Digital PCR for Single-Cell Analysis
Abstract
1. Introduction
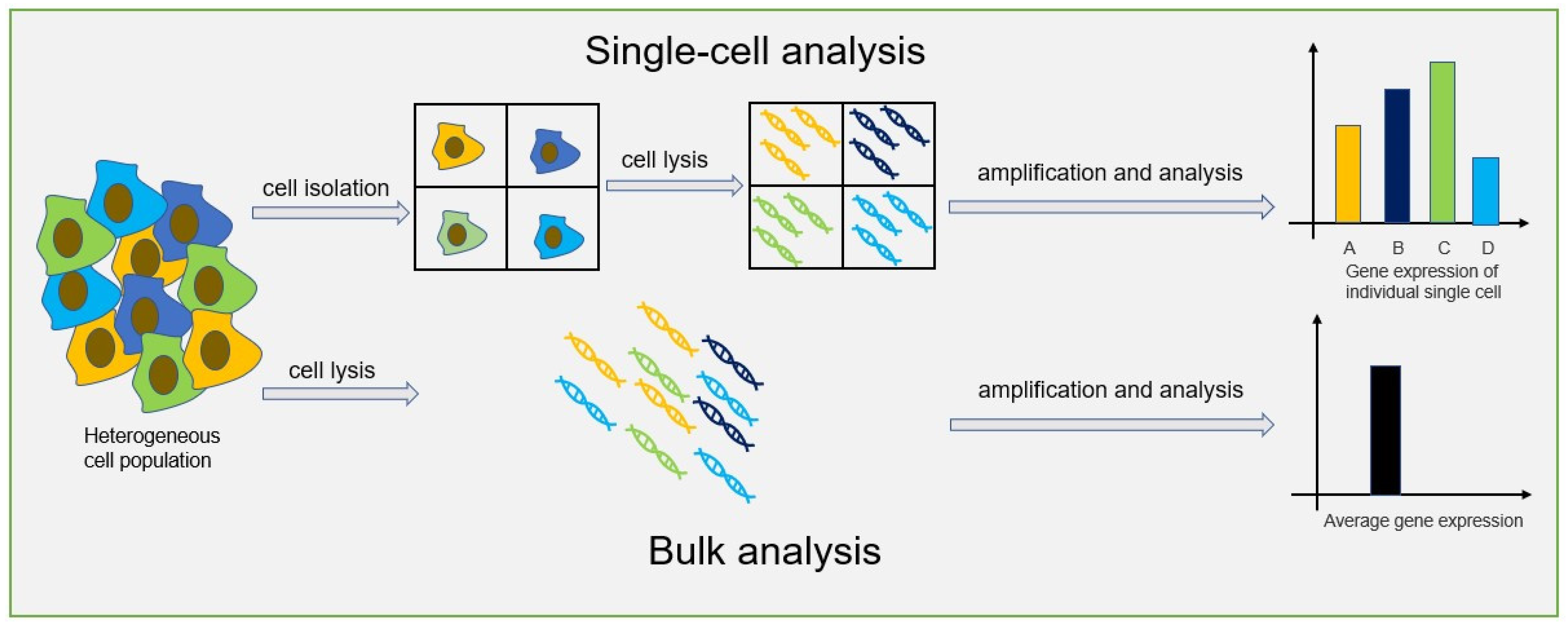
2. Single-Cell Analysis
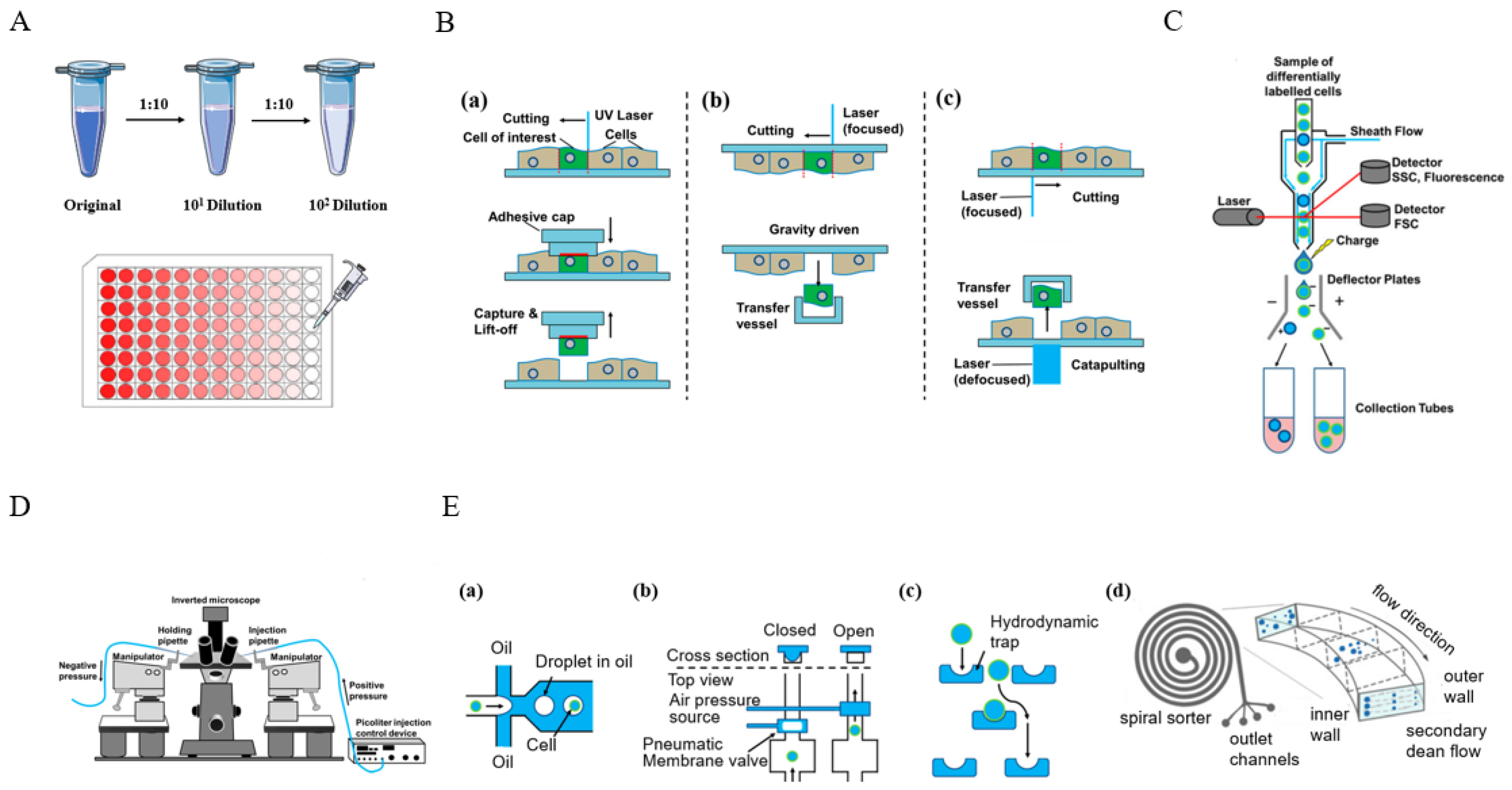
2.1. Single-Cell Isolation
2.2. Single-Cell Lysis
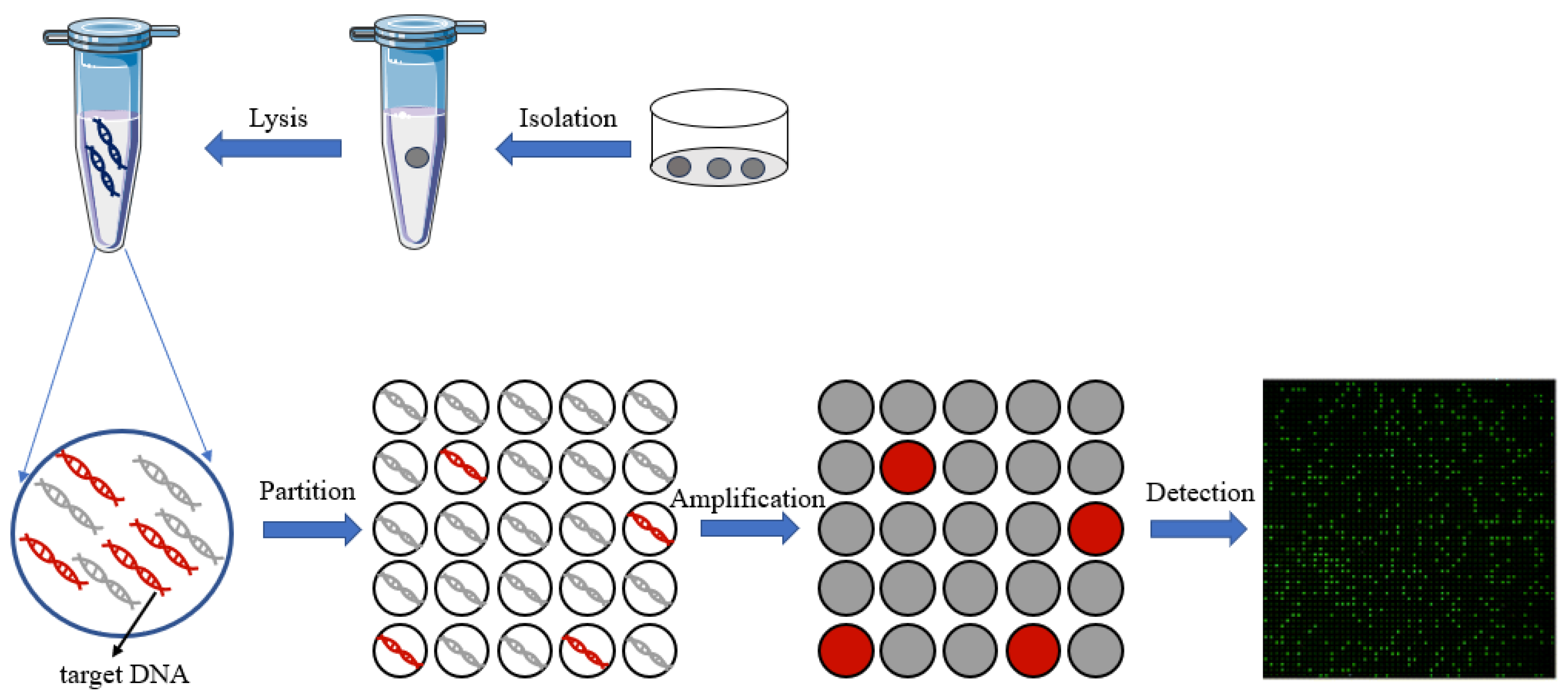
3. Digital PCR
4. Digital PCR for Single-Cell Analysis
4.1. Chamber-Based Digital PCR (cdPCR) for Single-Cell Analysis

4.2. Droplet-Based Digital PCR (ddPCR) for Single-Cell Analysis

5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodzic, E. Single-cell analysis: Advances and future perspectives. Bosn. J. Basic Med. Sci. 2016, 16, 313–314. [Google Scholar] [CrossRef] [PubMed]
- Pan, X. Single Cell Analysis: From Technology to Biology and Medicine. Single Cell Biol. 2015, 3, 106. [Google Scholar]
- Ren, X.; Zhang, L.; Zhang, Y.; Li, Z.; Siemers, N.; Zhang, Z. Insights Gained from Single-Cell Analysis of Immune Cells in the Tumor Microenvironment. Annu. Rev. Immunol. 2021, 39, 583–609. [Google Scholar] [CrossRef] [PubMed]
- Giladi, A.; Amit, I. Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell 2018, 172, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Dulken, B.W.; Buckley, M.T.; Navarro Negredo, P.; Saligrama, N.; Cayrol, R.; Leeman, D.S.; George, B.M.; Boutet, S.C.; Hebestreit, K.; Pluvinage, J.V.; et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 2019, 571, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Hao, S.; Zhang, S.; Zhu, C.; Cheng, H.; Yang, Z.; Hamey, F.K.; Wang, X.; Gao, A.; Wang, F.; et al. Differentiation of transplanted haematopoietic stem cells tracked by single-cell transcriptomic analysis. Nat. Cell Biol. 2020, 22, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; Vincent, M.; Pierredon, S.; Moutou, C.; Imbert-Bouteille, M.; Haquet, E.; Puechberty, J.; Willems, M.; Liautard-Haag, C.; Molinari, N.; et al. Single Circulating Fetal Trophoblastic Cells Eligible for Non Invasive Prenatal Diagnosis: The Exception Rather than the Rule. Sci. Rep. 2020, 10, 9861. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.; Liu, Z.; Qi, S.; Guo, X.; Wang, H.; Cheng, Y.; Tian, S.; Ma, M.; Peng, H.; et al. Single-cell Sequencing Reveals Clearance of Blastula Chromosomal Mosaicism in In Vitro Fertilization Babies. Genom. Proteom. Bioinform. 