Exploring the Interaction of Biotinylated FcGamma RI and IgG1 Monoclonal Antibodies on Streptavidin-Coated Plasmonic Sensor Chips for Label-Free VEGF Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. FcγRI Immobilization Procedure
2.3. Antibody Binding Assays
2.4. Sensor Surface Regeneration
2.5. Antibody/Antigen Interaction Assays
2.5.1. Antigen-Binding Assays
2.5.2. Antigen Concentration Analyses
2.6. Specificity Assays
3. Results
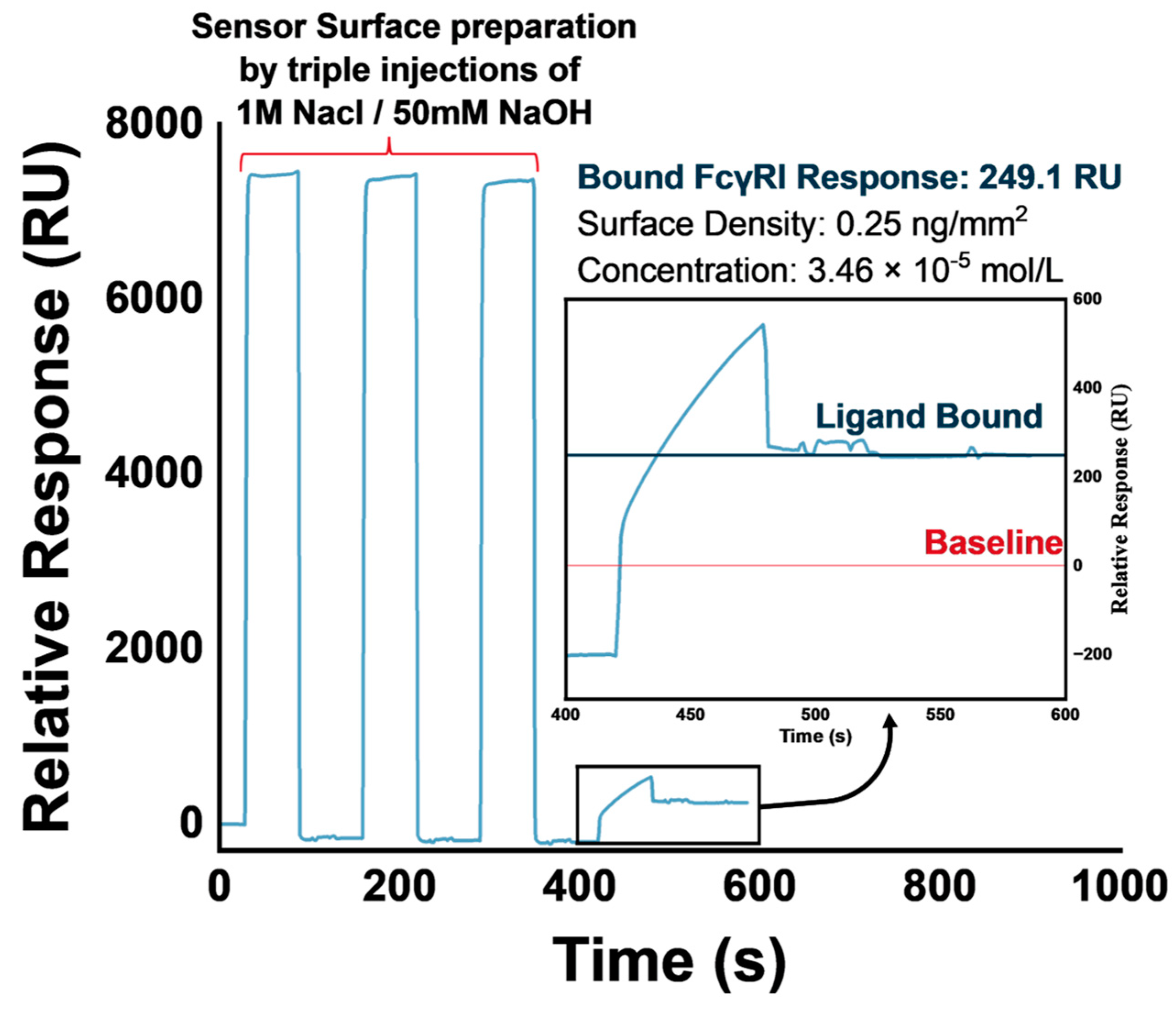
3.1. FcγRI Immobilization
3.2. Regeneration
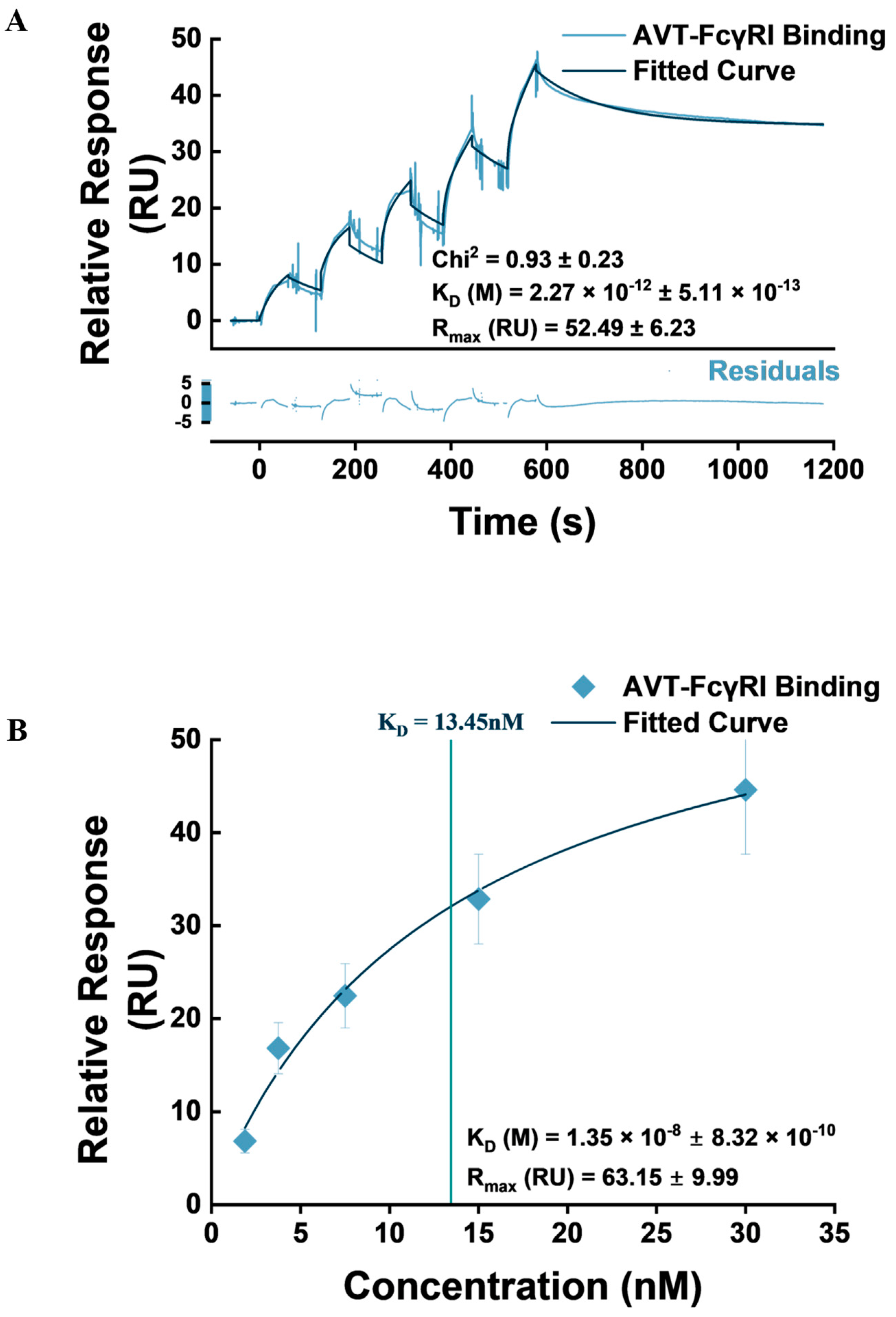
