The Construction and Application of a New Screening Method for Phosphodiesterase Inhibitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Solutions
2.2. Cell Culture
2.3. Cell Transfection
2.4. Live Cell cAMP/cGMP Measurement
2.5. In Vitro Enzymatic Activity Assay
2.6. Data Analysis and Statistics
3. Results
3.1. Construction of the PDE Inhibitor Screening Method
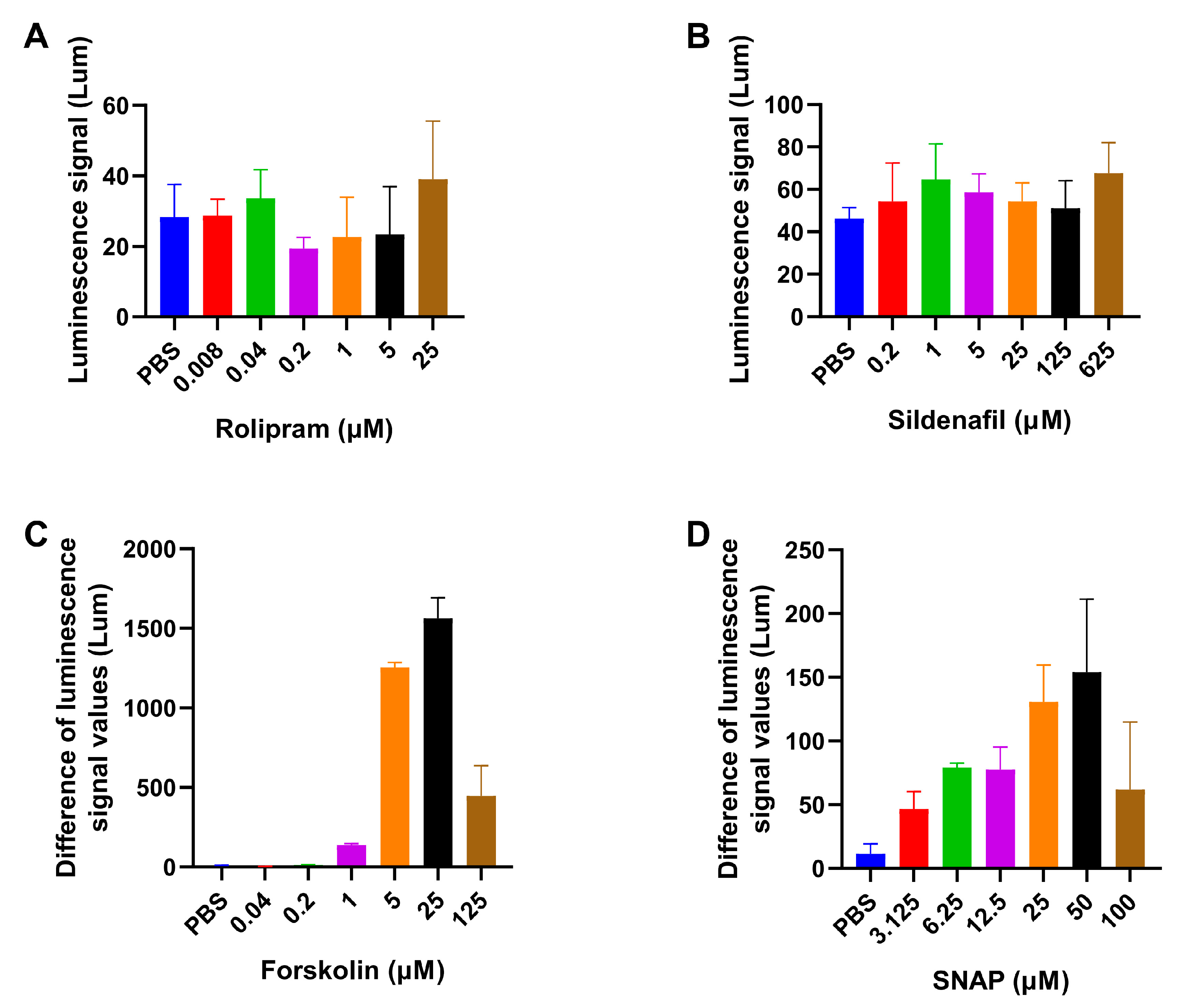
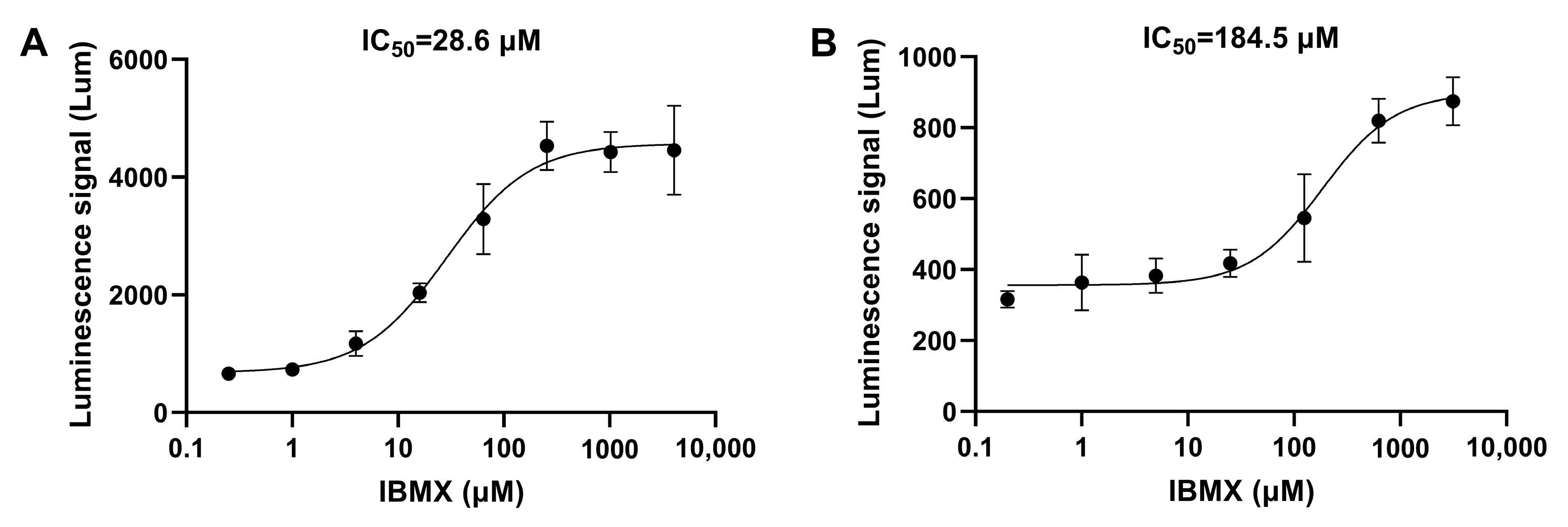
3.1.1. The Necessity and Optimal Concentration of AC/GC Agonists
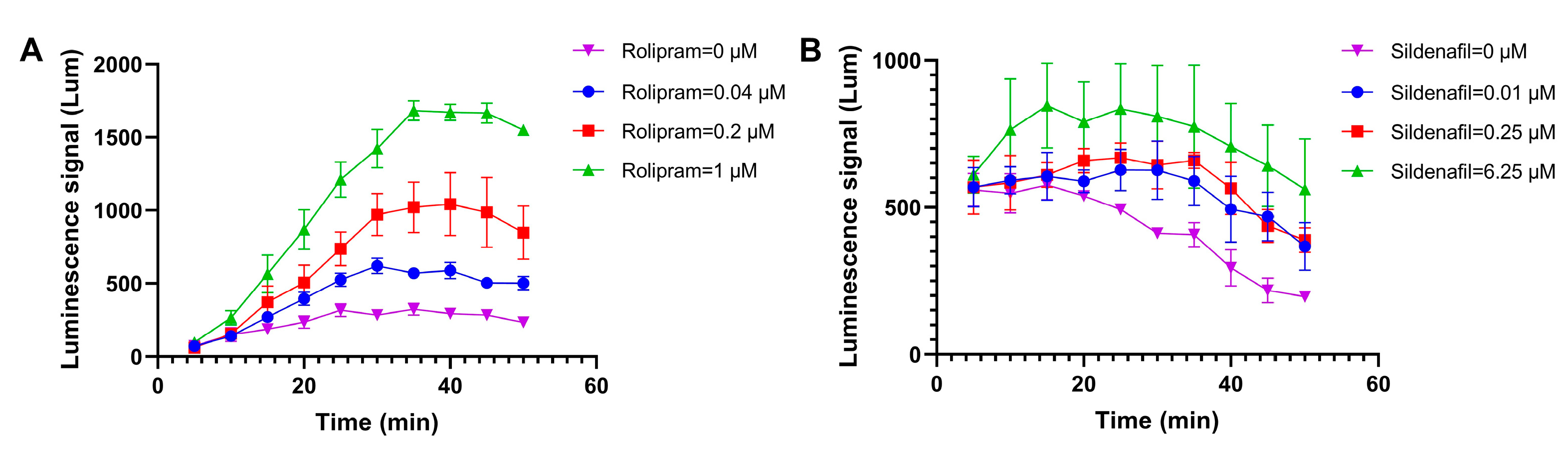
3.1.2. Determination of the Shortest Detection Time for Cell Screening Models
3.2. Validation of the Effectiveness of the PDE Inhibitor Screening Method
3.2.1. The Detection Results for Positive Control Drugs
3.2.2. The Detection Results for Reported Natural Products with PDE Inhibitory Activity
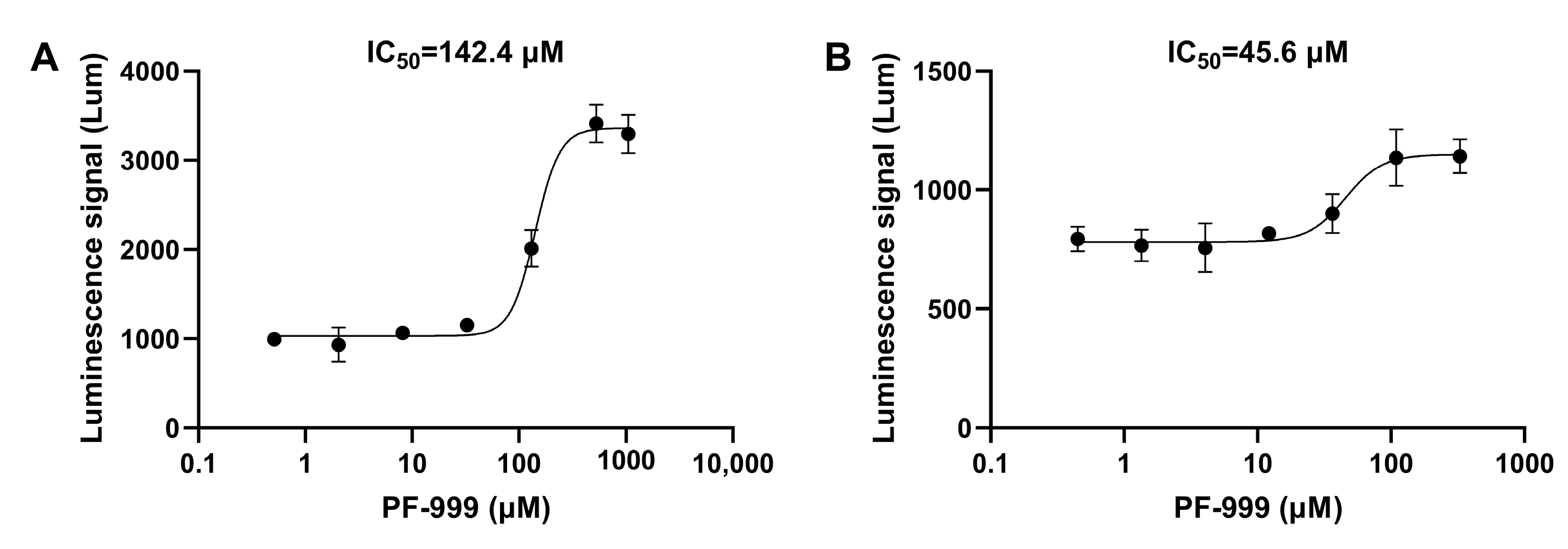
3.3. The PDE Inhibitory Activity of Compounds Obtained Using the New Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Wang, H.; Wang, W.Z.; Wang, D.; Skaggs, K.; Zhang, H.T. Phosphodiesterase 7(PDE7): A unique drug target for central nervous system diseases. Neuropharmacology 2021, 196, 108694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.T. Targeting phosphodiesterases (PDEs) for treatment of CNS diseases. Curr. Pharm. Des. 2015, 21, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Bollen, E.; Prickaerts, J. Phosphodiesterases in neurodegenerative disorders. IUBMB Life 2012, 64, 965–970. [Google Scholar] [CrossRef]