2022, 20, 1224–1231. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, L.; Liu, S.; Jiang, M.; Hu, B.; Zhou, K.; Guo, W.; Xu, Y.; Zhong, Y.; Zhou, X.; et al. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat. Commun. 2021, 12, 4091. [Google Scholar] [CrossRef]
- Liu, L.; Dong, X.; Tu, Y.; Miao, G.; Zhang, Z.; Zhang, L.; Wei, Z.; Yu, D.; Qiu, X. Methods and platforms for analysis of nucleic acids from single-cell based on microfluidics. Microfluid. Nanofluidics 2021, 25, 87. [Google Scholar] [CrossRef]
- Zhang, J.; Xue, J.; Luo, N.; Chen, F.; Chen, B.; Zhao, Y. Microwell array chip-based single-cell analysis. Lab. Chip. 2023, 23, 1066–1079. [Google Scholar] [CrossRef] [PubMed]
- Maioli, M.; Varadi, G.; Kurdi, R.; Caglioti, L.; Palyi, G. Limits of the Classical Concept of Concentration. J. Phys. Chem. B 2016, 120, 7438–7445. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A.; et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Hohnadel, M.; Maumy, M.; Chollet, R. Development of a micromanipulation method for single cell isolation of prokaryotes and its application in food safety. PLoS ONE 2018, 13, e198208. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Heiby, M.; Pierobon, M.; Liotta, L.A. Laser capture microdissection technology. Expert Rev. Mol. Diagn. 2007, 7, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Mao, S.; Khan, M.; Lin, J. Single-cell assay on microfluidic devices. Analyst 2019, 144, 808–823. [Google Scholar] [CrossRef]
- Pensold, D.; Zimmer-Bensch, G. Methods for Single-Cell Isolation and Preparation. Adv. Exp. Med. Biol. 2020, 1255, 7–27. [Google Scholar]
- Brunner, A.D.; Thielert, M.; Vasilopoulou, C.; Ammar, C.; Coscia, F.; Mund, A.; Hoerning, O.B.; Bache, N.; Apalategui, A.; Lubeck, M.; et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 2022, 18, e10798. [Google Scholar] [CrossRef]
- Gautam, V.; Chatterjee, S.; Sarkar, A.K. Single Cell Type Specific RNA Isolation and Gene Expression Analysis in Rice Using Laser Capture Microdissection (LCM)-Based Method. Methods Mol. Biol. 2021, 2238, 275–283. [Google Scholar]
