asian J. Androl. 2013, 15, 231–235. [Google Scholar] [CrossRef]
- Wei, J.; Lu, Y.; Wang, R.; Xu, X.; Liu, Q.; He, S.; Pan, H.; Liu, X.; Yuan, B.; Ding, Y.; et al. MicroRNA-375: Potential cancer suppressor and therapeutic drug. Biosci. Rep. 2021, 41, 1–14. [Google Scholar] [CrossRef]
- Selth, L.A.; Das, R.; Townley, S.L.; Coutinho, I.; Hanson, A.R.; Centenera, M.M.; Stylianou, N.; Sweeney, K.; Soekmadji, C.; Jovanovic, L.; et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Cai, S.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C.L.; Ladame, S.; Ivanov, A.P.; Edel, J.B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, G.A.; Shibakawa, A.; Patel, H.; Sita-Lumsden, A.; Zivi, A.; Rama, N.; Bevan, C.L.; Ladame, S. Amplification-free detection of circulating microRNA biomarkers from body fluids based on fluorogenic oligonucleotide-templated reaction between engineered peptide nucleic acid probes: Application to prostate cancer diagnosis. Anal. Chem. 2016, 88, 8091–8098. [Google Scholar] [CrossRef]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Zedan, A.H.; Osther, P.J.; Assenholt, J.; Madsen, J.S.; Hansen, T.F. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci. Rep. 2020, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, D.; Liston, M.; Patel, N.; Akoto, T.; Lui, B.; Yang, T.L.; To, D.M.; Majid, S.; Dahiya, R.; Tabatabai, Z.L.; et al. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene 2020, 39, 7209–7223. [Google Scholar] [CrossRef] [PubMed]
- Yaman, Y.T.; Vural, O.A.; Bolat, G.; Abaci, S. One-pot synthesized gold nanoparticle-peptide nanotube modified disposable sensor for impedimetric recognition of miRNA 410. Sens. Actuators Chem. 2020, 320, 128343. [Google Scholar] [CrossRef]
- Zhang, T.; Austin, R.G.; Park, S.E.; Runyambo, D.; Boominathan, R.; Rao, C.; Bronson, E.; Anand, M.; Healy, P.; George, D.J.; et al. Expression of immune checkpoints on circulating tumor cells in men with metastatic prostate cancer (mPC). J. Clin. Oncol. 2018, 36, 191. [Google Scholar] [CrossRef]
- Wang, J.; Ye, H.; Zhang, D.; Hu, Y.; Yu, X.; Wang, L.; Zuo, C.; Yu, Y.; Xu, G.; Liu, S. MicroRNA-410-5p as a potential serum biomarker for the diagnosis of prostate cancer. Cancer Cell Int. 2016, 16, 1–6. [Google Scholar] [CrossRef]
- Merriel, S.W.; Pocock, L.; Gilbert, E.; Creavin, S.; Walter, F.M.; Spencer, A.; Hamilton, W. Systematic review and meta-analysis of the diagnostic accuracy of prostate-specific antigen (PSA) for the detection of prostate cancer in symptomatic patients. BMC Med. 2022, 20, 54. [Google Scholar] [CrossRef]
- Moranova, L.; Stanik, M.; Hrstka, R.; Campuzano, S.; Bartosik, M. Electrochemical LAMP-based assay for detection of RNA biomarkers in prostate cancer. Talanta 2022, 238, 123064. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Salcedo, R.; Miranda-Castro, R.; de-los Santos-Álvarez, N.; Lobo-Castañón, M.J. Dual electrochemical genosensor for early diagnosis of prostate cancer through lncRNAs detection. Biosens. Bioelectron. 2021, 192, 113520. [Google Scholar] [CrossRef] [PubMed]
- Takita, S.; Nabok, A.; Mussa, M.; Kitchen, M.; Lishchuk, A.; Smith, D. Ultrasensitive prostate cancer marker PCA3 detection with impedimetric biosensor based on specific label-free aptamers. Biosens. Bioelectron. X 2024, 18, 100462. [Google Scholar] [CrossRef]