- Izquierdo, J.L.; Aparicio, J. Roflumilast for COPD. Drugs Today 2010, 46, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Krishnappa, P.; Fernandez-Pascual, E.; Carballido, J.; Martinez-Salamanca, J.I. Sildenafil/Viagra in the treatment of premature ejaculation. Int. J. Impot. Res. 2019, 31, 65–70. [Google Scholar] [CrossRef]
- Liang, J.; Huang, Y.Y.; Zhou, Q.; Gao, Y.; Li, Z.; Wu, D.; Yu, S.; Guo, L.; Chen, Z.; Huang, L.; et al. Discovery and Optimization of α-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia. J. Med. Chem. 2020, 63, 3370–3380. [Google Scholar] [CrossRef]
- Kwak, H.J.; Nam, J.Y.; Song, J.S.; No, Z.; Yang, S.D.; Cheon, H.G. Discovery of a novel orally active PDE-4 inhibitor effective in an ovalbumin-induced asthma murine model. Eur. J. Pharmacol. 2012, 685, 141–148. [Google Scholar] [CrossRef]
- Grammatika Pavlidou, N.; Dobrev, S.; Beneke, K.; Reinhardt, F.; Pecha, S.; Jacquet, E.; Abu-Taha, I.H.; Schmidt, C.; Voigt, N.; Kamler, M.; et al. Phosphodiesterase 8 governs cAMP/PKA-dependent reduction of L-type calcium current in human atrial fibrillation: A novel arrhythmogenic mechanism. Eur. Heart J. 2023, 44, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Matthiesen, K.; Nielsen, J. Cyclic AMP control measured in two compartments in HEK293 cells: Phosphodiesterase K(M) is more important than phosphodiesterase localization. PLoS ONE 2011, 6, e24392. [Google Scholar] [CrossRef]