3.3. IgG1-Type Monoclonal Antibody Capture Studies
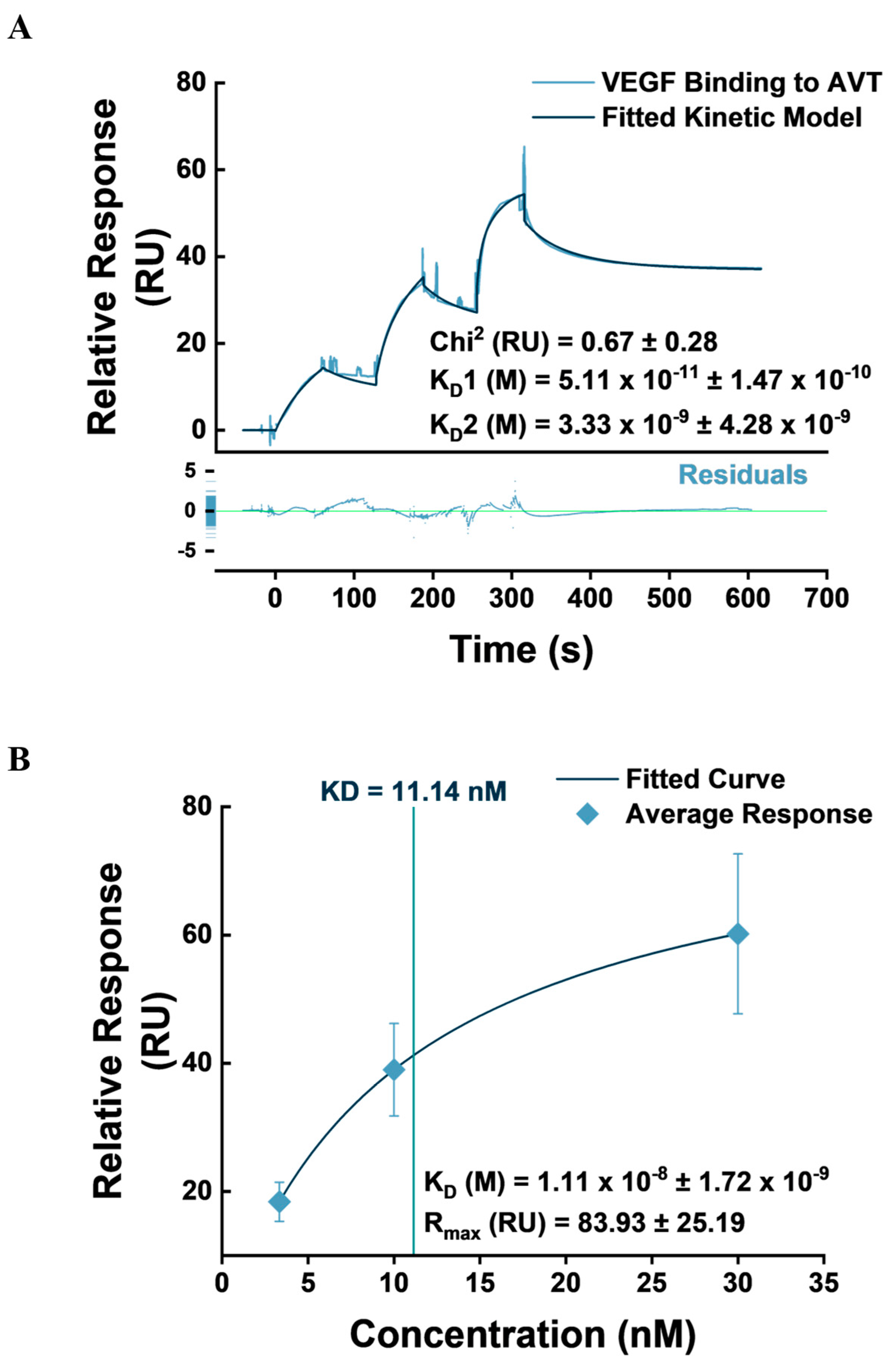
3.4. VEGF Binding Kinetics to IgG1-FcγRI Complex
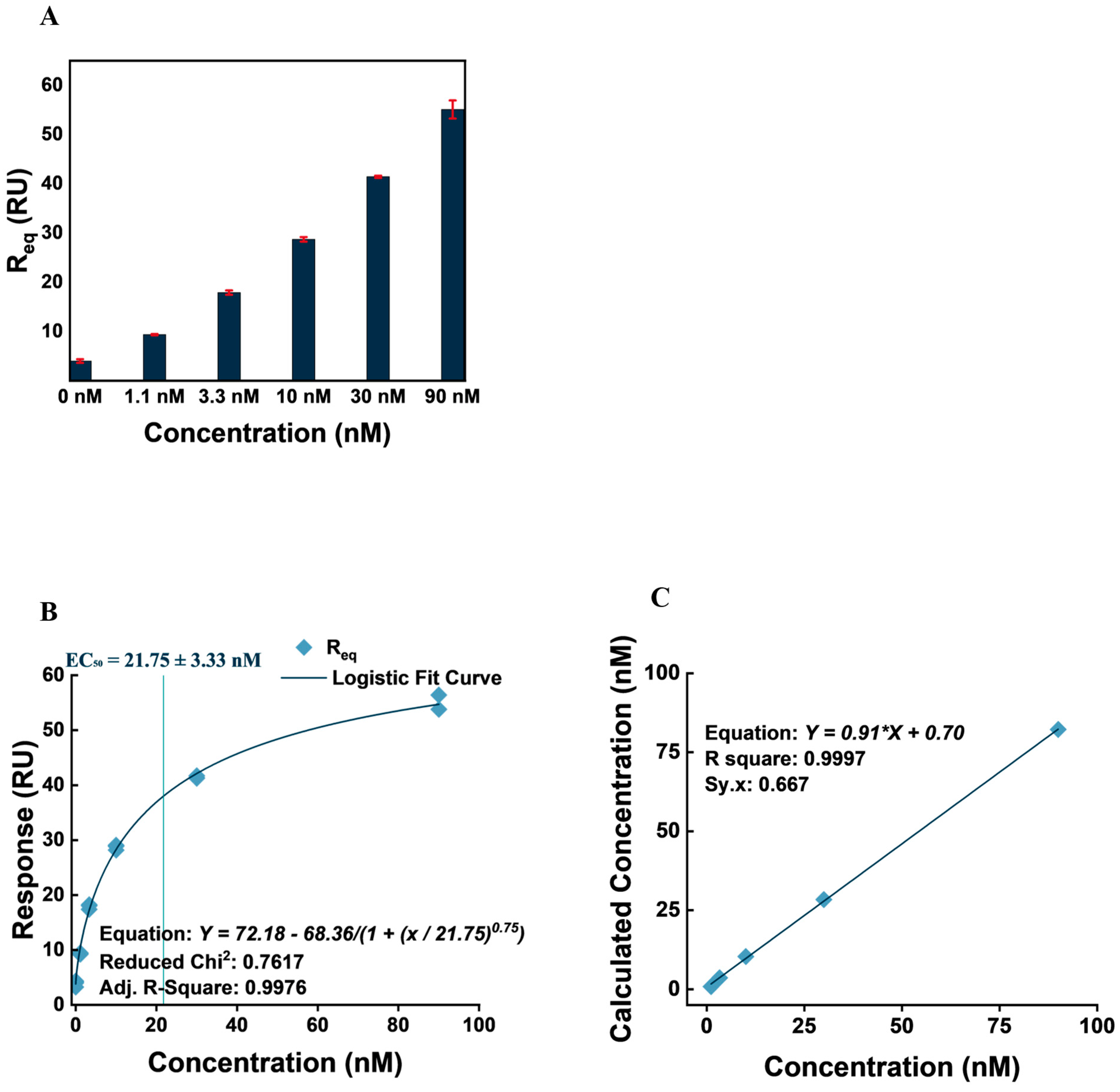
3.5. VEGF Detection and Quantification
- LOB = 59.1 pM (2.26 µg⋅mL−1);
- LOD = 129.9 pM (4.96 µg⋅mL−1);
- LOQ = 534.6 pM (20.42 µg⋅mL−1).