- Soares, R.R.; Neumann, F.; Caneira, C.R.; Madaboosi, N.; Ciftci, S.; Hernández-Neuta, I.; Pinto, I.F.; Santos, D.R.; Chu, V.; Russom, A.; et al. Silica bead-based microfluidic device with integrated photodiodes for the rapid capture and detection of rolling circle amplification products in the femtomolar range. BIosens. Bioelectron. 2019, 128, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Hägglöf, C.; Hammarsten, P.; Strömvall, K.; Egevad, L.; Josefsson, A.; Stattin, P.; Granfors, T.; Bergh, A. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS ONE 2014, 9, e86824. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.a.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.w.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Leshem, O.; Madar, S.; Kogan-Sakin, I.; Kamer, I.; Goldstein, I.; Brosh, R.; Cohen, Y.; Jacob-Hirsch, J.; Ehrlich, M.; Ben-Sasson, S.; et al. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS ONE 2011, 6, e21650. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Aubin, S.M.; Siddiqui, J.; Lonigro, R.J.; Sefton-Miller, L.; Miick, S.; Williamsen, S.; Hodge, P.; Meinke, J.; Blase, A.; et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 2011, 3, 94ra72. [Google Scholar] [CrossRef]
- Reig, Ò.; Marín-Aguilera, M.; Carrera, G.; Jiménez, N.; Paré, L.; García-Recio, S.; Gaba, L.; Pereira, M.V.; Fernández, P.; Prat, A.; et al. TMPRSS2-ERG in Blood and Docetaxel Resistance in Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2016, 70, 709–713. [Google Scholar] [CrossRef]
- Marín-Aguilera, M.; Reig, Ò.; Milà-Guasch, M.; Font, A.; Domènech, M.; Rodríguez-Vida, A.; Carles, J.; Suárez, C.; del Alba, A.G.; Jiménez, N.; et al. The influence of treatment sequence in the prognostic value of TMPRSS2-ERG as biomarker of taxane resistance in castration-resistant prostate cancer. Int. J. Cancer 2019, 145, 1970–1981. [Google Scholar] [CrossRef]
- Estébanez-Perpiñá, E.; Bevan, C.L.; McEwan, I.J. Eighty years of targeting androgen receptor activity in prostate cancer: The fight goes on. Cancers 2021, 13, 509. [Google Scholar] [CrossRef] [PubMed]
- Ferraldeschi, R.; Welti, J.; Luo, J.; Attard, G.; De Bono, J.S. Targeting the androgen receptor pathway in castration-resistant prostate cancer: Progresses and prospects. Oncogene 2014, 34, 1745–1757. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Silberstein, J.; Luber, B.; Wang, H.; Lu, C.; Chen, Y.; Zhu, Y.; Taylor, M.N.; Carducci, M.A.; Eisenberger, M.A.; Luo, J.; et al. Clinical significance of AR mRNA quantification from circulating tumor cells (CTCs) in men with metastatic castration-resistant prostate cancer (mCRPC) treated with abiraterone (Abi) or enzalutamide (Enza). J. Clin. Oncol. 2017, 35, 132. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Zhu, Y.; Silberstein, J.L.; Taylor, M.N.; Maughan, B.L.; Denmeade, S.R.; et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first & second-line Abiraterone & Enzalutamide. J. Clin. Oncol. 2017, 35, 2149–2156. [Google Scholar] [CrossRef] [PubMed]
- Stitz, R.; Stoiber, F.; Silye, R.; Vlachos, G.; Andaloro, S.; Rebhan, E.; Dunzinger, M. Clinical Implementation of a Noninvasive, Multi-Analyte Droplet Digital PCR Test to Screen for Androgen Receptor Alterations. J. Mol. Diagn. 2024, 26, 467–478. [Google Scholar] [CrossRef]
- Nimir, M.; Ma, Y.; Jeffreys, S.A.; Opperman, T.; Young, F.; Khan, T.; Ding, P.; Chua, W.; Balakrishnar, B.; Cooper, A.; et al. Detection of AR-v7 in liquid biopsies of castrate resistant prostate cancer patients: A comparison of AR-v7 analysis in circulating tumor cells, circulating tumor RNA and exosomes. Cells 2019, 8, 688. [Google Scholar] [CrossRef]
