Streptavidin-Conjugated DNA for the Boronate Affinity-Based Detection of Poly(ADP-Ribose) Polymerase-1 with Improved Sensitivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
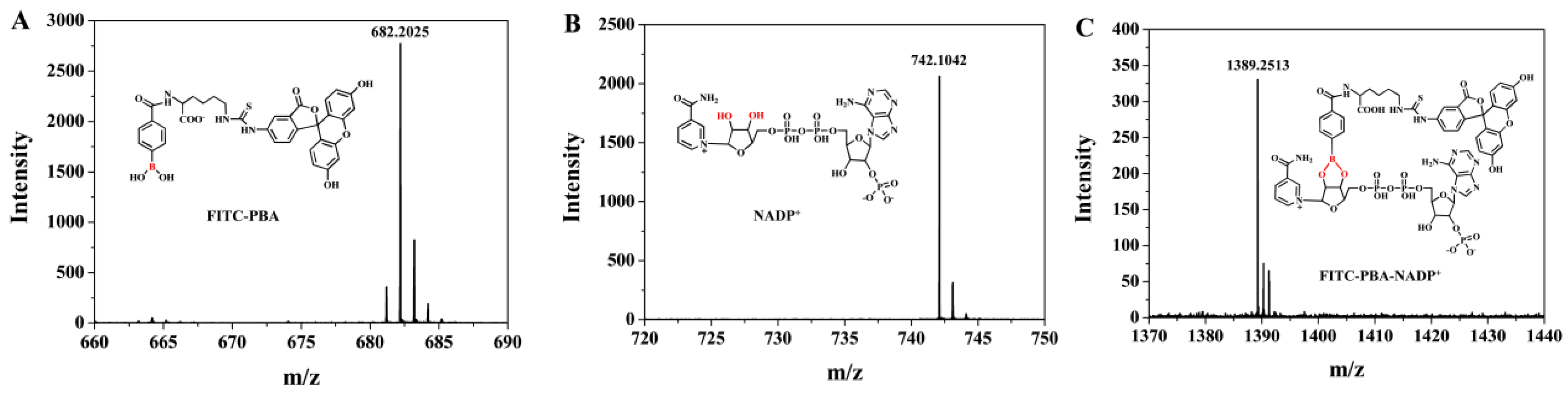
2.2. Mass Spectrometry
2.3. Procedures for PARP1 Detection
2.4. Probing of PARP1Activity via Surface Plasmon Resonance (SPR)
2.5. Extraction of Cytoplasms
2.6. Detection of Caspase-3 Activity in Cytoplasm Samples
