A Machine Learning Assisted Non-Enzymatic Electrochemical Biosensor to Detect Urea Based on Multi-Walled Carbon Nanotube Functionalized with Copper Oxide Micro-Flowers
Abstract
1. Introduction
2. Materials and Method
2.1. Chemicals Material Used
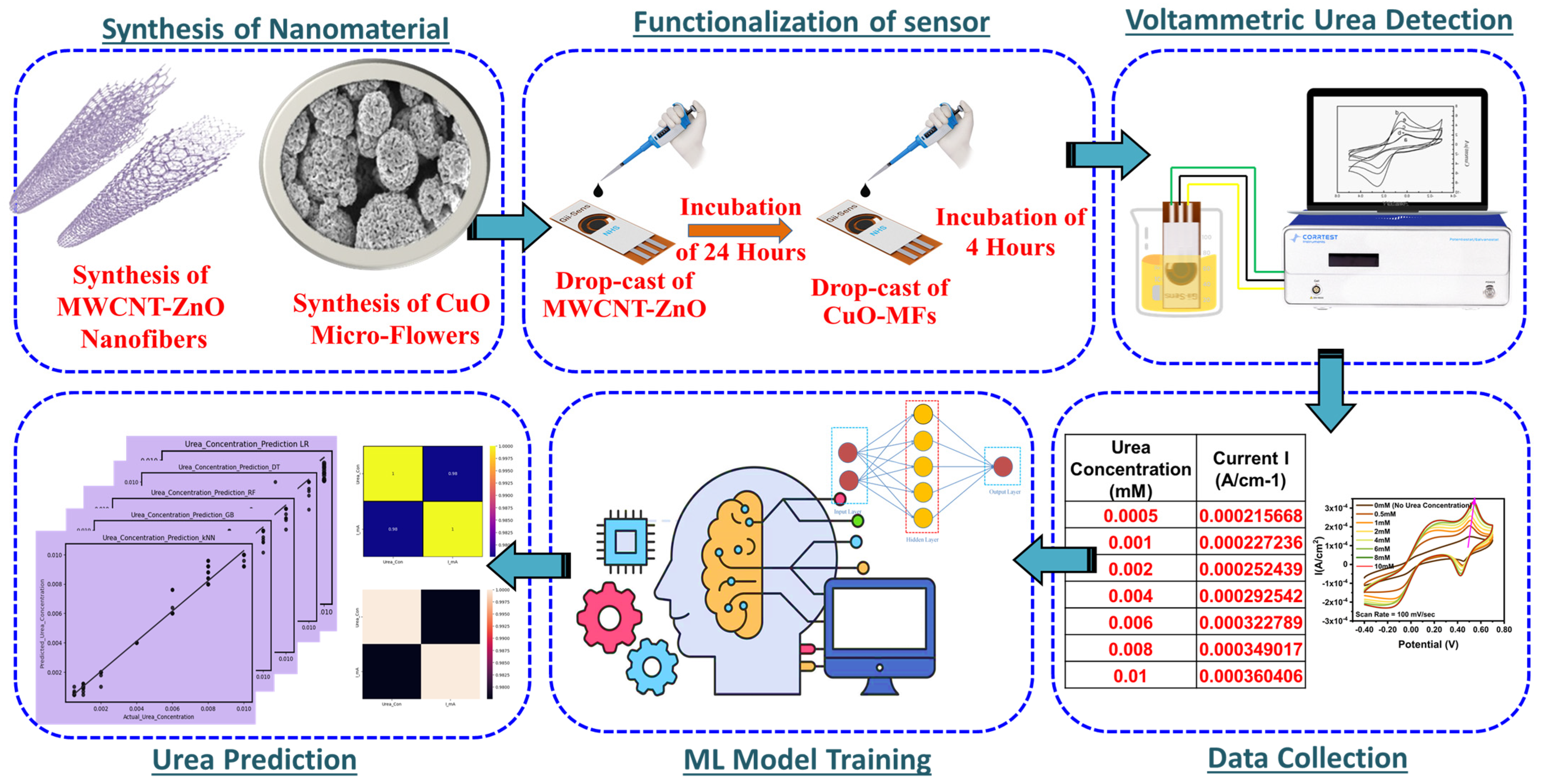
2.2. Methodology
2.2.1. Synthesis of MWCNT-ZnO Composite
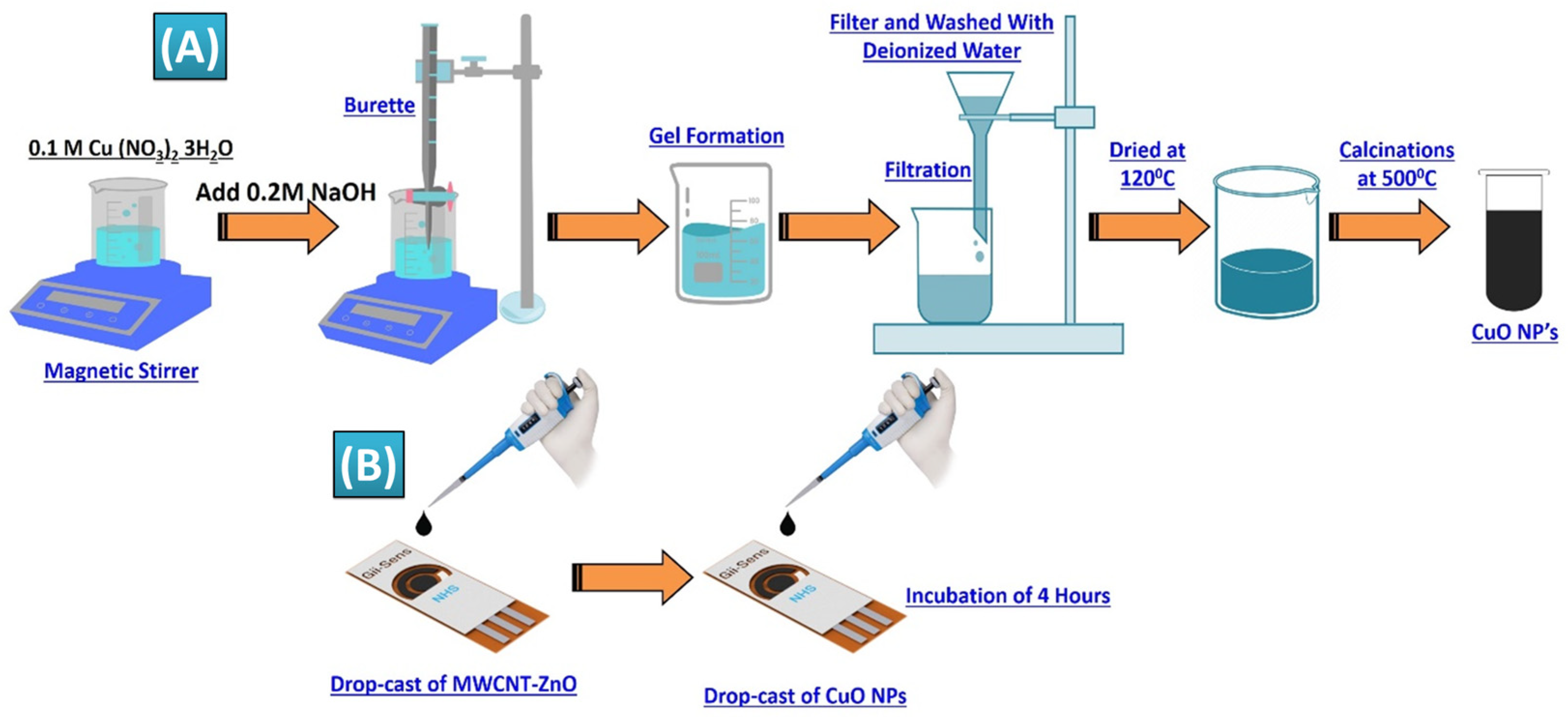
2.2.2. Preparation of CuO Micro-Flowers
2.2.3. Preparation and Functionalization of SPE
2.2.4. Preparation of Urea Stock Solution and Experimental Test Setup
3. Results and Discussion
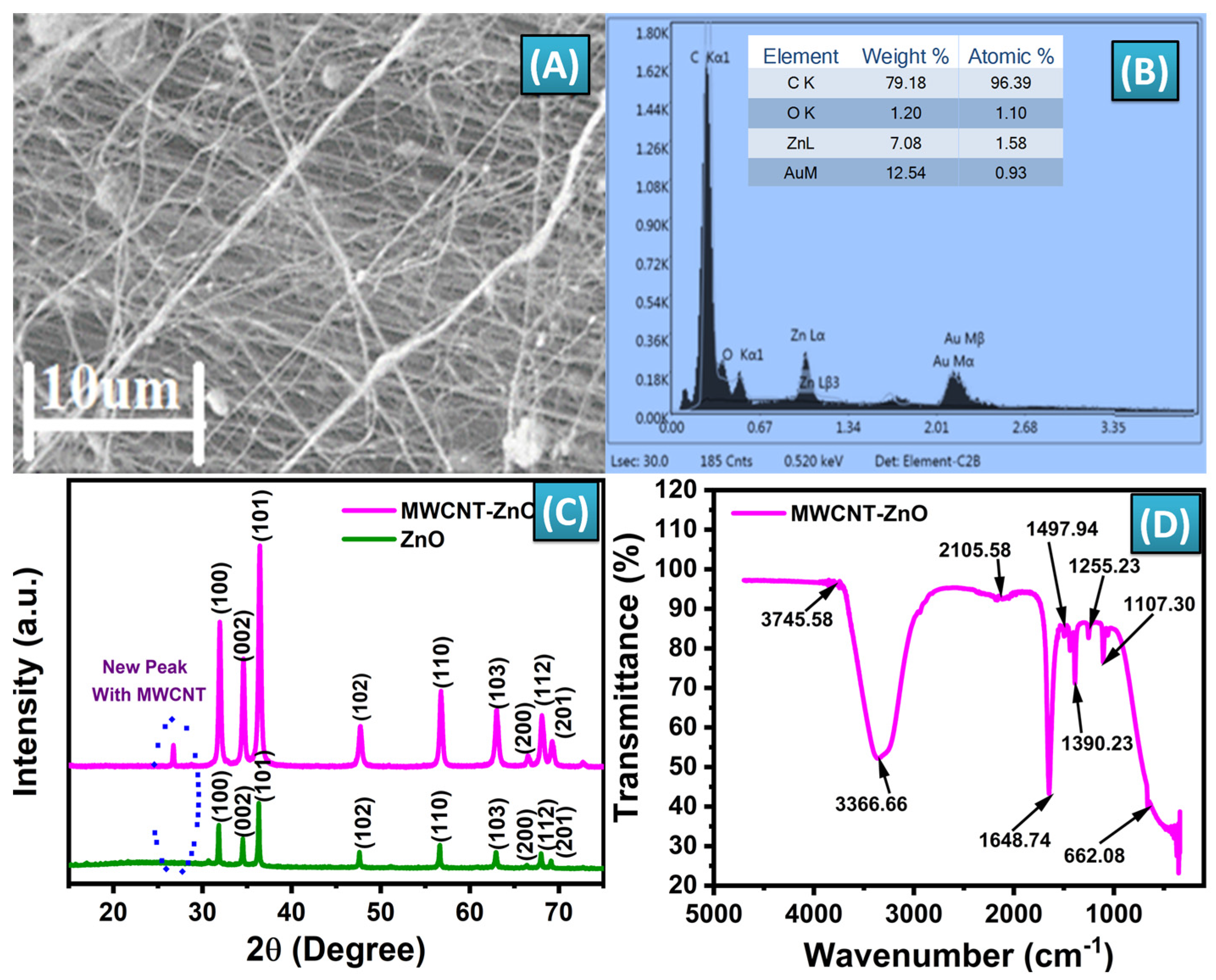
3.1. Characterization of MWCNT-ZnO Nanofibers
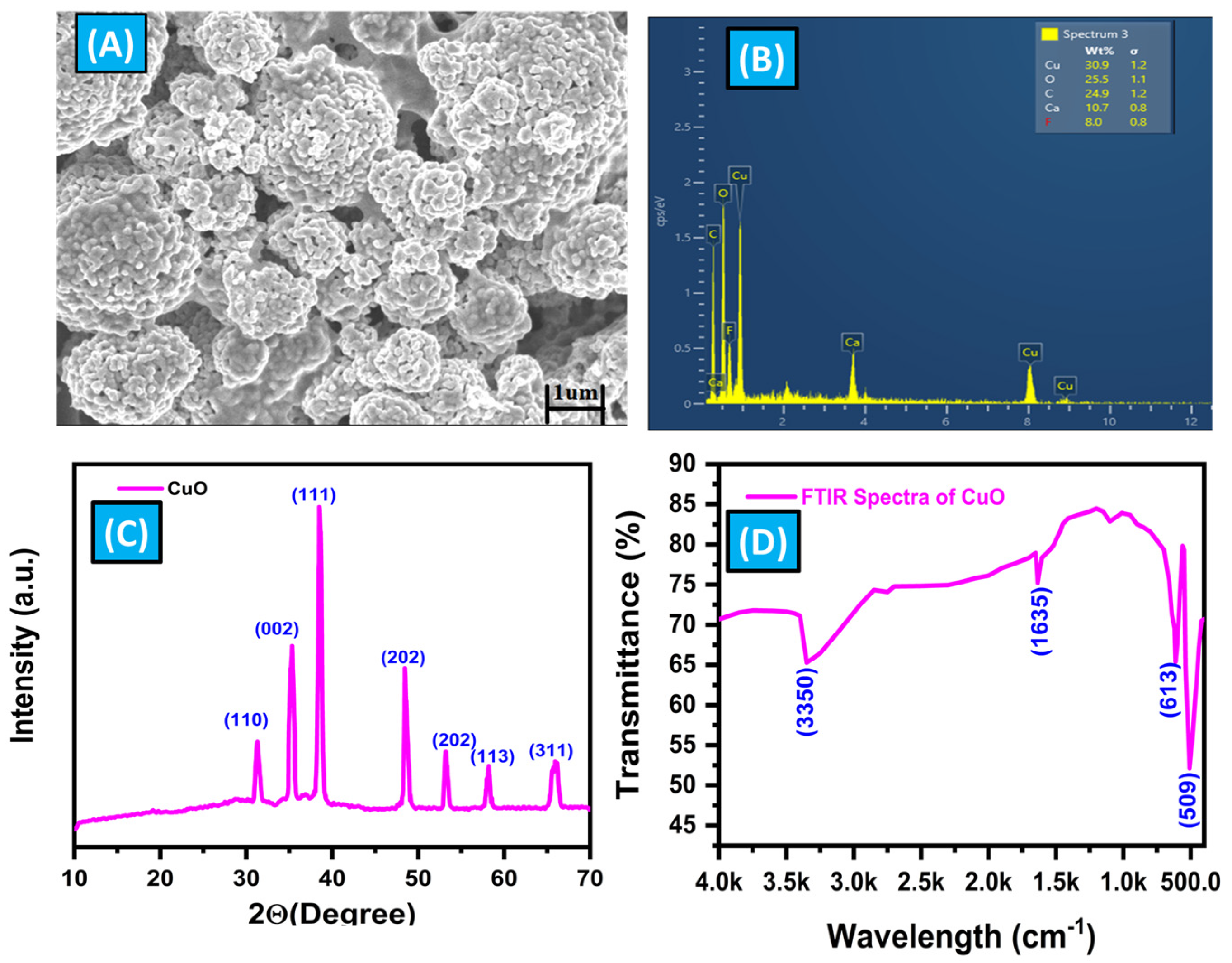
3.2. Characterization of CuO Micro-Flowers
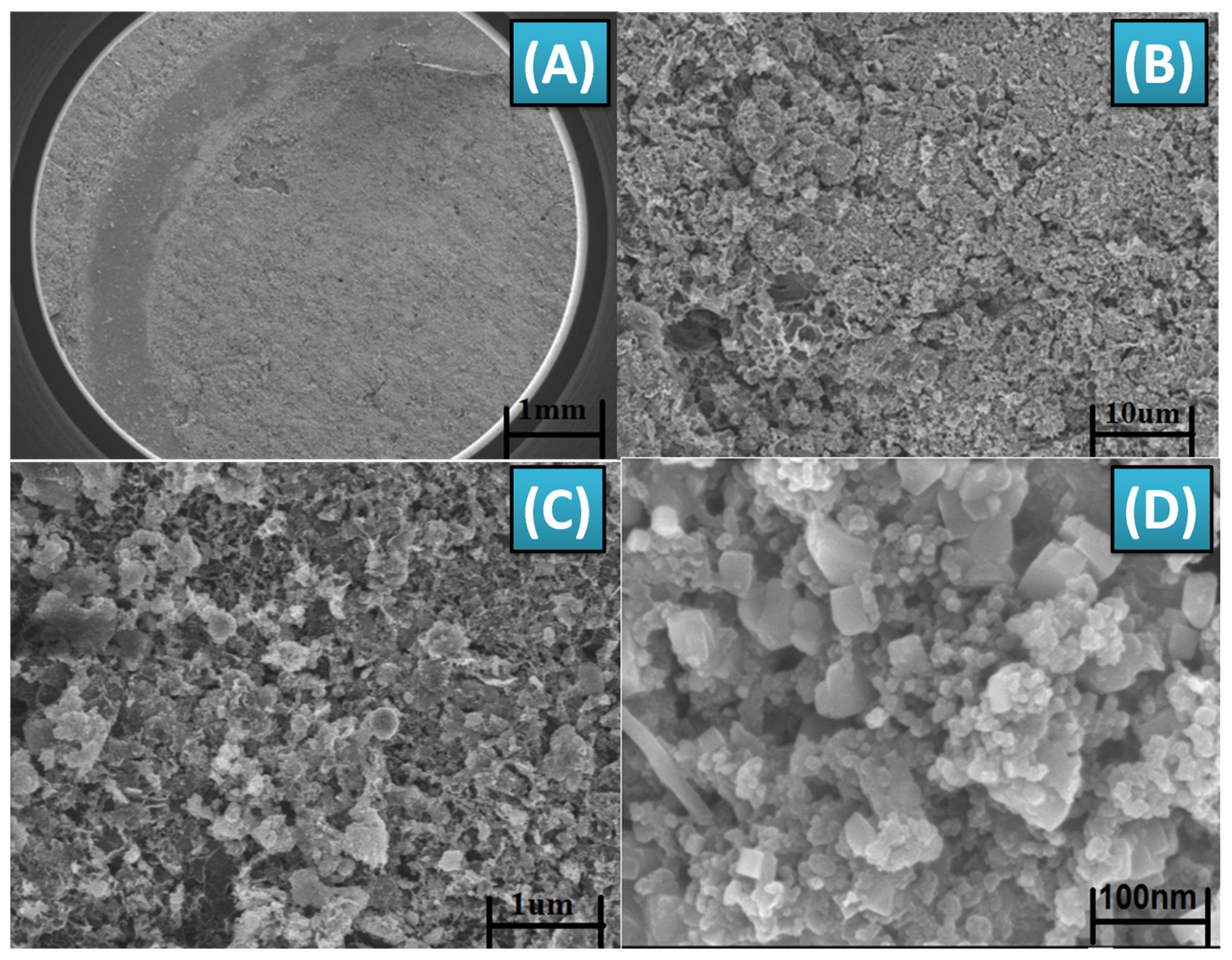
3.3. Morphology and Structural Studies of Functionalized Sensor
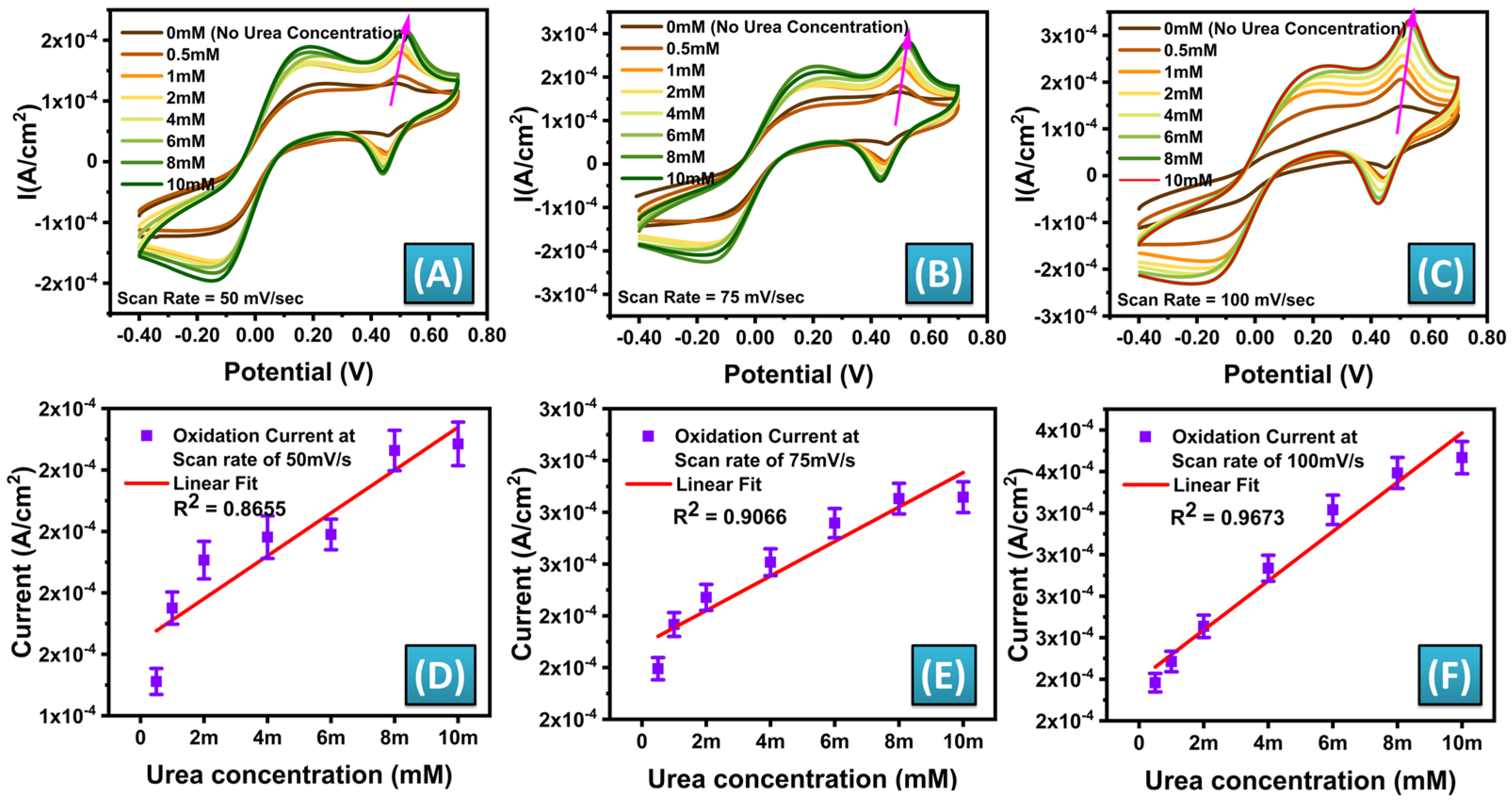
3.4. Electrochemical Cyclic Voltammetry Characterization of the MWCNT-ZnO/CuO-MFs Modified SPEs
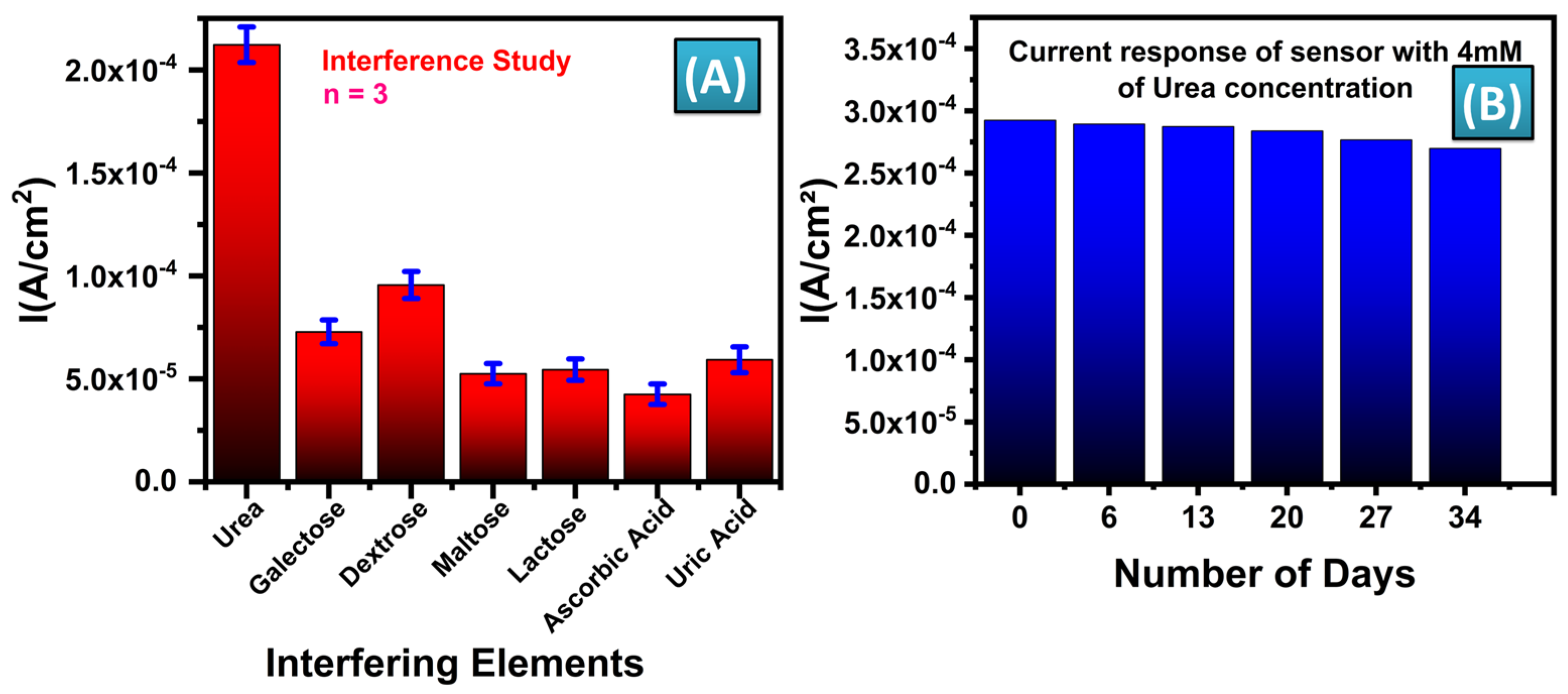
3.5. Selectivity and Stability Study of the Sensor
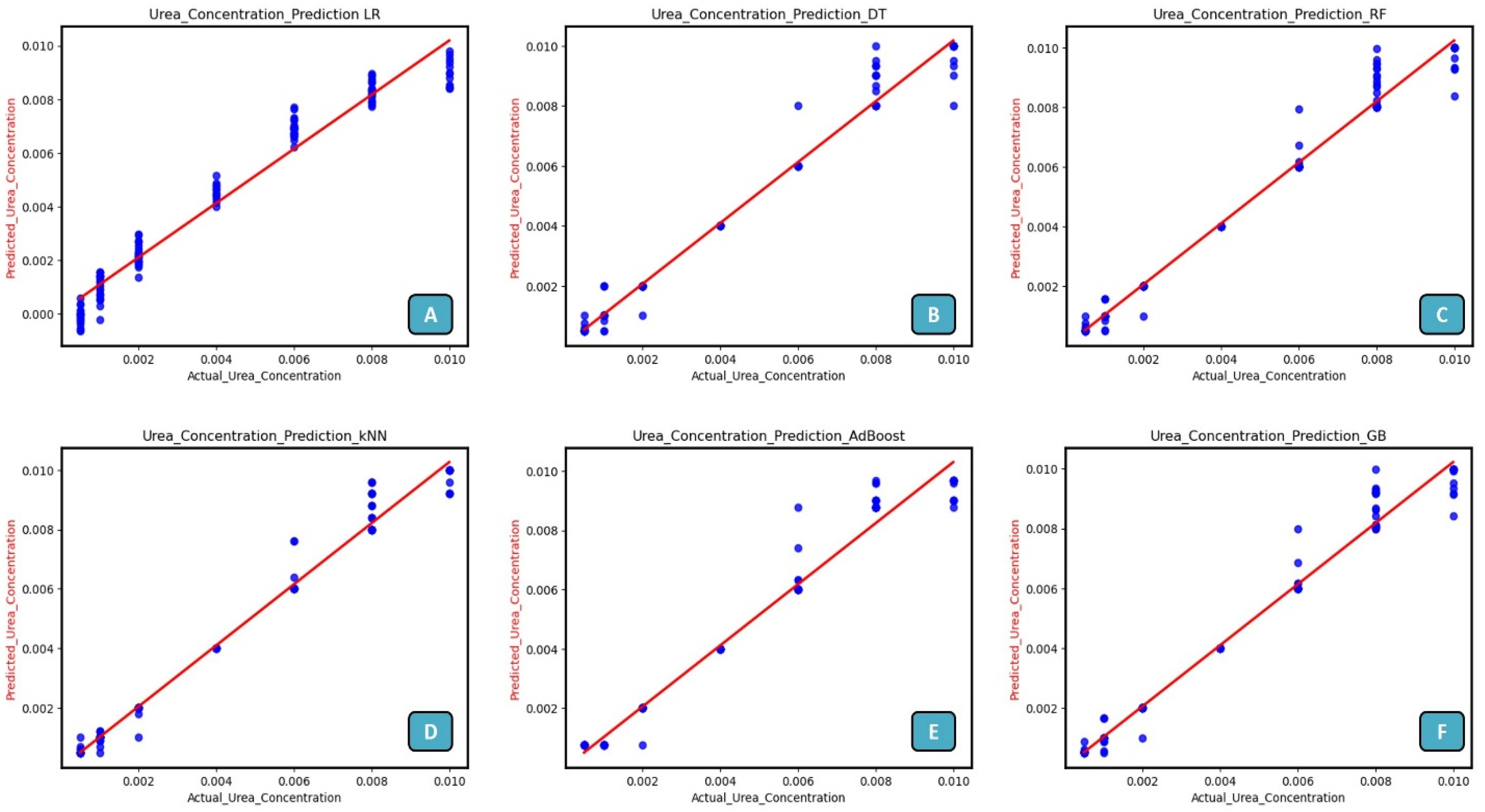
3.6. Machine Learning Approach for Prediction of Urea Concentration
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Umesawa, M.; Yamagishi, K.; Sawachi, S.; Ikeda, A.; Noda, H.; Ikehara, S.; Cui, R.; Sakurai, S.; Tanigawa, T.; Iso, H. Urea Nitrogen Concentrations in Spot Urine, Estimated Protein Intake and Blood Pressure Levels in a Japanese General Population. Am. J. Hypertens 2010, 23, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Das, G.; Yoon, H.H. Fabrication of an amperometric urea biosensor using urease and metal catalysts immobilized by a polyion complex. J. Electroanal. Chem. 2015, 747, 143–148. [Google Scholar] [CrossRef]
- Wang, F.S.; Goh, D.L.M.; Ong, H.T. Urea cycle disorder presenting as bilateral mesial temporal sclerosis—An unusual cause of seizures: A case report and review of the literature. J. Med. Case Rep. 2018, 12, 208. [Google Scholar] [CrossRef]
- Huang, C.-P.; Li, Y.-K.; Chen, T.-M. A highly sensitive system for urea detection by using CdSe/ZnS core-shell quantum dots. Biosens. Bioelectron. 2007, 22, 1835–1838. [Google Scholar] [CrossRef] [PubMed]
- Shaykhutdinov, R.A.; MacInnis, G.D.; Dowlatabadi, R.; Weljie, A.M.; Vogel, H.J. Quantitative analysis of metabolite concentrations in human urine samples using 13C{1H} NMR spectroscopy. Metabolomics 2009, 5, 307–317. [Google Scholar] [CrossRef]