- Cao, Y.; Lv, J.; Tan, Y.; Chen, R.; Jiang, X.; Meng, D.; Zou, K.; Pan, M.; Tang, L. Tribuloside acts on the PDE/cAMP/PKA pathway to enhance melanogenesis, melanocyte dendricity and melanosome transport. J. Ethnopharmacol. 2024, 323, 117673. [Google Scholar] [CrossRef]
- Salomon, Y.; Londos, C.; Rodbell, M. A highly sensitive adenylate cyclase assay. Anal. Biochem. 1974, 58, 541–548. [Google Scholar] [CrossRef]
- Hill, S.J.; Williams, C.; May, L.T. Insights into GPCR pharmacology from the measurement of changes in intracellular cyclic AMP; advantages and pitfalls of differing methodologies. Br. J. Pharmacol. 2010, 161, 1266–1275. [Google Scholar] [CrossRef]
- Cordell, R.L.; Hill, S.J.; Ortori, C.A.; Barrett, D.A. Quantitative profiling of nucleotides and related phosphate-containing metabolites in cultured mammalian cells by liquid chromatography tandem electrospray mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Comoglio, S.; Celada, F. An immuno-enzymatic assay of cortisol using E. coli beta-galactosidase as label. J. Immunol. Methods 1976, 10, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.S.; Woodward, E.E.; Hladik, M.L. Evaluation of ELISA for the analysis of imidacloprid in biological matrices: Cross-reactivities, matrix interferences, and comparison to LC-MS/MS. Chemosphere 2022, 286 Pt 3, 131746. [Google Scholar] [CrossRef]