3. Results and Discussion
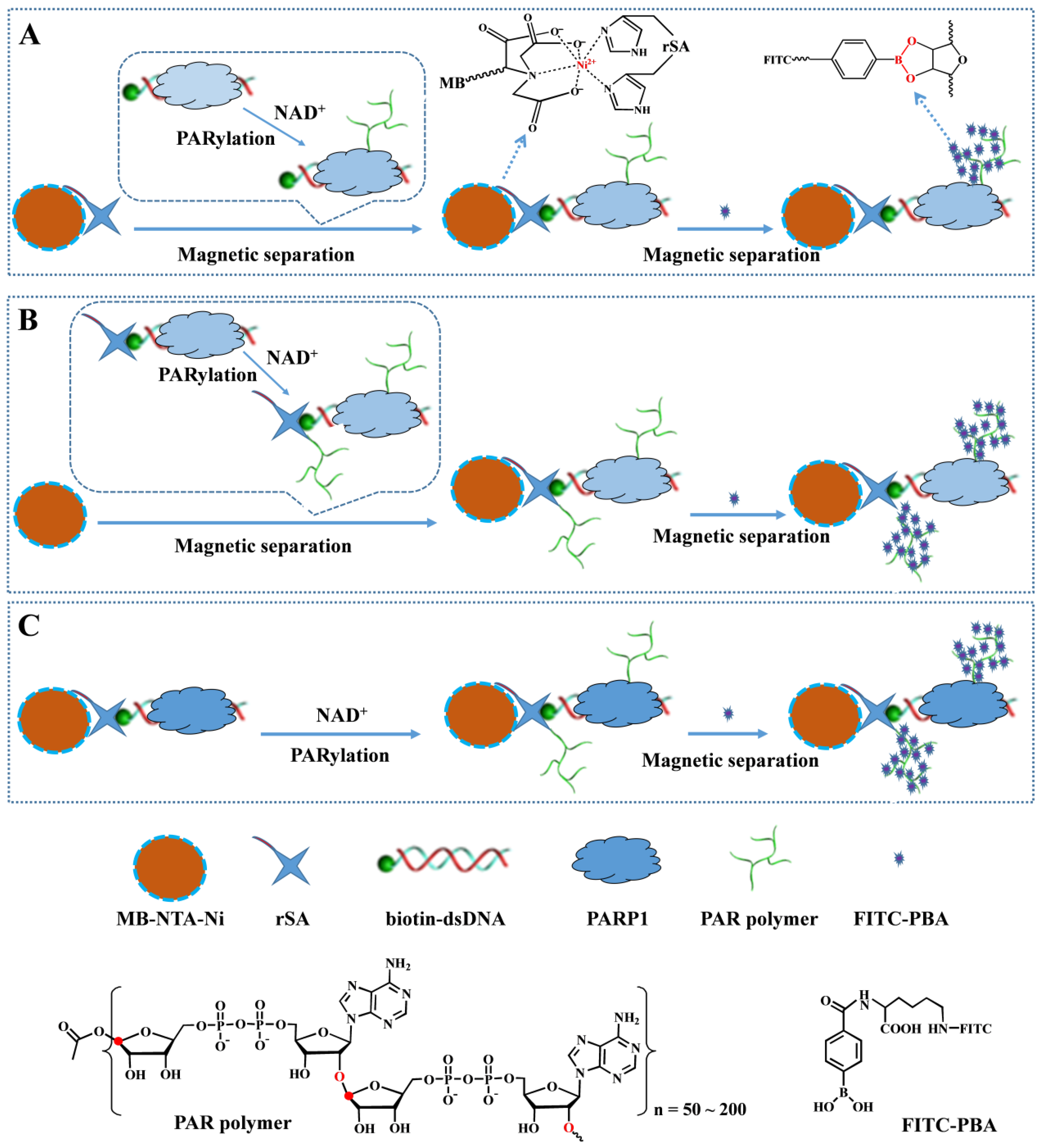
3.1. Strategies for PARP1 Detection
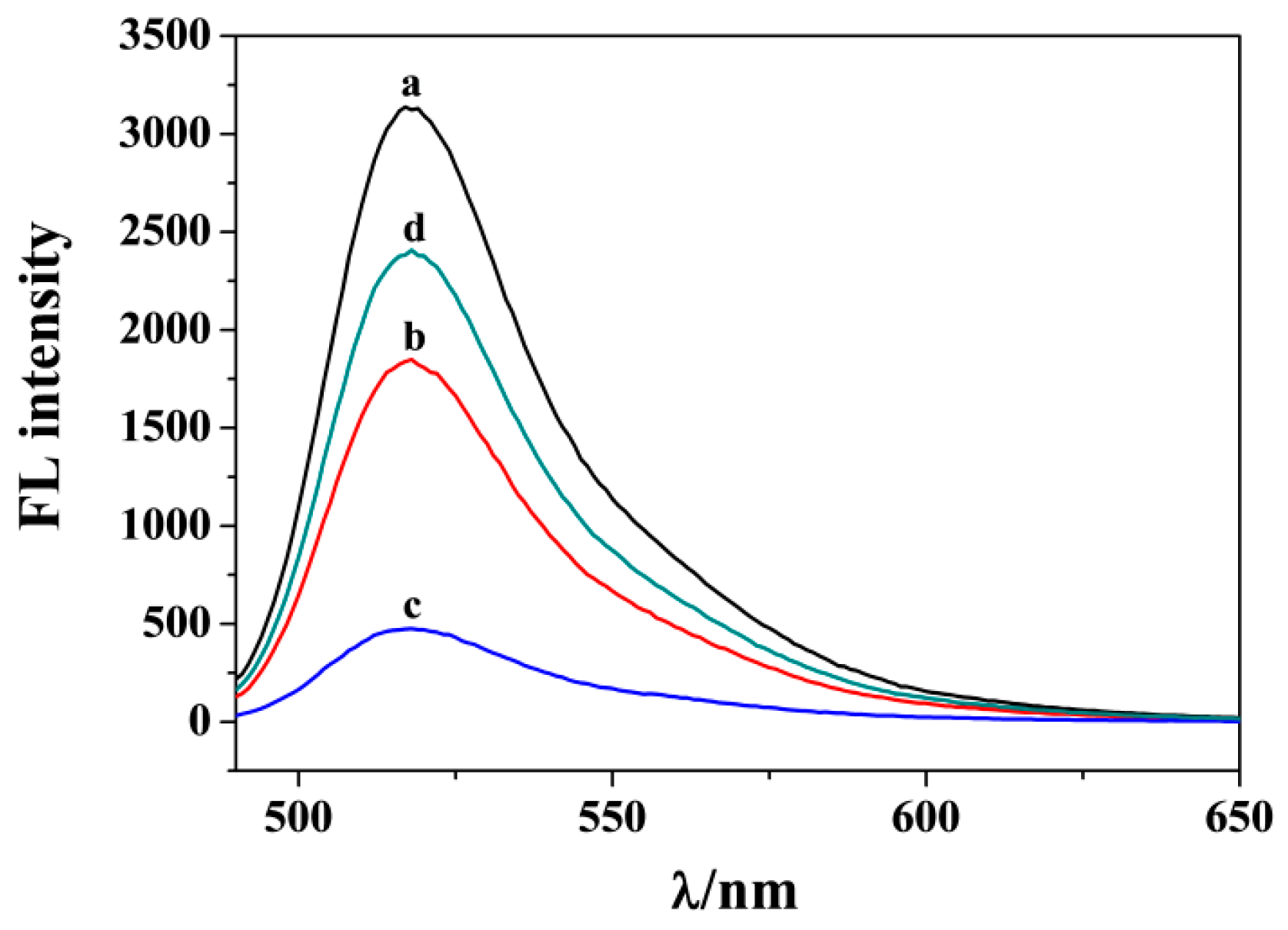
3.2. Feasibility for PARP1 Detection
3.3. SPR Analysis
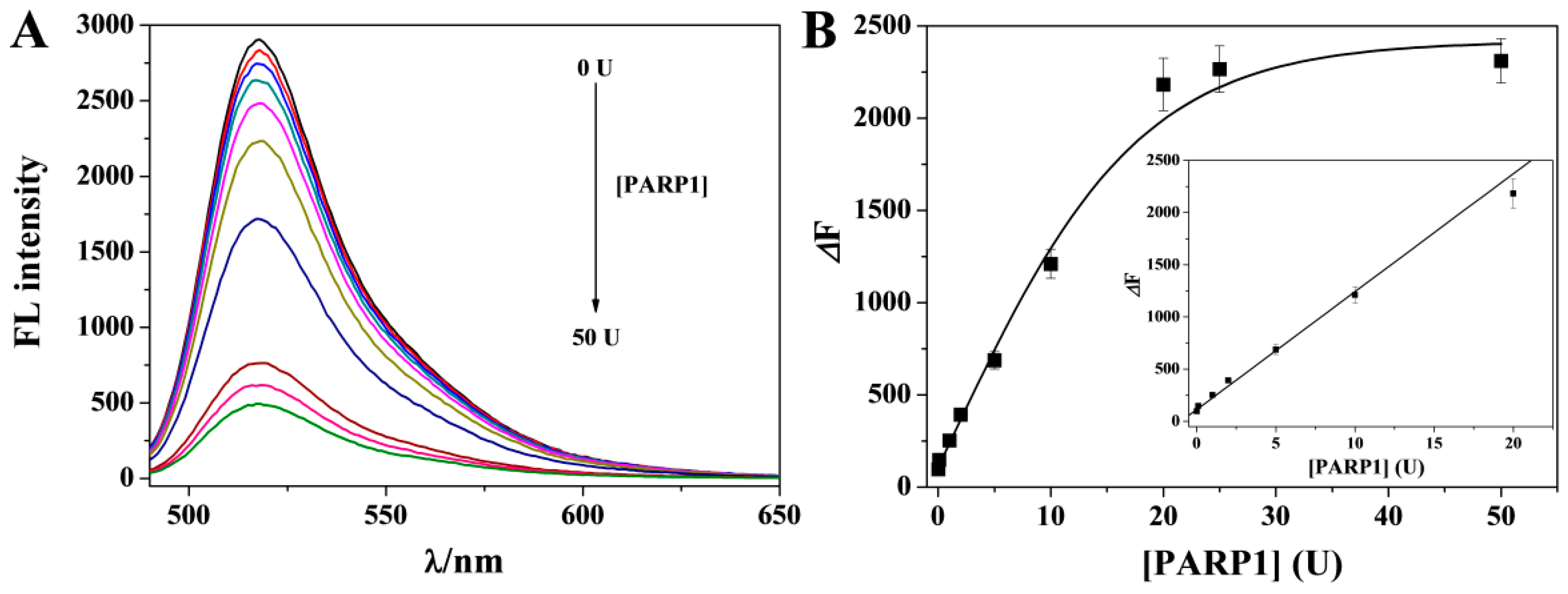
3.4. Analytical Performances
3.5. Evaluation of Inhibition Efficiency
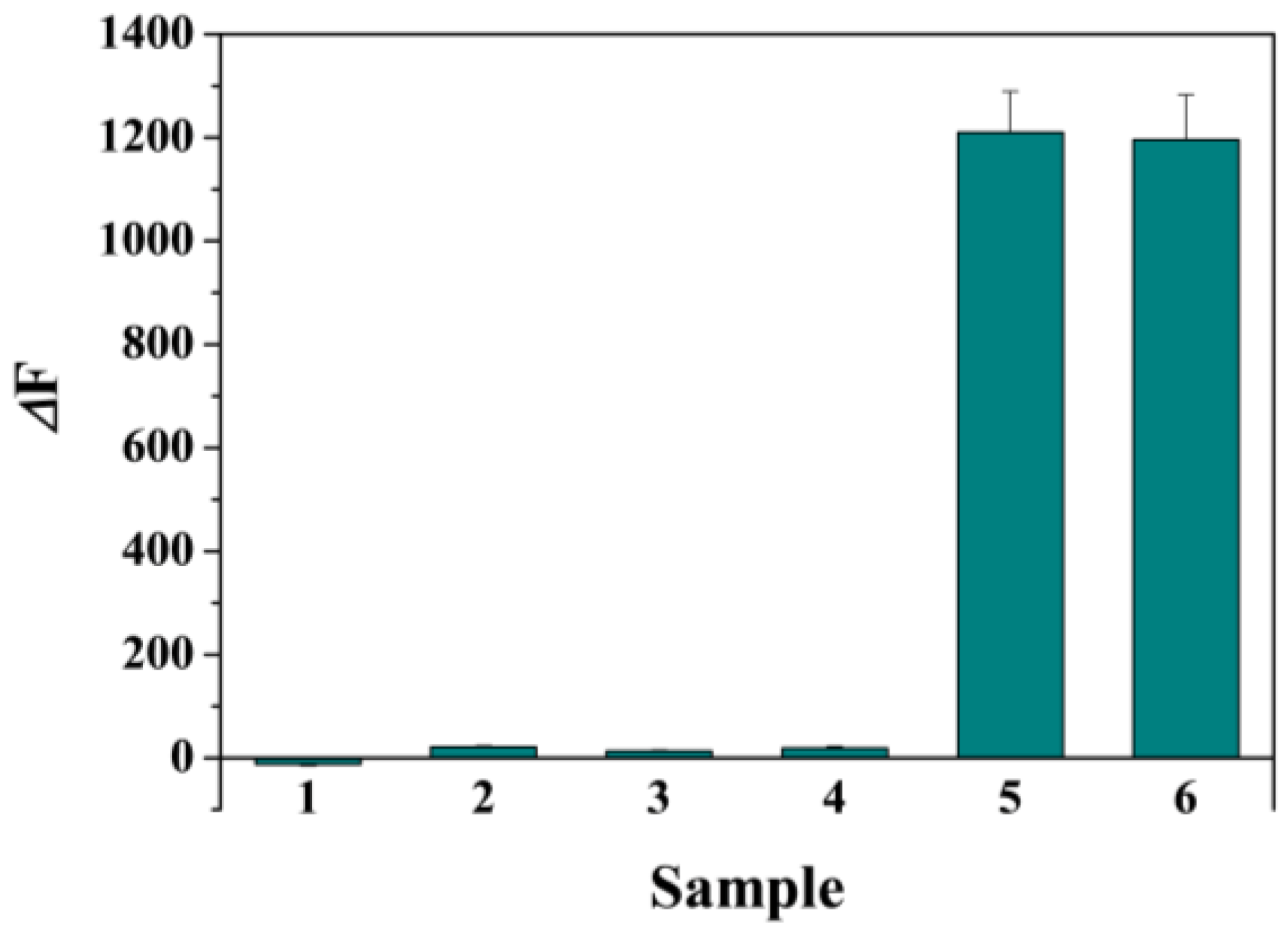
3.6. Selectivity
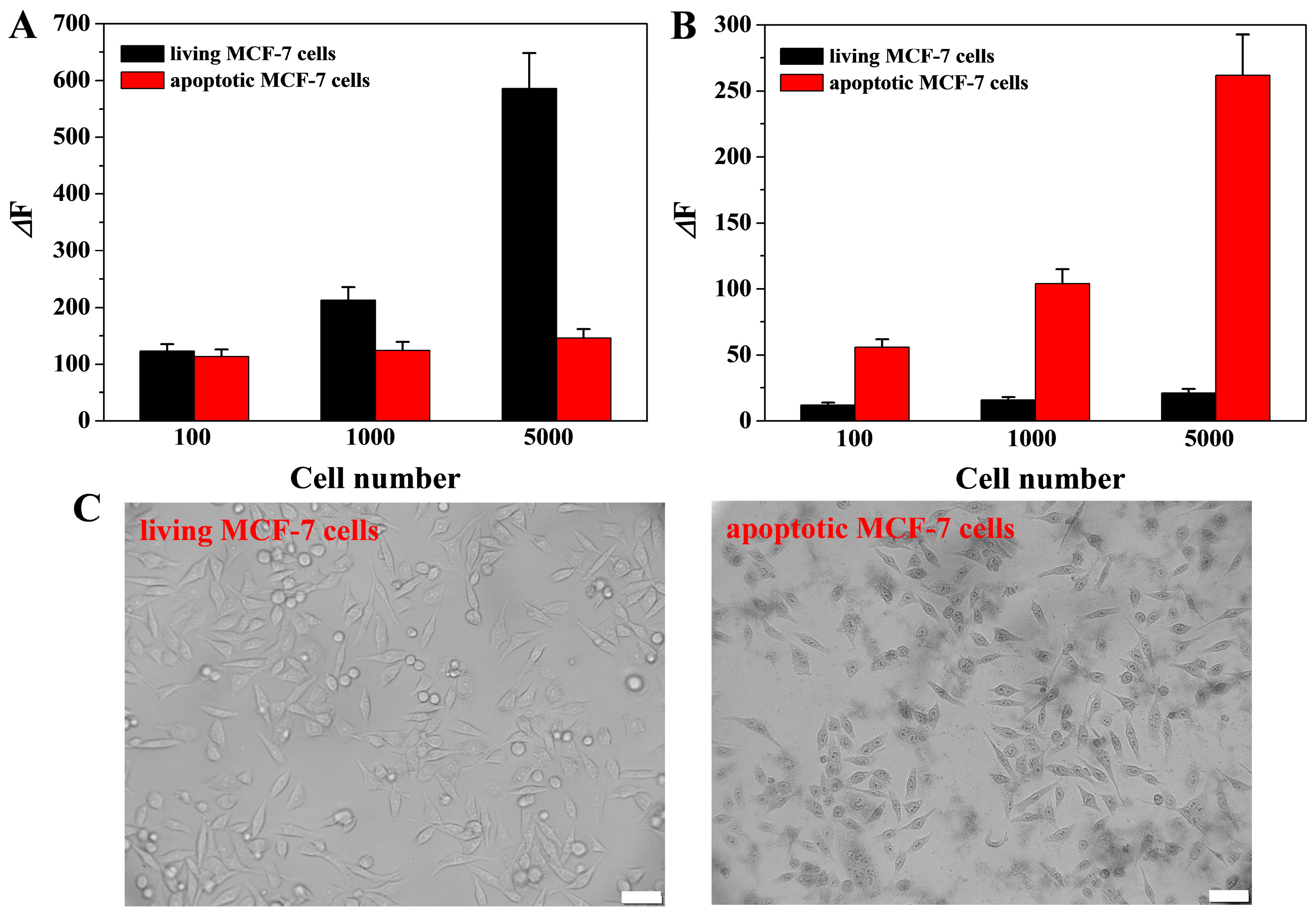
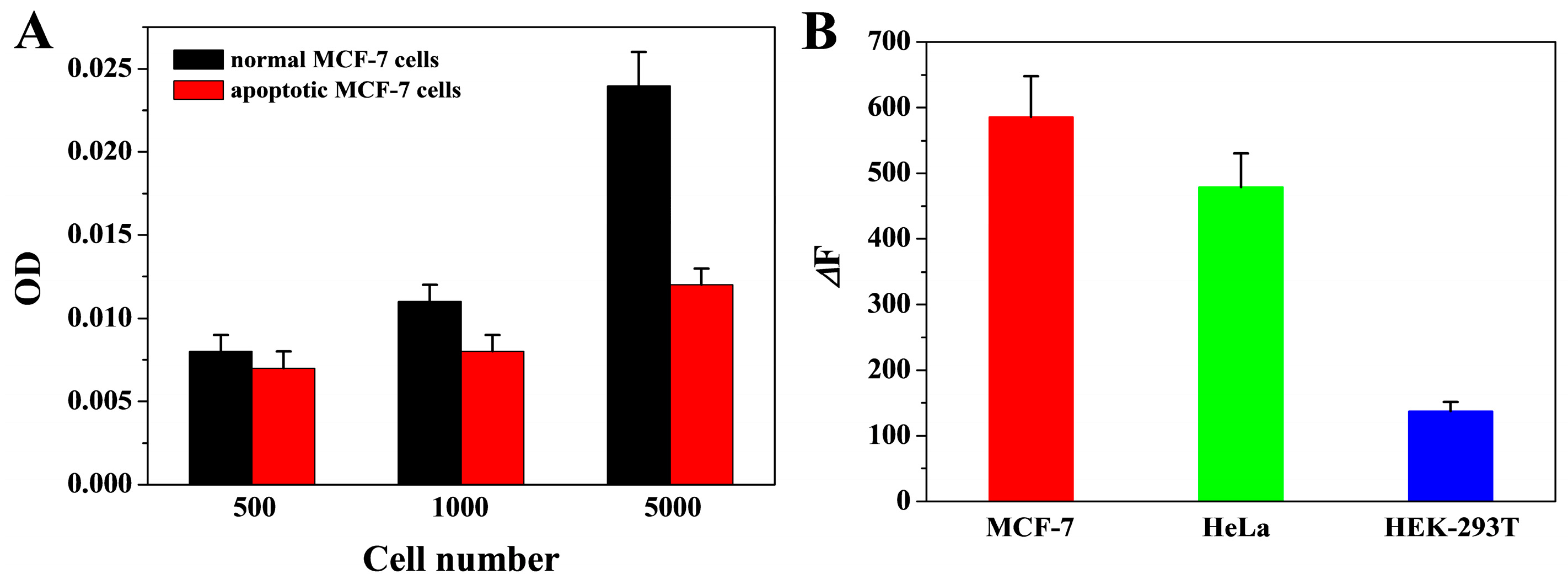