- Xu, X.; Wang, J.; Wu, L.; Guo, J.; Song, Y.; Tian, T.; Wang, W.; Zhu, Z.; Yang, C. Microfluidic Single-Cell Omics Analysis. Small 2019, 16, 1903905. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, K.; Huang, P.; Liu, D.; Guan, Y. Single-Cell Isolation Microfluidic Chip Based on Thermal Bubble Micropump Technology. Sensors 2023, 23, 3623. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.J.; Hill, A.S.; Love, J.C. Nanowell-Based Immunoassays for Measuring Single-Cell Secretion: Characterization of Transport and Surface Binding. Anal. Chem. 2014, 86, 11562–11569. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Pan, T.; Li, B.; Chu, J. Label-free single-cell isolation enabled by microfluidic impact printing and real-time cellular recognition. Lab. Chip. 2021, 21, 3695–3706. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Wang, J.; Potanina, A.; Quake, S.R. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011, 29, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.K.; Weingeist, D.M.; Bhatia, S.N.; Engelward, B.P. Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl. Acad. Sci. USA 2010, 107, 10008–10013. [Google Scholar] [CrossRef]
- Li, Z.G.; Liu, A.Q.; Klaseboer, E.; Zhang, J.B.; Ohl, C.D. Single cell membrane poration by bubble-induced microjets in a microfluidic chip. Lab. Chip. 2013, 13, 1144–1150. [Google Scholar] [CrossRef]
- Kim, J.; Hong, J.W.; Kim, D.P.; Shin, J.H.; Park, I. Nanowire-integrated microfluidic devices for facile and reagent-free mechanical cell lysis. Lab. Chip. 2012, 12, 2914–2921. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab. Chip. 2020, 20, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Qiu, L.; Yu, B.; Xu, Y.; Gao, Y.; Pan, T.; Tian, Q.; Song, Q.; Jin, W.; Jin, Q.; et al. Digital PCR on an integrated self-priming compartmentalization chip. Lab. Chip. 2014, 14, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gao, Y.; Yu, B.; Ren, H.; Qiu, L.; Han, S.; Jin, W.; Jin, Q.; Mu, Y. Self-priming compartmentalization digital LAMP for point-of-care. Lab. Chip. 2012, 12, 4755–4763. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, S.; Balducci, S.; Guerrini, F.; Del Re, M.; Cacciola, R. Digital Droplet PCR in Hematologic Malignancies: A New Useful Molecular Tool. Diagnostics 2022, 12, 1305. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Sho, S.; Court, C.M.; Kim, S.; Braxton, D.R.; Hou, S.; Muthusamy, V.R.; Watson, R.R.; Sedarat, A.; Tseng, H.; Tomlinson, J.S. Digital PCR Improves Mutation Analysis in Pancreas Fine Needle Aspiration Biopsy Specimens. PLoS ONE 2017, 12, e170897. [Google Scholar] [CrossRef]
- Brik, A.; Weber, D.G.; Casjens, S.; Rozynek, P.; Meier, S.; Behrens, T.; Stamatis, G.; Darwiche, K.; Theegarten, D.; Brüning, T.; et al. Digital PCR for the Analysis of MYC Copy Number Variation in Lung Cancer. Dis. Markers 2020, 2020, 4176376. [Google Scholar] [CrossRef]
- Tiwari, A.; Ahmed, W.; Oikarinen, S.; Sherchan, S.P.; Heikinheimo, A.; Jiang, G.; Simpson, S.L.; Greaves, J.; Bivins, A. Application of digital PCR for public health-related water quality monitoring. Sci. Total Environ. 2022, 837, 155663. [Google Scholar] [CrossRef]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef]
- Niu, C.; Xu, Y.; Zhang, C.; Zhu, P.; Huang, K.; Luo, Y.; Xu, W. Ultrasensitive Single Fluorescence-Labeled Probe-Mediated Single Universal Primer–Multiplex–Droplet Digital Polymerase Chain Reaction for High-Throughput Genetically Modified Organism Screening. Anal. Chem. 2018, 90, 5586–5593. [Google Scholar] [CrossRef]
- Denis, J.A.; Guillerm, E.; Coulet, F.; Larsen, A.K.; Lacorte, J. The Role of BEAMing and Digital PCR for Multiplexed Analysis in Molecular Oncology in the Era of Next-Generation Sequencing. Mol. Diagn. Ther. 2017, 21, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Lu, S.; Chapman, A.R.; Xie, X.S. Genome-Wide Detection of Single-Nucleotide and Copy-Number Variations of a Single Human Cell. Science 2012, 338, 1622–1626. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Yin, J.; Zou, Z.; Yin, F.; Liang, H.; Hu, Z.; Fang, W.; Lv, S.; Zhang, T.; Wang, B.; Mu, Y. A Self-Priming Digital Polymerase Chain Reaction Chip for Multiplex Genetic Analysis. Acs Nano 2020, 14, 10385–10393. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, M.; He, F.; Song, J.; Song, Q.; Xia, D.; Liu, R.; Yao, H.; Han, J. The Viral Load of Human Cytomegalovirus Infection in Children following Hematopoietic Stem Cell Transplant by Chip Digital PCR. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 1–6. [Google Scholar] [CrossRef]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643. [Google Scholar] [CrossRef]