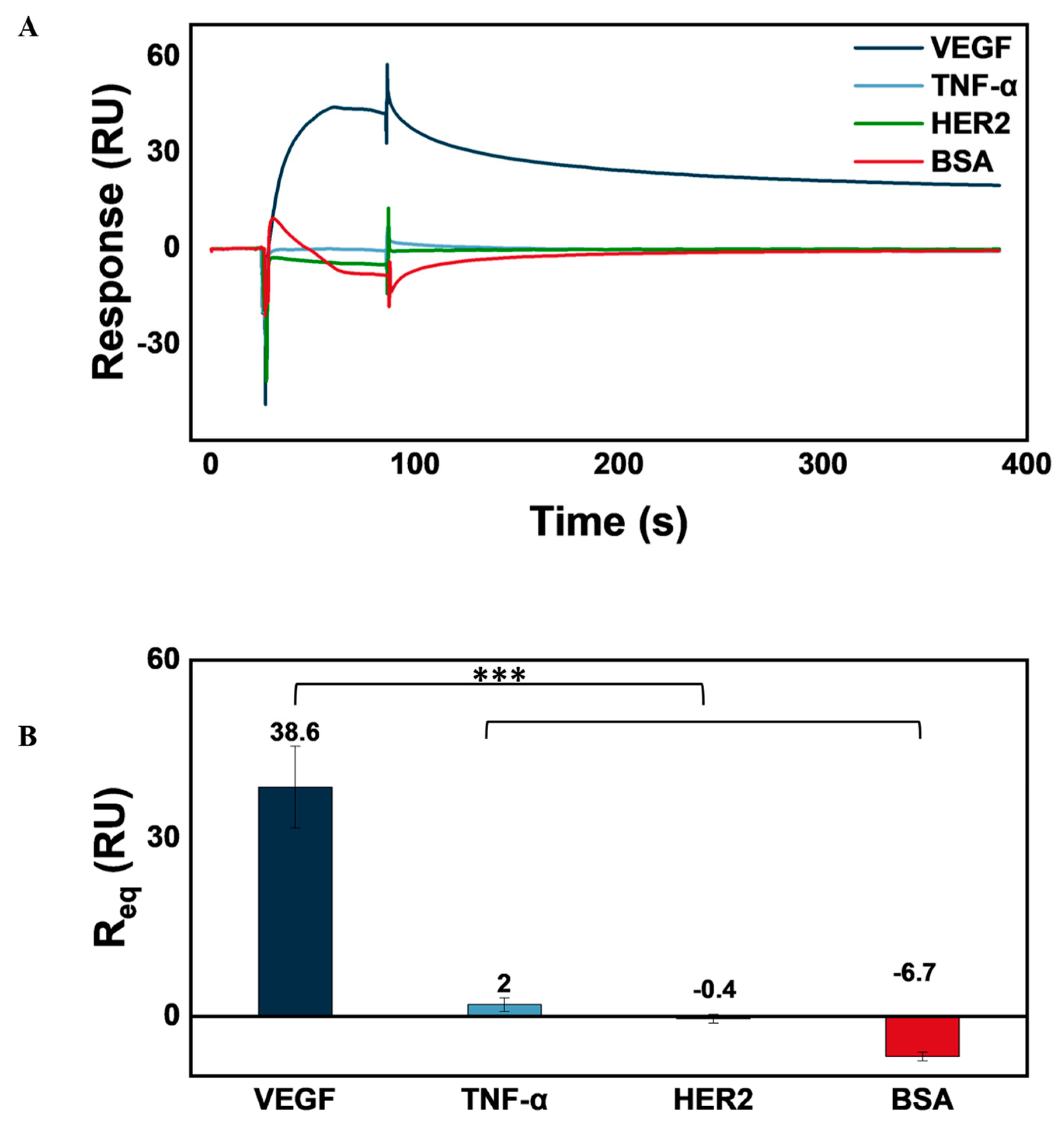
3.6. AVT Specificity for VEGF
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, H.; Ma, T.; Li, D.; He, M.; Wang, H.; Cui, Y. Circulating Vascular Endothelial Growth Factor and Cancer Risk: A Bidirectional Mendelian Randomization. Front. Genet. 2022, 13, 981032. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Hillan, K.J.; Gerber, H.-P.; Novotny, W. Discovery and Development of Bevacizumab, an Anti-VEGF Antibody for Treating Cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef]
- Ye, W. The Complexity of Translating Anti-Angiogenesis Therapy from Basic Science to the Clinic. Dev. Cell 2016, 37, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Nourizad, A.; Golmohammadi, S.; Aghanejad, A.; Tohidkia, M.R. Recent Trends in Aptamer-Based Nanobiosensors for Detection of Vascular Endothelial Growth Factors (VEGFs) Biomarker: A Review. Environ. Res. 2023, 236, 116726. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wang, X.; Zeng, T.; Xing, S.; Dai, S.; Wang, J.; Chen, S.; Li, X.; Xie, Y.; Zhu, Y.; et al. Serum VEGF Levels in the Early Diagnosis and Severity Assessment of Non-Small Cell Lung Cancer. J. Cancer 2018, 9, 1538. [Google Scholar] [CrossRef]
- Wang, R.; Chen, S.; Huang, L.; Zhou, Y.; Shao, Z. Monitoring Serum VEGF in Neoadjuvant Chemotherapy for Patients with Triple-Negative Breast Cancer: A New Strategy for Early Prediction of Treatment Response and Patient Survival. Oncologist 2019, 24, 753–761. [Google Scholar] [CrossRef]
- Maryam, N.; Ahmed, S.S.; Alam, R.; Hanif, M.U.; Saleem, M.; Gul, R. Role of Serum VEGF-A Biomarker for Early Diagnosis of Ovarian Cancer Instead of CA-125. J. Pak. Med. Assoc. 2021, 71, 2192–2197. [Google Scholar] [PubMed]
- Shatunova, E.A.; Korolev, M.A.; Omelchenko, V.O.; Kurochkina, Y.D.; Davydova, A.S.; Venyaminova, A.G.; Vorobyeva, M.A. Aptamers for Proteins Associated with Rheumatic Diseases: Progress, Challenges, and Prospects of Diagnostic and Therapeutic Applications. Biomedicines 2020, 8, 527. [Google Scholar] [CrossRef]
- Borrelli, E.; Grosso, D.; Barresi, C.; Lari, G.; Sacconi, R.; Senni, C.; Querques, L.; Bandello, F.; Querques, G. Long-Term Visual Outcomes and Morphologic Biomarkers of Vision Loss in Eyes with Diabetic Macular Edema Treated with Anti-VEGF Therapy. Am. J. Ophthalmol. 2022, 235, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.; Sharma, K.; Agyeah, G.; Krüger, R.; Grünewald, A.; Fitzgerald, J.C. Neurodegeneration and Neuroinflammation in Parkinson’s Disease: A Self-Sustained Loop. Curr. Neurol. Neurosci. Rep. 2022, 22, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Chattaraj, R.; Mohan, P.; Livingston, C.M.; Besmer, J.D.; Kumar, K.; Goodwin, A.P. Mutually-Reactive, Fluorogenic Hydrocyanine/Quinone Reporter Pairs for In-Solution Biosensing via Nanodroplet Association. ACS Appl. Mater. Interfaces 2016, 8, 802–808. [Google Scholar] [CrossRef]
- Lan, J.; Li, L.; Liu, Y.; Yan, L.; Li, C.; Chen, J.; Chen, X. Upconversion Luminescence Assay for the Detection of the Vascular Endothelial Growth Factor, a Biomarker for Breast Cancer. Microchim. Acta 2016, 183, 3201–3208. [Google Scholar] [CrossRef]