- Del Re, M.; Conteduca, V.; Crucitta, S.; Gurioli, G.; Casadei, C.; Restante, G.; Schepisi, G.; Lolli, C.; Cucchiara, F.; Danesi, R.; et al. Androgen receptor gain in circulating free DNA and splicing variant 7 in exosomes predict clinical outcome in CRPC patients treated with abiraterone and enzalutamide. Prostate Cancer Prostatic Dis. 2021, 24, 524–531. [Google Scholar] [CrossRef]
- Eskra, J.N.; Rabizadeh, D.; Pavlovich, C.P.; Catalona, W.J.; Luo, J. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019, 22, 362–381. [Google Scholar] [CrossRef]
- Wang, T.; Tang, L.; Lin, R.; He, D.; Wu, Y.; Zhang, Y.; Yang, P.; He, J. Individual variability in human urinary metabolites identifies age-related, body mass index-related, and sex-related biomarkers. Mol. Genet. Genom. Med. 2021, 9, e1738. [Google Scholar] [CrossRef]
- Welch, A.A.; Mulligan, A.; Bingham, S.A.; Khaw, K.T. Urine pH is an indicator of dietary acid-base load, fruit and vegetables and meat intakes: Results from the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk population study. Br. J. Nutr. 2008, 99, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Garimella, S.; Clay-Gilmour, A.; Vojtech, L.; Armstrong, B.; Bessonny, M.; Stamatikos, A. Comparison of Human Urinary Exosomes Isolated via Ultracentrifugation Alone versus Ultracentrifugation Followed by SEC Column-Purification. J. Pers. Med. 2022, 12, 340. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yuan, X.; Wang, J.; Wang, Y.; Li, S. The diagnostic value of miRNA-141 in prostate cancer: A systematic review and meta-analysis. Blood Genom. 2020, 4, 53–59. [Google Scholar] [CrossRef]
- Qin, Y.; Yao, J.; Wu, D.C.; Nottingham, R.M.; Mohr, S.; Hunicke-Smith, S.; Lambowitz, A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. Rna 2016, 22, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Sabahi, A.; Salahandish, R.; Ghaffarinejad, A.; Omidinia, E. Electrochemical nano-genosensor for highly sensitive detection of miR-21 biomarker based on SWCNT-grafted dendritic Au nanostructure for early detection of prostate cancer. Talanta 2020, 209, 120595. [Google Scholar] [CrossRef]
- Paul, R.; Ostermann, E.; Wei, Q. Advances in point-of-care nucleic acid extraction technologies for rapid diagnosis of human and plant diseases. Biosens. Bioelectron. 2020, 169, 112592. [Google Scholar] [CrossRef]
- Sriram, H.; Khanka, T.; Kedia, S.; Tyagi, P.; Ghogale, S.; Deshpande, N.; Chatterjee, G.; Rajpal, S.; Patkar, N.V.; Subramanian, P.G.; et al. Improved protocol for plasma microrna extraction and comparison of commercial kits. Biochem. Medica 2021, 31, 030705. [Google Scholar] [CrossRef]
- Lasserre, P.; Balansethupathy, B.; Vezza, V.J.; Butterworth, A.; Macdonald, A.; Blair, E.O.; McAteer, L.; Hannah, S.; Ward, A.C.; Hoskisson, P.A.; et al. SARS-CoV-2 Aptasensors Based on Electrochemical Impedance Spectroscopy and Low-Cost Gold Electrode Substrates. Anal. Chem. 2022, 94, 2126–2133. [Google Scholar] [CrossRef]
- Lazanas, A.C.; Prodromidis, M.I. Electrochemical Impedance Spectroscopy—A Tutorial. ACS Meas. Sci. Au 2023, 3, 162–193. [Google Scholar] [CrossRef]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical Impedance Spectroscopy (EIS): Principles, Construction, and Biosensing Applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef]