- Liu, L.; Mo, H.; Wei, S.; Raftery, D. Quantitative analysis of urea in human urine and serum by 1 H nuclear magnetic resonance. Analyst 2012, 137, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Vanholder, R.; Gryp, T.; Glorieux, G. Urea and chronic kidney disease: The comeback of the century? (in uraemia research). Nephrol. Dial. Transplant. 2018, 33, 4–12. [Google Scholar] [CrossRef]
- Piquero-Casals, J.; Morgado-Carrasco, D.; Granger, C.; Trullàs, C.; Jesús-Silva, A.; Krutmann, J. Urea in Dermatology: A Review of its Emollient, Moisturizing, Keratolytic, Skin Barrier Enhancing and Antimicrobial Properties. Dermatol. Ther. 2021, 11, 1905–1915. [Google Scholar] [CrossRef]
- Wong, I.Y. The History of Urea and Its Use in the Modern Fertilizer Industry. In Proceedings of the 4th International Conference on Biotechnology and Biomedicine, Nanjing, China, 25–27 March 2022; pp. 435–439. [Google Scholar] [CrossRef]
- Saha, B.K.; Rose, M.T.; Wong, V.N.L.; Cavagnaro, T.R.; Patti, A.F. Nitrogen Dynamics in Soil Fertilized with Slow Release Brown Coal-Urea Fertilizers. Sci. Rep. 2018, 8, 14577. [Google Scholar] [CrossRef]
- Raza, N.; Kim, K.-H. Quantification techniques for important environmental contaminants in milk and dairy products. TrAC Trends Anal. Chem. 2018, 98, 79–94. [Google Scholar] [CrossRef]
- Handford, C.E.; Campbell, K.; Elliott, C.T. Impacts of Milk Fraud on Food Safety and Nutrition with Special Emphasis on Developing Countries. Compr. Rev. Food Sci. Food Saf. 2016, 15, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Ezhilan, M.; Gumpu, M.B.; Ramachandra, B.L.; Nesakumar, N.; Babu, K.J.; Krishnan, U.M.; Rayappan, J.B.B. Design and development of electrochemical biosensor for the simultaneous detection of melamine and urea in adulterated milk samples. Sens. Actuators B Chem. 2017, 238, 1283–1292. [Google Scholar] [CrossRef]
- Oliver, K.V.; Maréchal, A.; Rich, P.R. Effects of the Hydration State on the Mid-Infrared Spectra of Urea and Creatinine in Relation to Urine Analyses. Appl. Spectrosc. 2016, 70, 983–994. [Google Scholar] [CrossRef]
- Clark, S.; Francis, P.S.; Conlan, X.A.; Barnett, N.W. Determination of urea using high-performance liquid chromatography with fluorescence detection after automated derivatisation with xanthydrol. J. Chromatogr. A 2007, 1161, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Gaddes, D.E.; Demirel, M.C.; Reeves, W.B.; Tadigadapa, S. Remote calorimetric detection of urea via flow injection analysis. Analyst 2015, 140, 8033–8040. [Google Scholar] [CrossRef]
- Abdel-Latif, M.S.; Guilbault, G.G. Fluorometric determination of urea by flow injection analysis. J. Biotechnol. 1990, 14, 53–61. [Google Scholar] [CrossRef]