- Russwurm, M.; Koesling, D. Measurement of cGMP-generating and -degrading activities and cGMP levels in cells and tissues: Focus on FRET-based cGMP indicators. Nitric Oxide 2018, 77, 44–52. [Google Scholar] [CrossRef]
- Jiang, L.I.; Collins, J.; Davis, R.; Lin, K.M.; DeCamp, D.; Roach, T.; Hsueh, R.; Rebres, R.A.; Ross, E.M.; Taussig, R.; et al. Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 2007, 282, 10576–10584. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jiang, T. Developments in FRET- and BRET-Based Biosensors. Micromachines 2022, 13, 1789. [Google Scholar] [CrossRef] [PubMed]
- Akuamoa, F.; Bovee, T.F.H.; van Dam, R.; Maro, L.; Wesseling, S.; Vervoort, J.; Rietjens, I.M.C.M.; Hoogenboom, R.L.A.P. Identification of phosphodiesterase type-5 (PDE-5) inhibitors in herbal supplements using a tiered approach and associated consumer risk. Food Addit. Contam. Part A 2022, 39, 1021–1032. [Google Scholar] [CrossRef]
- Akuamoa, F.; Hoogenboom, R.L.A.P.; Hamers, A.; Rietjens, I.M.C.M.; Bovee, T.F.H. PDE-5 inhibitors in selected herbal supplements from the Ghanaian market for better erectile function as tested by a bioassay. Toxicol. In Vitro 2021, 73, 105130. [Google Scholar] [CrossRef]
- Fan, F.; Binkowski, B.F.; Butler, B.L.; Stecha, P.F.; Lewis, M.K.; Wood, K.V. Novel genetically encoded biosensors using firefly luciferase. ACS Chem. Biol. 2008, 3, 346–351. [Google Scholar] [CrossRef] [PubMed]