- Zhao, S.; Yang, W.; Lai, R.Y. A Folding-Based Electrochemical Aptasensor for Detection of Vascular Endothelial Growth Factor in Human Whole Blood. Biosens. Bioelectron. 2011, 26, 2442–2447. [Google Scholar] [CrossRef]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-Invasive Cancer Diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Spasic, D.; Delport, F.; Van Stappen, T.; Detrez, I.; Daems, D.; Vermeire, S.; Gils, A.; Lammertyn, J. Immunoassay for Detection of Infliximab in Whole Blood Using a Fiber-Optic Surface Plasmon Resonance Biosensor. Anal. Chem. 2017, 89, 3664–3671. [Google Scholar] [CrossRef]
- Vashist, S.K.; Schneider, E.M.; Luong, J.H.T. Surface Plasmon Resonance-Based Immunoassay for Human C-Reactive Protein. Analyst 2015, 140, 4445–4452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kukkar, D.; Hashemi, B.; Kim, K.; Deep, A. Advanced Functional Structure-Based Sensing and Imaging Strategies for Cancer Detection: Possibilities, Opportunities, Challenges, and Prospects. Adv. Funct. Mater. 2019, 29, 1807859. [Google Scholar] [CrossRef]
- Patil, P.O.; Pandey, G.R.; Patil, A.G.; Borse, V.B.; Deshmukh, P.K.; Patil, D.R.; Tade, R.S.; Nangare, S.N.; Khan, Z.G.; Patil, A.M.; et al. Graphene-Based Nanocomposites for Sensitivity Enhancement of Surface Plasmon Resonance Sensor for Biological and Chemical Sensing: A Review. Biosens. Bioelectron. 2019, 139, 111324. [Google Scholar] [CrossRef]
- Kurt, H.; Pishva, P.; Pehlivan, Z.S.; Arsoy, E.G.; Saleem, Q.; Bayazıt, M.K.; Yüce, M. Nanoplasmonic Biosensors: Theory, Structure, Design, and Review of Recent Applications. Anal. Chim. Acta 2021, 1185, 338842. [Google Scholar] [CrossRef]
- Karlsson, R.; Pol, E.; Frostell, Å. Comparison of Surface Plasmon Resonance Binding Curves for Characterization of Protein Interactions and Analysis of Screening Data. Anal. Biochem. 2016, 502, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Anantha, P.; Raj, P.; Zheng, P.; Tanwar, S.; Barman, I. Gold Nanoprism Enhanced SERS Aptasensor for Simultaneous Detection of Thrombin and VEGF. Sens. Actuators B Chem. 2025, 423, 136811. [Google Scholar] [CrossRef]
- Khan, I.M.; Niazi, S.; Mohsin, A.; Zhou, Y. Soft Scaffold Aided Plasmon-Enhanced Upconversion Luminescence and Its Application in Vascular Endothelial Growth Factor (VEGF) Detection. Sens. Actuators B Chem. 2024, 410, 135657. [Google Scholar] [CrossRef]
- Stuart, D.D.; Van Zant, W.; Valiulis, S.; Malinick, A.S.; Hanson, V.; Cheng, Q. Trends in Surface Plasmon Resonance Biosensing: Materials, Methods, and Machine Learning. Anal. Bioanal. Chem. 2024, 416, 5221–5232. [Google Scholar] [CrossRef]
- Kumar, V.R.; Kampan, N.C.; Abd Aziz, N.H.; Teik, C.K.; Shafiee, M.N.; Menon, P.S. Recent Advances in Surface Plasmon Resonance (SPR) Technology for Detecting Ovarian Cancer Biomarkers. Cancers 2023, 15, 5607. [Google Scholar] [CrossRef]
- Das, S.; Devireddy, R.; Gartia, M.R. Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection. Biosensors 2023, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Lopez Baltazar, J.M.; Gu, W.; Bocková, M.; Yu, Q. Immunoassays for Extracellular Vesicle Detection via Transmembrane Proteins Using Surface Plasmon Resonance Biosensors. ACS Sens. 2024, 9, 3594–3603. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Li, M.C.; Chen, K.R.; Cheng, Y.J.; Wu, X.Y.; Chen, S.A.; Youh, M.J.; Kuo, C.C.; Lin, Y.X.; Lin, C.Y.; et al. Facile and Unplugged Surface Plasmon Resonance Biosensor with NIR-Emitting Perovskite Nanocomposites for Fast Detection of SARS-CoV-2. Anal. Chem. 2023, 95, 7186–7194. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Yim, B.; Park, J.; Kim, N.G.; Kim, B.S.; Park, Y.; Yoon, Y.K.; Kim, J. Method for the Rapid Detection of SARS-CoV-2-Neutralizing Antibodies Using a Nanogel-Based Surface Plasmon Resonance Biosensor. ACS Appl. Polym. Mater. 2023, 5, 2195–2202. [Google Scholar] [CrossRef]