- Nemati, A.; Chaichi, M.J.; Hosseinkhani, S.; Lakouraj, M.M.; Seyedalipour, B. Sensitive determination of urea in luciferin chemiluminescence system using an experimental design. Chem. Pap. 2023, 77, 2571–2580. [Google Scholar] [CrossRef]
- Nie, F.; Wang, N.; Xu, P.; Zheng, J. Determination of urea in milk based on N -bromosuccinimide–dichlorofluorescein postchemiluminescence method. J. Food Drug Anal. 2017, 25, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Bhaiyya, M.; Pattnaik, P.K.; Goel, S. Multiplexed and simultaneous biosensing in a 3D-printed portable six-well smartphone operated electrochemiluminescence standalone point-of-care platform. Microchim. Acta 2022, 189, 79. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Kulkarni, M.B.; Pattnaik, P.K.; Goel, S. Internet of things-enabled photomultiplier tube- and smartphone-based electrochemiluminescence platform to detect choline and dopamine using 3D-printed closed bipolar electrodes. Luminescence 2022, 37, 357–365. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Kumar, P.S.; Pattnaik, P.K.; Shankar, K.; Goel, S. Stereolithography 3-D Printed Electrochemiluminescence Platform with Random Grade Graphite Electrodes: Detection of HO and Cholesterol Using a Smartphone. IEEE Sens. J. 2023, 23, 750–757. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, M.; Singh, G. Recent advancements in urea biosensors for biomedical applications. IET Nanobiotechnol. 2021, 15, 358–379. [Google Scholar] [CrossRef] [PubMed]
- Magar, H.S.; Hassan, R.Y.A.; Abbas, M.N. Non-enzymatic disposable electrochemical sensors based on CuO/Co3O4@MWCNTs nanocomposite modified screen-printed electrode for the direct determination of urea. Sci. Rep. 2023, 13, 2034. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Couto, R.A.S.; Lima, J.L.F.C.; Quinaz, M.B. Recent developments, characteristics and potential applications of screen-printed electrodes in pharmaceutical and biological analysis. Talanta 2016, 146, 801–814. [Google Scholar] [CrossRef]
- Foster, C.W.; Kadara, R.O.; Banks, C.E. Fundamentals of Screen-Printing Electrochemical Architectures; Springer: Berlin/Heidelberg, Germany, 2016; pp. 13–23. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.-T.; Li, D.-W.; Long, Y.-T. Recent developments and applications of screen-printed electrodes in environmental assays—A review. Anal. Chim. Acta 2012, 734, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Mincu, N.-B.; Lazar, V.; Stan, D.; Mihailescu, C.M.; Iosub, R.; Mateescu, A.L. Screen-Printed Electrodes (SPE) for In Vitro Diagnostic Purpose. Diagnostics 2020, 10, 517. [Google Scholar] [CrossRef]
- Ferrari, A.G.-M.; Rowley-Neale, S.J.; Banks, C.E. Screen-printed electrodes: Transitioning the laboratory in-to-the field. Talanta Open 2021, 3, 100032. [Google Scholar] [CrossRef]