- DiRaddo, J.O.; Miller, E.J.; Hathaway, H.A.; Grajkowska, E.; Wroblewska, B.; Wolfe, B.B.; Liotta, D.C.; Wroblewski, J.T. A real-time method for measuring cAMP production modulated by Gαi/o-coupled metabotropic glutamate receptors. J. Pharmacol. Exp. Ther. 2014, 349, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Shi, X.; Han, C.; Rao, C.; Wang, J. A rapid reporter assay for recombinant human brain natriuretic peptide (rhBNP) by GloSensor technology. J. Pharm. Anal. 2018, 8, 297–301. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.J.; Houslay, M.D. Action of rolipram on specific PDE4 cAMP phosphodiesterase isoforms and on the phosphorylation of cAMP-response-element-binding protein (CREB) and p38 mitogen-activated protein (MAP) kinase in U937 monocytic cells. Biochem. J. 2000, 347 Pt. 2, 571–578. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, D.; Yang, X.; Li, J.; Jiang, X.; Tian, G.; Terrett, N.K.; Jin, J.; Wu, H.; He, Q.; et al. The selectivity and potency of the new PDE5 inhibitor TPN729MA. J. Sex. Med. 2013, 10, 2790–2797. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Zeller, M.; Guan, K.; Wunder, F.; Wagner, M.; El-Armouche, A. PDE2 at the crossway between cAMP and cGMP signalling in the heart. Cell. Signal. 2017, 38, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Helal, C.J.; Arnold, E.; Boyden, T.; Chang, C.; Chappie, T.A.; Fisher, E.; Hajos, M.; Harms, J.F.; Hoffman, W.E.; Humphrey, J.M.; et al. Identification of a Potent, Highly Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor Clinical Candidate. J. Med. Chem. 2018, 61, 1001–1018. [Google Scholar] [CrossRef] [PubMed]