- White, A.K.; Heyries, K.A.; Doolin, C.; VanInsberghe, M.; Hansen, C.L. High-Throughput Microfluidic Single-Cell Digital Polymerase Chain Reaction. Anal. Chem. 2013, 85, 7182–7190. [Google Scholar] [CrossRef]
- Thompson, A.M.; Gansen, A.; Paguirigan, A.L.; Kreutz, J.E.; Radich, J.P.; Chiu, D.T. Self-Digitization Microfluidic Chip for Absolute Quantification of mRNA in Single Cells. Anal. Chem. 2014, 86, 12308–12314. [Google Scholar] [CrossRef]
- Yu, T.; Tang, C.; Zhang, Y.; Zhang, R.; Yan, W. Microfluidics-based digital quantitative PCR for single-cell small RNA quantification. Biol. Reprod 2017, 97, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Zaimy, M.A.; Saffarzadeh, N.; Mohammadi, A.; Pourghadamyari, H.; Izadi, P.; Sarli, A.; Moghaddam, L.K.; Paschepari, S.R.; Azizi, H.; Torkamandi, S.; et al. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017, 24, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Qiu, L.; Xu, Y.; Li, G.; Mu, Y. Single cell digital polymerase chain reaction on self-priming compartmentalization chip. Biomicrofluidics 2017, 11, 14109. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Xu, Y.; Qiu, L.; Ma, C.; Yu, B.; Song, Q.; Jin, W.; Jin, Q.; Liu, J.; Mu, Y. A scalable self-priming fractal branching microchannel net chip for digital PCR. Lab. Chip. 2017, 17, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, J.; Zhu, Q.; Song, Q.; Mu, Y. Co-detection of ALDH1A1, ABCG2, ALCAM and CD133 in three A549 subpopulations at the single cell level by one-step digital RT-PCR. Integr. Biol. 2018, 10, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Mau-Hsu, D.; Chen, K.; Wei, C.; Chiu, C.; Young, T. Evaluation of digital real-time PCR assay as a molecular diagnostic tool for single-cell analysis. Sci. Rep. 2018, 8, 3432. [Google Scholar] [CrossRef] [PubMed]