- Zalke, J.B.; Narkhede, N.P.; Rotake, D.R.; Singh, S.G. Facile chemiresistive biosensor functionalized with PANI/GOx and novel green synthesized silver nanoparticles for glucose sensing. Microchem. J. 2024, 200, 110339. [Google Scholar] [CrossRef]
- Zalke, J.; Narkhede, N.; Pandhurnekar, C.P.; Rotake, D.R.; Singh, S.G. Non-enzymatic glucose detection with Screen-Printed Chemiresistive sensor using green synthesised silver nanoparticle and multi-walled carbon nanotubes-zinc oxide nanofibers. Nanotechnology 2023, 35, 065502. [Google Scholar] [CrossRef]
- Banks, C.E.; Foster, C.W.; Kadara, R.O. Screen-Printing Electrochemical Architectures; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Hassan, R.Y.A. Advances in Electrochemical Nano-Biosensors for Biomedical and Environmental Applications: From Current Work to Future Perspectives. Sensors 2022, 22, 7539. [Google Scholar] [CrossRef]
- Yuan, T.; Voznyy, O. Guidelines for reliable urea detection in electrocatalysis. Cell Rep. Phys. Sci. 2023, 4, 101521. [Google Scholar] [CrossRef]
- Yoon, J.; Lim, J.; Shin, M.; Lee, J.-Y.; Choi, J.-W. Recent progress in nanomaterial-based bioelectronic devices for biocomputing system. Biosens. Bioelectron. 2022, 212, 114427. [Google Scholar] [CrossRef]
- Ramesh, M.; Janani, R.; Deepa, C.; Rajeshkumar, L. Nanotechnology-Enabled Biosensors: A Review of Fundamentals, Design Principles, Materials, and Applications. Biosensors 2022, 13, 40. [Google Scholar] [CrossRef]
- Li, L.; Wang, T.; Zhong, Y.; Li, R.; Deng, W.; Xiao, X.; Xu, Y.; Zhang, J.; Hu, X.; Wang, Y. A review of nanomaterials for biosensing applications. J. Mater. Chem. B 2024, 12, 1168–1193. [Google Scholar] [CrossRef]
- Yuan, C.; Wu, H.B.; Xie, Y.; Lou, X.W. Mixed Transition-Metal Oxides: Design, Synthesis, and Energy-Related Applications. Angew. Chem. Int. Ed. 2014, 53, 1488–1504. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.I.; Jamshaid, U.; Jamshaid, T.; Zafar, N.; Fessi, H.; Elaissari, A. Carbon nanotubes from synthesis to in vivo biomedical applications. Int. J. Pharm. 2016, 501, 278–299. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Flahaut, E.; Golzio, M. Overview of Carbon Nanotubes for Biomedical Applications. Materials 2019, 12, 624. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Rewatkar, P.; Pimpalkar, A.; Jain, D.; Srivastava, S.K.; Zalke, J.; Kalambe, J.; Balpande, S.; Kale, P.; Kalantri, Y.; et al. Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform. Micromachines 2024, 15, 1059. [Google Scholar] [CrossRef]