- Crosswhite, P.; Sun, Z. Inhibition of phosphodiesterase-1 attenuates cold-induced pulmonary hypertension. Hypertension 2013, 61, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.M.; Meyer, M.R.; Wink, C.S.; Zapp, J.; Maurer, H.H. Studies on the in vivo contribution of human cytochrome P450s to the hepatic metabolism of glaucine, a new drug of abuse. Biochem. Pharmacol. 2013, 86, 1497–1506. [Google Scholar] [CrossRef]
- Xin, Z.C.; Kim, E.K.; Lin, C.S.; Liu, W.J.; Tian, L.; Yuan, Y.M.; Fu, J. Effects of icariin on cGMP-specific PDE5 and cAMP-specific PDE4 activities. Asian J. Androl. 2003, 5, 15–18. [Google Scholar]
- Chen, P.; Chai, Y. Dynamics of cAMP/cGMP in patients under a stress state. Chin. J. Traumatol. 2002, 5, 115–117. [Google Scholar] [PubMed]
- Ding, Y.; Yu, B.; Zhou, S.; Ding, C.; Zhang, Z.; Xu, S.; Xu, Z. Improvement of solubility and pharmacokinetic profile of hepatoprotector icariin through complexation with HP-γ-cyclodextrin. Front. Pharmacol. 2023, 14, 1138686. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, L.; Han, Y.; Pan, L.; Yang, J.; Sun, M.; Zhou, P.; Sun, Y.; Bi, Y.; Qiu, H.J. Adaptation of African swine fever virus to HEK293T cells. Transbound. Emerg. Dis. 2021, 68, 2853–2866. [Google Scholar] [CrossRef] [PubMed]

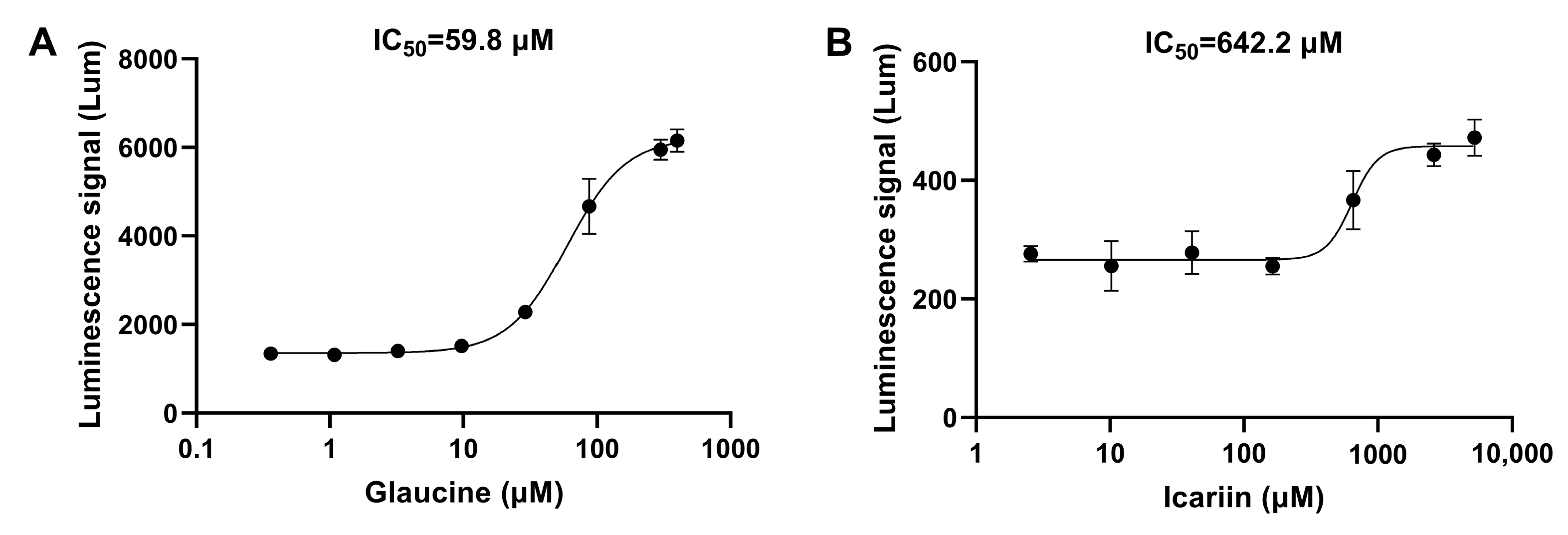
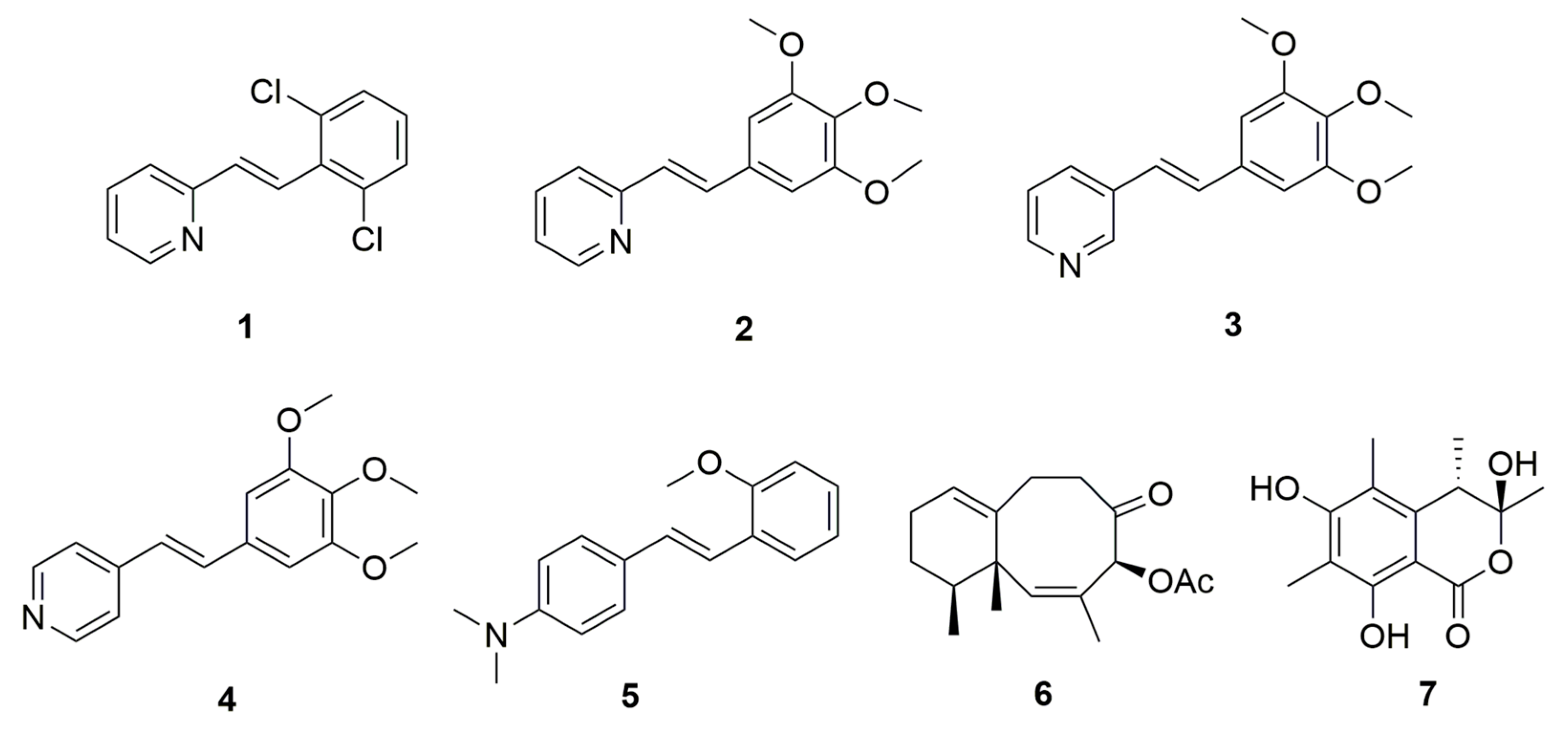