- Garg, S.; Sachdeva, A.; Peeters, M.; McClements, J. Point-of-Care Prostate Specific Antigen Testing: Examining Translational Progress toward Clinical Implementation. ACS Sens. 2023, 8, 3643–3658. [Google Scholar] [CrossRef]
- Mwanza, D.; Adeniyi, O.; Tesfalidet, S.; Nyokong, T.; Mashazi, P. Capacitive label-free ultrasensitive detection of PSA on a covalently attached monoclonal anti-PSA antibody gold surface. J. Electroanal. Chem. 2022, 927, 116983. [Google Scholar] [CrossRef]
- Soares, J.C.; Soares, A.C.; Rodrigues, V.C.; Melendez, M.E.; Santos, A.C.; Faria, E.F.; Reis, R.M.; Carvalho, A.L.; Oliveira, O.N. Detection of the Prostate Cancer Biomarker PCA3 with Electrochemical and Impedance-Based Biosensors. ACS Appl. Mater. Interfaces 2019, 11, 46645–46650. [Google Scholar] [CrossRef]
- Weng, W.H.; Jhou, C.H.; Xie, H.X.; Pan, T.M. Label-Free Detection of AR-V7 mRNA in Prostate Cancer Using Yb2Ti2O7–Based Electrolyte-Insulator-Semiconductor Biosensors. J. Electrochem. Soc. 2016, 163, B710–B717. [Google Scholar] [CrossRef]
- Kinnamon, D.; Ghanta, R.; Lin, K.C.; Muthukumar, S.; Prasad, S. Portable biosensor for monitoring cortisol in low-volume perspired human sweat. Sci. Rep. 2017, 7, 13312. [Google Scholar] [CrossRef] [PubMed]
- Mugoni, V.; Ciani, Y.; Nardella, C.; Demichelis, F. Circulating RNAs in prostate cancer patients. Cancer Lett. 2022, 524, 57–69. [Google Scholar] [CrossRef]
- Rodrigues, V.C.; Soares, J.C.; Soares, A.C.; Braz, D.C.; Melendez, M.E.; Ribas, L.C.; Scabini, L.F.; Bruno, O.M.; Carvalho, A.L.; Reis, R.M.; et al. Electrochemical and optical detection and machine learning applied to images of genosensors for diagnosis of prostate cancer with the biomarker PCA3. Talanta 2021, 222, 121444. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, K.; Neves, A.F.; Rocha, R.M.; Faria, P.R.; Alves, P.T.; Souza, A.G.; Fujimura, P.T.; Santos, F.A.; Araújo, T.G.; Ward, L.S.; et al. Prostate-specific RNA aptamer: Promising nucleic acid antibody-like cancer detection. Sci. Rep. 2015, 5, 12090. [Google Scholar] [CrossRef]
- Abdelbaset, R.; Shawky, S.M.; Abdullah, M.A.; Morsy, O.E.; Yahia, Y.A.; Ghallab, Y.H.; Matboli, M.; Ismail, Y. A new label free spiral sensor using impedance spectroscopy to characterize hepatocellular carcinoma in tissue and serum samples. Sci. Rep. 2024, 14, 13155. [Google Scholar] [CrossRef]
- Farokhi, S.; Roushani, M. Flower-like core-shell nanostructures based on natural asphalt coated with Ni-LDH nanosheets as an electrochemical platform for prostate cancer biomarker sensing. Microchim. Acta 2023, 190, 198. [Google Scholar] [CrossRef]
- Takita, S.; Nabok, A.; Lishchuk, A.; Mussa, M.H.; Smith, D. Detection of Prostate Cancer Biomarker PCA3 with Electrochemical Apta-Sensor. Eng. Proc. 2022, 16, 8. [Google Scholar] [CrossRef]
- Feng, X.; Gan, N.; Zhang, H.; Li, T.; Cao, Y.; Hu, F.; Jiang, Q. Ratiometric biosensor array for multiplexed detection of microRNAs based on electrochemiluminescence coupled with cyclic voltammetry. Biosens. Bioelectron. 2016, 75, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Chiou, Y.E.; Yu, K.J.; Pang, S.N.; Yang, Y.L.; Pang, S.T.; Weng, W.H. A Highly Sensitive Urinary Exosomal miRNAs Biosensor Applied to Evaluation of Prostate Cancer Progression. Bioengineering 2022, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Broomfield, J.; Kalofonou, M.; Pataillot-Meakin, T.; Powell, S.M.; Fernandes, R.C.; Moser, N.; L. Bevan, C.; Georgiou, P. Detection of YAP1 and AR-V7 mRNA for Prostate Cancer Prognosis Using an ISFET Lab-On-Chip Platform. ACS Sens. 2022, 7, 3389–3398. [Google Scholar] [CrossRef] [PubMed]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Chooto, P. Cyclic Voltammetry and Its Applications; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar] [CrossRef]