- Luo, X.; Morrin, A.; Killard, A.J.; Smyth, M.R. Application of Nanoparticles in Electrochemical Sensors and Biosensors. Electroanalysis 2006, 18, 319–326. [Google Scholar] [CrossRef]
- Kaushik, A.; Solanki, P.R.; Ansari, A.A.; Sumana, G.; Ahmad, S.; Malhotra, B.D. Iron oxide-chitosan nanobiocomposite for urea sensor. Sens. Actuators B Chem. 2009, 138, 572–580. [Google Scholar] [CrossRef]
- Tak, M.; Gupta, V.; Tomar, M. Zinc oxide–multiwalled carbon nanotubes hybrid nanocomposite based urea biosensor. J. Mater. Chem. B 2013, 1, 6392. [Google Scholar] [CrossRef] [PubMed]
- Dervisevic, E.; Dervisevic, M.; Nyangwebah, J.N.; Şenel, M. Development of novel amperometric urea biosensor based on Fc-PAMAM and MWCNT bio-nanocomposite film. Sens. Actuators B Chem. 2017, 246, 920–926. [Google Scholar] [CrossRef]
- Tyagi, M.; Tomar, M.; Gupta, V. NiO nanoparticle-based urea biosensor. Biosens. Bioelectron. 2013, 41, 110–115. [Google Scholar] [CrossRef]
- Baabu, P.R.S.; Gumpu, M.B.; Nesakumar, N.; Rayappan, J.B.B.; Kulandaisamy, A.J. Electroactive Manganese Oxide–Reduced Graphene Oxide Interfaced Electrochemical Detection of Urea. Water Air Soil Pollut. 2020, 231, 545. [Google Scholar] [CrossRef]
- Sha, R.; Komori, K.; Badhulika, S. Graphene–Polyaniline composite based ultra-sensitive electrochemical sensor for non-enzymatic detection of urea. Electrochim. Acta 2017, 233, 44–51. [Google Scholar] [CrossRef]
- Mondal, S.; Sangaranarayanan, M.V. A novel non-enzymatic sensor for urea using a polypyrrole-coated platinum electrode. Sens. Actuators B Chem. 2013, 177, 478–486. [Google Scholar] [CrossRef]
- Nguyen, N.S.; Das, G.; Yoon, H.H. Nickel/cobalt oxide-decorated 3D graphene nanocomposite electrode for enhanced electrochemical detection of urea. Biosens. Bioelectron. 2016, 77, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.-J.; Yang, N.-N.; Gao, E.-Q. Making metal–organic frameworks electron-deficient for ultrasensitive electrochemical detection of dopamine. Electrochem. Commun. 2018, 89, 32–37. [Google Scholar] [CrossRef]
- Kumar, T.H.V.; Sundramoorthy, A.K. Non-Enzymatic Electrochemical Detection of Urea on Silver Nanoparticles Anchored Nitrogen-Doped Single-Walled Carbon Nanotube Modified Electrode. J. Electrochem. Soc. 2018, 165, B3006–B3016. [Google Scholar] [CrossRef]
- Bao, C.; Niu, Q.; Chen, Z.-A.; Cao, X.; Wang, H.; Lu, W. Ultrathin nickel-metal–organic framework nanobelt based electrochemical sensor for the determination of urea in human body fluids. RSC Adv. 2019, 9, 29474–29481. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Tahira, A.; Solangi, A.; Beni, V.; Morante, J.R.; Liu, X.; Falhman, M.; Mazzaro, R.; Ibupoto, Z.H.; Vomiero, A. A practical non-enzymatic urea sensor based on NiCo2 O4 nanoneedles. RSC Adv. 2019, 9, 14443–14451. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Yoon, Y.S.; Kim, D.-J. Silver-Nanoparticle-Decorated NiOOH Nanorods for Electrocatalytic Urea Sensing. ACS Appl. Nano Mater. 2020, 3, 7651–7658. [Google Scholar] [CrossRef]