- Singh, A.K.; Anwar, M.; Pradhan, R.; Ashar, M.S.; Rai, N.; Dey, S. Surface Plasmon Resonance Based-Optical Biosensor: Emerging Diagnostic Tool for Early Detection of Diseases. J. Biophotonics 2023, 16, e202200380. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ross, G.; Nielen, M.W.F.; Eriksson, J.; Salentijn, G.I.; Mak, W.C. A Portable Smartphone-Based Imaging Surface Plasmon Resonance Biosensor for Allergen Detection in Plant-Based Milks. Talanta 2023, 257, 124366. [Google Scholar] [CrossRef]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The Role of IgG Fc Receptors in Antibody-Dependent Enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Gürel, B.; Çapkın, E.; Parlar, A.; Özkan, A.; Çorbacıoğlu, M.; Dağlikoca, D.E.; Yüce, M. Optimized Methods for Analytical and Functional Comparison of Biosimilar Mab Drugs: A Case Study for Avastin, Mvasi, and Zirabev. Sci. Pharm. 2022, 90, 36. [Google Scholar] [CrossRef]
- Forest-Nault, C.; Gaudreault, J.; Henry, O.; Durocher, Y.; De Crescenzo, G. On the Use of Surface Plasmon Resonance Biosensing to Understand IgG-FcγR Interactions. Int. J. Mol. Sci. 2021, 22, 6616. [Google Scholar] [CrossRef] [PubMed]
- Kutlu, A.; Çapkın, E.; Adacan, K.; Yüce, M. Fc–FcγRI Complexes: Molecular Dynamics Simulations Shed Light on Ectodomain D3′s Potential Role in IgG Binding. ACS Omega 2024, 9, 49272–49282. [Google Scholar] [CrossRef]
- Dorion-Thibaudeau, J.; Durocher, Y.; De Crescenzo, G. Quantification and Simultaneous Evaluation of the Bioactivity of Antibody Produced in CHO Cell Culture—The Use of the Ectodomain of FcγRI and Surface Plasmon Resonance-Based Biosensor. Mol. Immunol. 2017, 82, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Capkin, E.; Kurt, H.; Gurel, B.; Bicak, D.; Akgun Bas, S.; Daglikoca, D.E.; Yuce, M. Characterization of FcγRIa (CD64) as a Ligand Molecule for Site-Specific IgG1 Capture: A Side-By-Side Comparison with Protein A. Langmuir 2022, 38, 14623–14634. [Google Scholar] [CrossRef]
- Sadeghalvad, M.; Rezaei, N. Introduction on Monoclonal Antibodies. In Monoclonal Antibodies; Rezaei, N., Ed.; IntechOpen: London, UK, 2021; ISBN 978-1-83968-369-5. [Google Scholar]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Garcia, J.; Hurwitz, H.I.; Sandler, A.B.; Miles, D.; Coleman, R.L.; Deurloo, R.; Chinot, O.L. Bevacizumab (Avastin®) in Cancer Treatment: A Review of 15 Years of Clinical Experience and Future Outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar] [CrossRef]
- Cytiva Series S Sensor Chip, SA. Available online: https://cdn.cytivalifesciences.com/api/public/content/digi-16861-original (accessed on 15 November 2024).
- Suárez, I.; Salmerón-García, A.; Cabeza, J.; Capitán-Vallvey, L.F.; Navas, N. Development and Use of Specific ELISA Methods for Quantifying the Biological Activity of Bevacizumab, Cetuximab and Trastuzumab in Stability Studies. J. Chromatogr. B 2016, 1032, 155–164. [Google Scholar] [CrossRef]
- Sensor Surface Handbook; Biacore: Uppsala, Sweden, 2003.