- Simões, F.R.; Xavier, M.G. 6-Electrochemical Sensors. In Nanoscience and its Applications; Micro and Nano Technologies; Da Róz, A.L., Ferreira, M., de Lima Leite, F., Oliveira, O.N.B.T.N., Eds.; William Andrew Publishing: Norwich, NY, USA, 2017; pp. 155–178. [Google Scholar] [CrossRef]
- Venton, B.J.; DiScenza, D.J. Chapter 3—Voltammetry. In Electrochemistry for Bioanalysis; Patel, B.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 27–50. [Google Scholar] [CrossRef]
- Mirceski, V.; Skrzypek, S.; Stojanov, L. Square-wave voltammetry. ChemTexts 2018, 4, 17. [Google Scholar] [CrossRef]
- Li, F.; Han, X.; Liu, S. Development of an electrochemical DNA biosensor with a high sensitivity of fM by dendritic gold nanostructure modified electrode. Biosens. Bioelectron. 2011, 26, 2619–2625. [Google Scholar] [CrossRef]
- Yang, F.K.; Tian, C.; Zhou, L.X.; Guan, T.Y.; Chen, G.L.; Zheng, Y.Y.; Cao, Z.G. The value of urinary exosomal microRNA-21 in the early diagnosis and prognosis of bladder cancer. Kaohsiung J. Med. Sci. 2024, 40, 660–670. [Google Scholar] [CrossRef]
- Yang, C.; Dou, B.; Shi, K.; Chai, Y.; Xiang, Y.; Yuan, R. Multiplexed and amplified electronic sensor for the detection of microRNAs from cancer cells. Anal. Chem. 2014, 86, 11913–11918. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.M.; Dey, S.; Trau, M. A Sample-to-Targeted Gene Analysis Biochip for Nanofluidic Manipulation of Solid-Phase Circulating Tumor Nucleic Acid Amplification in Liquid Biopsies. ACS Sens. 2018, 3, 2597–2603. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Li, L.; Li, J.; Lin, M.; Liu, G.; Liang, W.; Xu, L.; Li, Y.; Zuo, X.; Ren, S.; et al. DNA Framework-Mediated Electrochemical Biosensing Platform for Amplification-Free MicroRNA Analysis. Anal. Chem. 2020, 92, 4498–4503. [Google Scholar] [CrossRef] [PubMed]
- Broomfield, J.; Kalofonou, M.; Franklin, S.; Powell, S.M.; Pataillot-Meakin, T.; Moser, N.; Bevan, C.L.; Georgiou, P. Handheld ISFET Lab-on-Chip Detection of TMPRSS2-ERG and AR mRNA for Prostate Cancer Prognostics. IEEE Sens. Lett. 2023, 7, 1–4. [Google Scholar] [CrossRef]
- Paimard, G.; Ghasali, E.; Baeza, M. Screen-Printed Electrodes: Fabrication, Modification, and Biosensing Applications. Chemosensors 2023, 11, 113. [Google Scholar] [CrossRef]
- Moser, N.; Keeble, L.; Rodriguez-Manzano, J.; Georgiou, P. ISFET arrays for lab-on-chip technology: A review. In Proceedings of the 2019 26th IEEE International Conference on Electronics, Circuits and Systems, ICECS 2019, Genoa, Italy, 27–29 November 2019; pp. 57–60. [Google Scholar] [CrossRef]
- Georgiou, P.; Toumazou, C. ISFET characteristics in CMOS and their application to weak inversion operation. Sens. Actuators B Chem. 2009, 143, 211–217. [Google Scholar] [CrossRef]
- Rodriguez-Manzano, J.; Malpartida-Cardenas, K.; Moser, N.; Pennisi, I.; Cavuto, M.; Miglietta, L.; Moniri, A.; Penn, R.; Satta, G.; Randell, P.; et al. Handheld point-of-care system for rapid detection of SARS-CoV-2 extracted RNA in under 20 min. ACS Cent. Sci. 2021, 7, 307–317. [Google Scholar] [CrossRef]
- Chen, C.Y. DNA polymerases drive DNA sequencing-by-synthesis technologies: Both past and present. Front. Microbiol. 2014, 5, 305. [Google Scholar] [CrossRef]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef]