- Yoon, J.; Lee, E.; Lee, D.; Oh, T.-S.; Yoon, Y.S.; Kim, D.-J. Communication—Highly Sensitive Ag/ZnO Nanorods Composite Electrode for Non-Enzymatic Urea Detection. J. Electrochem. Soc. 2017, 164, B558–B560. [Google Scholar] [CrossRef]
- Tran, T.Q.N.; Das, G.; Yoon, H.H. Nickel-metal organic framework/MWCNT composite electrode for non-enzymatic urea detection. Sens. Actuators B Chem. 2017, 243, 78–83. [Google Scholar] [CrossRef]
- Tran, T.Q.N.; Yoon, S.W.; Park, B.J.; Yoon, H.H. CeO2-modified LaNi0.6Fe0.4O3 perovskite and MWCNT nanocomposite for electrocatalytic oxidation and detection of urea. J. Electroanal. Chem. 2018, 818, 76–83. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, F.; Halder, A.; Zhang, M. Free-Standing NiO Nanosheets as Non-Enzymatic Electrochemical Sensors. ChemistrySelect 2020, 5, 2424–2429. [Google Scholar] [CrossRef]
- Babitha, K.B.; Soorya, P.S.; Mohamed, P.; Rakhi, R.B.; Ananthakumar, S. Development of ZnO@rGO nanocomposites for the enzyme free electrochemical detection of urea and glucose. Mater. Adv. 2020, 1, 1939–1951. [Google Scholar] [CrossRef]
- Padmalaya, G.; Rathi, B.S.; Kumar, P.S.; Rangasamy, G. Electrochemical sensor for urea determination using structural c-multiwall carbon nanotubes decorated CuO hybrid nanocomposite: Application in rice water samples. Desalination Water Treat. 2024, 320, 100701. [Google Scholar] [CrossRef]
- Kahar, K.; Dhekekar, R.; Bhaiyya, M.; Srivastava, S.K.; Rewatkar, P.; Balpande, S.; Goel, S. Optimization of MEMS-based Energy Scavengers and output prediction with machine learning and synthetic data approach. Sens. Actuators A Phys. 2023, 358, 114429. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Bhaiyya, M.; Dudala, S.; Hota, C.; Goel, S. A machine learning approach for electrochemiluminescence based point of care testing device to detect multiple biomarkers. Sens. Actuators A Phys. 2023, 350, 114135. [Google Scholar] [CrossRef]
- Bhaiyya, M.L.; Srivastava, S.K.; Pattnaik, P.K.; Goel, S. Closed-Bipolar Mini Electrochemiluminescence Sensor to Detect Various Biomarkers: A Machine Learning Approach. IEEE Trans. Instrum. Meas. 2023, 72, 1–8. [Google Scholar] [CrossRef]