- Hanson, E.K.; Whelan, R.J. Combining the Benefits of Biotin–Streptavidin Aptamer Immobilization with the Versatility of Ni-NTA Regeneration Strategies for SPR. Sensors 2024, 24, 2805. [Google Scholar] [CrossRef]
- Gunnarsson, A.; Stubbs, C.J.; Rawlins, P.B.; Taylor-Newman, E.; Lee, W.C.; Geschwindner, S.; Hytönen, V.; Holdgate, G.; Jha, R.; Dahl, G. Regenerable Biosensors for Small-Molecule Kinetic Characterization Using SPR. SLAS Discov. 2021, 26, 730–739. [Google Scholar] [CrossRef]
- Dorion-Thibaudeau, J.; Raymond, C.; Lattová, E.; Perreault, H.; Durocher, Y.; De Crescenzo, G. Towards the Development of a Surface Plasmon Resonance Assay to Evaluate the Glycosylation Pattern of Monoclonal Antibodies Using the Extracellular Domains of CD16a and CD64. J. Immunol. Methods 2014, 408, 24–34. [Google Scholar] [CrossRef]
- Anderson, K.W.; Bergonzo, C.; Scott, K.; Karageorgos, I.L.; Gallagher, E.S.; Tayi, V.S.; Butler, M.; Hudgens, J.W. HDX-MS and MD Simulations Provide Evidence for Stabilization of the IgG1—FcγRIa (CD64a) Immune Complex Through Intermolecular Glycoprotein Bonds. J. Mol. Biol. 2022, 434, 167391. [Google Scholar] [CrossRef]
- Hearty, S.; Leonard, P.; Ma, H.; O’Kennedy, R. Measuring Antibody-Antigen Binding Kinetics Using Surface Plasmon Resonance. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1827, pp. 421–455. ISBN 978-1-4939-8648-4. [Google Scholar]
- Arney, J.W.; Weeks, K.M. RNA-Ligand Interactions Quantified by Surface Plasmon Resonance with Reference Subtraction. Biochemistry 2022, 61, 1625–1632. [Google Scholar] [CrossRef]
- Cytiva Kinetics and Affinity Measurements with Biacore Systems. Available online: https://cdn.cytivalifesciences.com/api/public/content/digi-33041-pdf (accessed on 6 November 2024).
- Çapkın, E.; Kutlu, A.; Yüce, M. Repurposing Fc Gamma Receptor I (FcγRI, CD64) for Site-Oriented Monoclonal Antibody Capture: A Proof-of-Concept Study for Real-Time Detection of Tumor Necrosis Factor-Alpha (TNF-α). Heliyon 2023, 9, e19469. [Google Scholar] [CrossRef] [PubMed]
- Crowley, A.R.; Ackerman, M.E. Mind the Gap: How Interspecies Variability in IgG and Its Receptors May Complicate Comparisons of Human and Non-Human Primate Effector Function. Front. Immunol. 2019, 10, 434608. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Q. Antigen Improves Binding of IgGs to FcγRs in SPR Analysis. Anal. Biochem. 2022, 640, 114411. [Google Scholar] [CrossRef]
- Wang, Y.; Fei, D.; Vanderlaan, M.; Song, A. Biological Activity of Bevacizumab, a Humanized Anti-VEGF Antibody in Vitro. Angiogenesis 2004, 7, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Deng, C.; Wang, G.; Mei, X.; Xie, J.; Liu, Y.; Liu, Y.; Yang, Y.; Li, S.; Liu, C. Physicochemical and Functional Similarity Assessment Between Proposed Bevacizumab Biosimilar BAT1706 and Reference Bevacizumab. Drugs R D 2023, 23, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, X.; Fuh, G.; Yu, L.; Wakshull, E.; Khosraviani, M.; Day, E.S.; Demeule, B.; Liu, J.; Shire, S.J.; et al. Comparison of Binding Characteristics and in Vitro Activities of Three Inhibitors of Vascular Endothelial Growth Factor A. Mol. Pharm. 2014, 11, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.; Martin, J.; Ruan, Q.; Rafique, A.; Rosconi, M.P.; Shi, E.; Pyles, E.A.; Yancopoulos, G.D.; Stahl, N.; Wiegand, S.J. Binding and Neutralization of Vascular Endothelial Growth Factor (VEGF) and Related Ligands by VEGF Trap, Ranibizumab and Bevacizumab. Angiogenesis 2012, 15, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Reusch, J.; Andersen, J.T.; Rant, U.; Schlothauer, T. Insight into the Avidity–Affinity Relationship of the Bivalent, PH-Dependent Interaction between IgG and FcRn. mAbs 2024, 16, 2361585. [Google Scholar] [CrossRef] [PubMed]
- Dagogo, J.; Orumie, U.C.; BIU, E.O. Mathematical Modeling Properties of Weibull, Logistic Gompertz, Hills and Richards Models with Four Parameters. Int. J. Appl. Sci. Math. 2023, 10, 2394–2894. [Google Scholar]
- Moulahoum, H.; Ghorbanizamani, F. The LOD Paradox: When Lower Isn’t Always Better in Biosensor Research and Development. Biosens. Bioelectron. 2024, 264, 116670. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, S.; Nosrati, R.; Yousefi, M.; Nezami, A.; Soltani, F.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Aptamer-Based Biosensors and Nanosensors for the Detection of Vascular Endothelial Growth Factor (VEGF): A Review. Biosens. Bioelectron. 2018, 110, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Oldak, L.; Leśniewska, A.; Zelazowska-Rutkowska, B.; Latoch, E.; Lukaszewski, Z.; Krawczuk-Rybak, M.; Gorodkiewicz, E. An Array SPRi Biosensor for Simultaneous VEGF-A and FGF-2 Determination in Biological Samples. Appl. Sci. 2022, 12, 12699. [Google Scholar] [CrossRef]
- Ni, S.; Shen, Z.; Zhang, P.; Liu, G. Enhanced Performance of an Electrochemical Aptasensor for Real-Time Detection of Vascular Endothelial Growth Factor (VEGF) by Nanofabrication and Ratiometric Measurement. Anal. Chim. Acta 2020, 1121, 74–82. [Google Scholar] [CrossRef]
- Tabasi, O.; Falamaki, C. Recent Advancements in the Methodologies Applied for the Sensitivity Enhancement of Surface Plasmon Resonance Sensors. Anal. Methods 2018, 10, 3906–3925. [Google Scholar] [CrossRef]
- Biacore Concentration Analysis Handbook; Cytiva: Marlborough, MA, USA, 2001.