- Lokhandwala, P.M.; Riel, S.L.; Haley, L.; Lu, C.; Chen, Y.; Silberstein, J.; Zhu, Y.; Zheng, G.; Lin, M.T.; Gocke, C.D.; et al. Analytical Validation of Androgen Receptor Splice Variant 7 Detection in a Clinical Laboratory Improvement Amendments (CLIA) Laboratory Setting. J. Mol. Diagn. 2017, 19, 115–125. [Google Scholar] [CrossRef]
- Sokoll, L.J.; Ellis, W.; Lange, P.; Noteboom, J.; Elliott, D.J.; Deras, I.L.; Blase, A.; Koo, S.; Sarno, M.; Rittenhouse, H.; et al. A multicenter evaluation of the PCA3 molecular urine test: Pre-analytical effects, analytical performance, and diagnostic accuracy. Clin. Chim. Acta 2008, 389, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Canter, D.J.; Freedland, S.; Rajamani, S.; Latsis, M.; Variano, M.; Halat, S.; Tward, J.; Cohen, T.; Stone, S.; Schlomm, T.; et al. Analysis of the prognostic utility of the cell cycle progression (CCP) score generated from needle biopsy in men treated with definitive therapy. Prostate Cancer Prostatic Dis. 2020, 23, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Fizazi, K.; Piulats, J.M.; Reaume, M.N.; Ostler, P.; McDermott, R.; Gingerich, J.R.; Pintus, E.; Sridhar, S.S.; Bambury, R.M.; Emmenegger, U.; et al. Rucaparib or Physician’s Choice in Metastatic Prostate Cancer. N. Engl. J. Med. 2023, 388, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Su, J.; Zhao, Z.; Shao, Y.; Dou, Y.; Li, F.; Deng, W.; Shi, J.; Li, Q.; Zuo, X.; et al. DNA Framework-Supported Electrochemical Analysis of DNA Methylation for Prostate Cancers. Nano Lett. 2020, 20, 7028–7035. [Google Scholar] [CrossRef]
- Mantikas, K.T.; Moser, N.; Gulli, C.; Cunningham, D.; Georgiou, P.; Simillis, C.; Kalofonou, M. Detection of the Colorectal Cancer TP53 p.R248W Mutation on a Lab-on-Chip ISFET Platform. In Proceedings of the 2023 IEEE BioSensors Conference, BioSensors 2023—Proceedings, London, UK, 30 July–1 August 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Alexandrou, G.; Moser, N.; Mantikas, K.T.; Rodriguez-Manzano, J.; Ali, S.; Coombes, R.C.; Shaw, J.; Georgiou, P.; Toumazou, C.; Kalofonou, M. Detection of Multiple Breast Cancer ESR1 Mutations on an ISFET Based Lab-on-Chip Platform. IEEE Trans. Biomed. Circuits Syst. 2021, 15, 380–389. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broomfield, J.; Kalofonou, M.; Bevan, C.L.; Georgiou, P. Recent Electrochemical Advancements for Liquid-Biopsy Nucleic Acid Detection for Point-of-Care Prostate Cancer Diagnostics and Prognostics. Biosensors 2024, 14, 443. https://doi.org/10.3390/bios14090443
Broomfield J, Kalofonou M, Bevan CL, Georgiou P. Recent Electrochemical Advancements for Liquid-Biopsy Nucleic Acid Detection for Point-of-Care Prostate Cancer Diagnostics and Prognostics. Biosensors. 2024; 14(9):443. https://doi.org/10.3390/bios14090443
Chicago/Turabian StyleBroomfield, Joseph, Melpomeni Kalofonou, Charlotte L. Bevan, and Pantelis Georgiou. 2024. "Recent Electrochemical Advancements for Liquid-Biopsy Nucleic Acid Detection for Point-of-Care Prostate Cancer Diagnostics and Prognostics" Biosensors 14, no. 9: 443. https://doi.org/10.3390/bios14090443
APA StyleBroomfield, J., Kalofonou, M., Bevan, C. L., & Georgiou, P. (2024). Recent Electrochemical Advancements for Liquid-Biopsy Nucleic Acid Detection for Point-of-Care Prostate Cancer Diagnostics and Prognostics. Biosensors, 14(9), 443. https://doi.org/10.3390/bios14090443