- Dörner, L.; Cancellieri, C.; Rheingans, B.; Walter, M.; Kägi, R.; Schmutz, P.; Kovalenko, M.V.; Jeurgens, L.P. Cost-effective sol-gel synthesis of porous CuO nanoparticle aggregates with tunable specific surface area. Sci. Rep. 2019, 9, 11758. [Google Scholar] [CrossRef] [PubMed]
- Che, H.; Liu, A.; Mu, J.; Zhang, X. Synthesis of Novel Hollow Copper Oxide Micro-Flowers Assembled by Nanoparticles and Their Improved Catalytic Performances for the Synthesis of Organosilane. Nano 2016, 11, 1650032. [Google Scholar] [CrossRef]
- Patel, M.; Mishra, S.; Verma, R.; Shikha, D. Synthesis of ZnO and CuO nanoparticles via Sol gel method and its characterization by using various technique. Discov. Mater. 2022, 2, 1. [Google Scholar] [CrossRef]
- Kayani, Z.N.; Umer, M.; Riaz, S.; Naseem, S. Characterization of Copper Oxide Nanoparticles Fabricated by the Sol–Gel Method. J. Electron. Mater. 2015, 44, 3704–3709. [Google Scholar] [CrossRef]
- Supraja, P.; Singh, V.; Vanjari, S.R.K.; Singh, S.G. Electrospun CNT embedded ZnO nanofiber based biosensor for electrochemical detection of Atrazine: A step closure to single molecule detection. Microsyst. Nanoeng. 2020, 6, 3. [Google Scholar] [CrossRef]
- Abbas, S.M.; Hussain, S.T.; Ali, S.; Ahmad, N.; Ali, N.; Abbas, S. Structure and electrochemical performance of ZnO/CNT composite as anode material for lithium-ion batteries. J. Mater. Sci. 2013, 48, 5429–5436. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, X.; Pan, L.; Li, H.; Sun, Z.; Sun, C.; Tay, B.K. Carbon nanotube–zinc oxide electrode and gel polymer electrolyte for electrochemical supercapacitors. J. Alloys Compd. 2009, 480, L17–L19. [Google Scholar] [CrossRef]
- Samadi, M.; Shivaee, H.A.; Zanetti, M.; Pourjavadi, A.; Moshfegh, A. Visible light photocatalytic activity of novel MWCNT-doped ZnO electrospun nanofibers. J. Mol. Catal. A Chem. 2012, 359, 42–48. [Google Scholar] [CrossRef]
- Helli, M.; Sadrnezhaad, S.K.; Hosseini-Hosseinabad, S.M.; Vahdatkhah, P. Synthesis and characterization of CuO micro-flowers/PPy nanowires nanocomposites as high-capacity anode material for lithium-ion batteries. J. Appl. Electrochem. 2024, 54, 1–11. [Google Scholar] [CrossRef]
- Haarindraprasad, R.; Hashim, U.; Gopinath, S.C.B.; Perumal, V.; Liu, W.-W.; Balakrishnan, S.R. Fabrication of interdigitated high-performance zinc oxide nanowire modified electrodes for glucose sensing. Anal. Chim. Acta 2016, 925, 70–81. [Google Scholar] [CrossRef] [PubMed]
| Electrode Material Used for Urea Detection | Machine Learning | Sensitivity | Limit of Detection | Linear Range | Reference |
|---|---|---|---|---|---|
| Ag/ZnO nanorod | No | 0.1622 μAμM−1 cm−2 | 13.98 μM | 26.3 to 427 μM | [57] |
| Gr-PANi | No | −226.9 μA/μM cm2 | 5.88 μM | 10 μM–200 μM | [49] |