- Lloréns-Rico, V.; Simcock, J.A.; Huys, G.R.B.; Raes, J. Single-cell approaches in human microbiome research. Cell. 2022, 185, 2725–2738. [Google Scholar] [CrossRef]
- Liu, X.; Noll, L.; Shi, X.; Porter, E.; Wang, Y.; Stoy, C.; Lu, N.; Nagaraja, T.G.; Anderson, G.; Bai, J. Single-Cell-Based Digital PCR Detection and Association of Shiga Toxin-Producing Escherichia coli Serogroups and Major Virulence Genes. J. Clin. Microbiol. 2020, 58, e01684-19. [Google Scholar] [CrossRef]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- Maeda, R.; Kami, D.; Maeda, H.; Shikuma, A.; Gojo, S. High throughput single cell analysis of mitochondrial heteroplasmy in mitochondrial diseases. Sci. Rep. 2020, 10, 10821. [Google Scholar] [CrossRef]
- O’Hara, R.; Tedone, E.; Ludlow, A.; Huang, E.; Arosio, B.; Mari, D.; Shay, J.W. Quantitative mitochondrial DNA copy number determination using droplet digital PCR with single-cell resolution. Genome Res. 2019, 29, 1878–1888. [Google Scholar] [CrossRef] [PubMed]
- Burr, S.P.; Chinnery, P.F. Measuring Single-Cell Mitochondrial DNA Copy Number and Heteroplasmy using Digital Droplet Polymerase Chain Reaction. J. Vis. Exp. 2022, 185, e63870. [Google Scholar]
- Albayrak, C.; Jordi, C.A.; Zechner, C.; Lin, J.; Bichsel, C.A.; Khammash, M.; Tay, S. Digital Quantification of Proteins and mRNA in Single Mammalian Cells. Mol. Cell 2016, 61, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Jordi, C.; Son, M.; Van Phan, H.; Drayman, N.; Abasiyanik, M.F.; Vistain, L.; Tu, H.; Tay, S. Ultra-sensitive digital quantification of proteins and mRNA in single cells. Nat. Commun. 2019, 10, 3544. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tian, H.; Liu, C.; Yang, D.; Li, Z. A Clamp-Based One-Step Droplet Digital Reverse Transcription PCR (ddRT-PCR) for Precise Quantitation of Messenger RNA Mutation in Single Cells. ACS Sens. 2018, 3, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Sun, Y.; Liu, C.; Duan, X.; Tang, W.; Li, Z. Precise Quantitation of MicroRNA in a Single Cell with Droplet Digital PCR Based on Ligation Reaction. Anal. Chem. 2016, 88, 11384–11389. [Google Scholar] [CrossRef] [PubMed]