| Binding Models | Chi2 | Visual Inspection Comments |
|---|---|---|
| 1:1 Langmuir Binding | 2.82 ± 0.64 | Poor fit with significant systemic deviations in the dissociation phase. Residuals’ distribution spans −30 to +10 RU. |
| Heterogenous Ligand | 0.93 ± 0.23 | Relatively good fit with significant overlap across association and dissociation phases. Residuals’ distribution is non-systemic and spans −5 to +5 RU. |
| Bivalent Analyte | 3.34 ± 0.91 | Poor fit with significant deviations, especially in the dissociation phase. Residuals’ distribution is non-systemic but spans −30 to +10 RU. |
| Two-State Reaction | 2.78 ± 0.34 | It’s a relatively poor fit. There are significant deviations in the dissociation phase at lower concentrations. Residuals’ distribution is systemic, spanning −30 to +10 RU |
| S/N | Chi2 RU | Set 1: Rmax = 60.21 − 44.96 | Set 2: Rmax = 11.12 − 8.504 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ka M−1s−1 E+5 | kd s−1 E-7 | KD pM | Rmax RU | ka M−1s−1 E+5 | kd s−1 E-3 | KD nM | Rmax RU | ||
| 1 | 1.25 | 3.33 | 8.90 | 2.67 | 60.21 | 97 | 5.9 | 0.60 | 11.12 |
| 2 | 0.81 | 2.76 | 4.27 | 1.55 | 52.29 | 96 | 6.8 | 0.71 | 10.42 |
| 3 | 0.74 | 3.35 | 8.67 | 2.59 | 44.96 | 159 | 11 | 0.72 | 8.504 |
| Mean | 0.93 | 3.15 | 7.28 | 2.27 | 52.49 | 117 | 8.03 | 0.68 | 10.01 |
| SD | 0.23 | 0.27 | 2.13 | 0.51 | 6.23 | 29.3 | 2.4 | 0.05 | 1.11 |
| Flow Channel | AVT Capture Level (RU) | Binding Profile 1 | Binding Profile 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ka M−1s−1 E+5 | kd s−1 E-7 | KD pM | Rmax RU | ka M−1s−1 E+5 | kd s−1 E-3 | KD nM | Rmax RU | ||
| Fc 2-1 | 1888.17 ± 2.15 | 11.3 ± 0.45 | 20 ± 9.36 | 1.79 ± 0.87 | 30.67 ± 2.31 | 73.8 ± 5.62 | 15.5 ± 0.82 | 2.12 ± 0.27 | 16.23 ± 0.23 |
| Fc 3-1 | 986.37 ± 3.65 | 13 ± 0.61 | 12.2 ± 4.22 | 0.96 ± 0.38 | 64.81 ± 5.25 | 108 ± 50.8 | 14.6 ± 12.5 | 1.12 ± 0.98 | 5.78 ± 0.97 |
| Fc 4-1 | 3020.87 ± 5.15 | 12.3 ± 0.19 | 2140 ± 3610 | 151 ± 252 | 19.55 ± 3.57 | 131 ± 156 | 43.6 ± 24.9 | 6.76 ± 6.71 | 15.78 ± 1.9 |
| Mean | 12.2 | 724 | 51.1 | 38.35 | 104 | 24.6 | 3.33 | 12.6 | |
| SD | 1.25 | 209 | 147 | 20.7 | 85.7 | 19.9 | 4.28 | 5.23 | |
| Concentration (nM) | Mean Calculated Concentration (nM) | Standard Deviation | Accuracy/Recovery (%) | CV (%) |
|---|---|---|---|---|
| 1.1 | 0.8667 | 0.0577 | 78.7879 | 6.6617 |
| 3.3 | 3.6333 | 0.2082 | 110.101 | 5.7294 |
| 10 | 10.3667 | 0.4163 | 103.6667 | 4.0161 |
| 30 | 28.4 | 0.5292 | 94.6667 | 1.8632 |
| 90 | 82.21 | 0.1556 | 91.3444 | 0.1892 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khaligh, S.S.; Khalid-Salako, F.; Kurt, H.; Yüce, M. Exploring the Interaction of Biotinylated FcGamma RI and IgG1 Monoclonal Antibodies on Streptavidin-Coated Plasmonic Sensor Chips for Label-Free VEGF Detection. Biosensors 2024, 14, 634. https://doi.org/10.3390/bios14120634
Khaligh SS, Khalid-Salako F, Kurt H, Yüce M. Exploring the Interaction of Biotinylated FcGamma RI and IgG1 Monoclonal Antibodies on Streptavidin-Coated Plasmonic Sensor Chips for Label-Free VEGF Detection. Biosensors. 2024; 14(12):634. https://doi.org/10.3390/bios14120634
Chicago/Turabian StyleKhaligh, Soodeh Salimi, Fahd Khalid-Salako, Hasan Kurt, and Meral Yüce. 2024. "Exploring the Interaction of Biotinylated FcGamma RI and IgG1 Monoclonal Antibodies on Streptavidin-Coated Plasmonic Sensor Chips for Label-Free VEGF Detection" Biosensors 14, no. 12: 634. https://doi.org/10.3390/bios14120634
APA StyleKhaligh, S. S., Khalid-Salako, F., Kurt, H., & Yüce, M. (2024). Exploring the Interaction of Biotinylated FcGamma RI and IgG1 Monoclonal Antibodies on Streptavidin-Coated Plasmonic Sensor Chips for Label-Free VEGF Detection. Biosensors, 14(12), 634. https://doi.org/10.3390/bios14120634