| Ni-MOF/MWCNT | No | 685 μAmM−1 cm−2 | 3 μM | 0.01–1.12 mM | [58] |
| Ag-N-SWCNTs | No | 141 μAmM−1 cm−2 | 4.7 nM | 66 nM to 20.6 mM | [53] |
| LaNi0.6Fe0.4O3-CeO2 (LNF-C)/MWCNT/ITO | No | 195.6 μAmM−1 cm−2 | 1 μM | 25 to 670 μM | [59] |
| NiO Nanosheets | 3.4 A/M cm2 | 2 μM | 4.4 μM to 181.6 μM | [60] | |
| ZnO@rGO | No | 682.8 μA mM−1 cm−2 | 0.012 μM | 0.02 × 10−3 mM to 7.2 × 10−3 mM | [61] |
| CuO/Co3O4@ MWCNTs | No | -- | 0.223 pM | 10−12 to 10−2 M | [24] |
| CuO/c-MWCNT/GCE | No | 23.8983 µA/mM | 0.16 mM/L | 2 mM–8 mM | [62] |
| MWCNT-ZnO/CuO-MFs | Yes | 117.98 mA mM−1 cm−2 | 78.479 nM | 0.5 mM to 8 mM | This Work |
| ML Models | Various ML Model Regression Accuracy Parameters | |||
|---|---|---|---|---|
| MAE | MSE | RMSE | R2 Score | |
| LR | 0.0005 | 4.81 | 0.0006 | 0.953 |
| DT | 0.0001 | 2.20 | 0.0004 | 0.978 |
| RF | 0.0001 | 2.12 | 0.0004 | 0.979 |
| KNN | 0.0001 | 1.93 | 0.0004 | 0.981 |
| AdaBoost | 0.0003 | 2.84 | 0.0005 | 0.972 |
| GB | 0.0001 | 2.13 | 0.0004 | 0.979 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalke, J.B.; Bhaiyya, M.L.; Jain, P.A.; Sakharkar, D.N.; Kalambe, J.; Narkhede, N.P.; Thakre, M.B.; Rotake, D.R.; Kulkarni, M.B.; Singh, S.G. A Machine Learning Assisted Non-Enzymatic Electrochemical Biosensor to Detect Urea Based on Multi-Walled Carbon Nanotube Functionalized with Copper Oxide Micro-Flowers. Biosensors 2024, 14, 504. https://doi.org/10.3390/bios14100504
Zalke JB, Bhaiyya ML, Jain PA, Sakharkar DN, Kalambe J, Narkhede NP, Thakre MB, Rotake DR, Kulkarni MB, Singh SG. A Machine Learning Assisted Non-Enzymatic Electrochemical Biosensor to Detect Urea Based on Multi-Walled Carbon Nanotube Functionalized with Copper Oxide Micro-Flowers. Biosensors. 2024; 14(10):504. https://doi.org/10.3390/bios14100504
Chicago/Turabian StyleZalke, Jitendra B., Manish L. Bhaiyya, Pooja A. Jain, Devashree N. Sakharkar, Jayu Kalambe, Nitin P. Narkhede, Mangesh B. Thakre, Dinesh R. Rotake, Madhusudan B. Kulkarni, and Shiv Govind Singh. 2024. "A Machine Learning Assisted Non-Enzymatic Electrochemical Biosensor to Detect Urea Based on Multi-Walled Carbon Nanotube Functionalized with Copper Oxide Micro-Flowers" Biosensors 14, no. 10: 504. https://doi.org/10.3390/bios14100504
APA StyleZalke, J. B., Bhaiyya, M. L., Jain, P. A., Sakharkar, D. N., Kalambe, J., Narkhede, N. P., Thakre, M. B., Rotake, D. R., Kulkarni, M. B., & Singh, S. G. (2024). A Machine Learning Assisted Non-Enzymatic Electrochemical Biosensor to Detect Urea Based on Multi-Walled Carbon Nanotube Functionalized with Copper Oxide Micro-Flowers. Biosensors, 14(10), 504. https://doi.org/10.3390/bios14100504