- McKelvey, B.A.; Umbricht, C.B.; Zeiger, M.A. Telomerase Reverse Transcriptase (TERT) Regulation in Thyroid Cancer: A Review. Front. Endocrinol. 2020, 11, 485. [Google Scholar] [CrossRef]
- Ludlow, A.T.; Robin, J.D.; Sayed, M.; Litterst, C.M.; Shelton, D.N.; Shay, J.W.; Wright, W.E. Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res 2014, 42, e104. [Google Scholar] [CrossRef]
- Ko, J.; Wang, Y.; Carlson, J.C.T.; Marquard, A.; Gungabeesoon, J.; Charest, A.; Weitz, D.; Pittet, M.J.; Weissleder, R. Single Extracellular Vesicle Protein Analysis Using Immuno-Droplet Digital Polymerase Chain Reaction Amplification. Adv. Biosyst. 2020, 4, 1900307. [Google Scholar] [CrossRef]
- Ma, J.; Tran, G.; Wan, A.M.D.; Young, E.W.K.; Kumacheva, E.; Iscove, N.N.; Zandstra, P.W. Microdroplet-based one-step RT-PCR for ultrahigh throughput single-cell multiplex gene expression analysis and rare cell detection. Sci. Rep. 2021, 11, 6777. [Google Scholar] [CrossRef]
| Method | Sample Type | Throughput | Cost | Isolation Efficiency | Automation | Reference |
|---|---|---|---|---|---|---|
| Limited serial dilution | Dissociated cell suspension | Low | Low | Low | No | [13] |
| FACS | Dissociated cell suspension | High | Very high | Low | Yes | [14] |
| Micromanipulation | Dissociated cell suspension | Low | Moderate | High | No | [15] |
| LCM | Tissue | Low | Moderate | High | No | [16] |
| Microfluidic | Dissociated cell suspension | Very high | High | Moderate | Yes | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, W.; Liu, X.; Maiga, M.; Cao, W.; Mu, Y.; Yan, Q.; Zhu, Q. Digital PCR for Single-Cell Analysis. Biosensors 2024, 14, 64. https://doi.org/10.3390/bios14020064
Fang W, Liu X, Maiga M, Cao W, Mu Y, Yan Q, Zhu Q. Digital PCR for Single-Cell Analysis. Biosensors. 2024; 14(2):64. https://doi.org/10.3390/bios14020064
Chicago/Turabian StyleFang, Weibo, Xudong Liu, Mariam Maiga, Wenjian Cao, Ying Mu, Qiang Yan, and Qiangyuan Zhu. 2024. "Digital PCR for Single-Cell Analysis" Biosensors 14, no. 2: 64. https://doi.org/10.3390/bios14020064
APA StyleFang, W., Liu, X., Maiga, M., Cao, W., Mu, Y., Yan, Q., & Zhu, Q. (2024). Digital PCR for Single-Cell Analysis. Biosensors, 14(2), 64. https://doi.org/10.3390/bios14020064