| Compound | Luminescence Signal (%) |
|---|---|
| Rolipram | 300 |
| 1 | 109 |
| 2 | 51 |
| 3 | 87 |
| 4 | 77 |
| 5 | 55 |
| 6 | 51 |
| 7 | 53 |
| Compound | PDE4B | PDE7A |
|---|---|---|
| Rolipram | 0.1 | - |
| BRL-50481 | - | 0.2 |
| 1 | 63.7 | 28.7 |
| 2 | 25.5 | 47.4 |
| 3 | 5.7 | 4.5 |
| 4 | 12.6 | 8.6 |
| 5 | 14.1 | 2.1 |
| 6 | - | 36.2 |
| 7 | 28.2 | 5.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Wang, Z.; Liu, X.; Sun, R.; Ma, S.; Ma, Z.; Wang, Q.; Li, G.; Zhang, H.-T. The Construction and Application of a New Screening Method for Phosphodiesterase Inhibitors. Biosensors 2024, 14, 252. https://doi.org/10.3390/bios14050252
Gao C, Wang Z, Liu X, Sun R, Ma S, Ma Z, Wang Q, Li G, Zhang H-T. The Construction and Application of a New Screening Method for Phosphodiesterase Inhibitors. Biosensors. 2024; 14(5):252. https://doi.org/10.3390/bios14050252
Chicago/Turabian StyleGao, Chunhua, Zhe Wang, Xiaojing Liu, Rongzhen Sun, Shengyao Ma, Zongchen Ma, Qi Wang, Guoqiang Li, and Han-Ting Zhang. 2024. "The Construction and Application of a New Screening Method for Phosphodiesterase Inhibitors" Biosensors 14, no. 5: 252. https://doi.org/10.3390/bios14050252
APA StyleGao, C., Wang, Z., Liu, X., Sun, R., Ma, S., Ma, Z., Wang, Q., Li, G., & Zhang, H.-T. (2024). The Construction and Application of a New Screening Method for Phosphodiesterase Inhibitors. Biosensors, 14(5), 252. https://doi.org/10.3390/bios14050252