3.7. Real Sample Assays
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rhine, K.; Odeh, H.M.; Shorter, J.; Myong, S. Regulation of biomolecular condensates by poly(ADP-ribose). Chem. Rev. 2023. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Yu, S.-W.; Wang, H.; Poitras, M.F.; Coombs, C.; Bowers, W.J.; Federoff, H.J.; Poirier, G.G.; Dawson, T.M.; Dawson, V.L. Mediation of Poly(ADP-Ribose) Polymerase-1–Dependent Cell Death by Apoptosis-Inducing Factor. Science 2002, 297, 259–263. [Google Scholar] [CrossRef]
- Wang, Y.; An, R.; Umanah, G.K.; Park, H.; Nambiar, K.; Eacker, S.M.; Kim, B.W.; Bao, L.; Harraz, M.M.; Chang, C.; et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 2016, 354, aad6872. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, Z.; Ołdak, Ł.; Gorodkiewicz, E. Methods of PARP-1 determination and its importance in living organisms. Protein Peptide Lett. 2022, 29, 496–504. [Google Scholar]
- Teyssonneau, D.; Margot, H.; Cabart, M.; Anonnay, M.; Sargos, P.; Vuong, N.-S.; Soubeyran, I.; Sevenet, N.; Roubaud, G. Prostate cancer and PARP inhibitors: Progress and challenges. J. Hematol. Oncol. 2021, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, Y.; Zhao, F. Electrochemical methods for the detection of poly(ADP-ribose) polymerase-1. Int. J. Electrochem. Sci. 2021, 16, 21098. [Google Scholar] [CrossRef]
- Wallrodt, S.; Buntz, A.; Wang, Y.; Zumbusch, A.; Marx, A. Bioorthogonally functionalized NAD+ analogues for in-cell visualization of poly(ADP-ribose) formation. Angew. Chem. Int. Ed. 2016, 55, 7660–7664. [Google Scholar] [CrossRef]
- Shuhendler, A.J.; Cui, L.; Chen, Z.; Shen, B.; Chen, M.; James, M.L.; Witney, T.H.; Bazalova-Carter, M.; Gambhir, S.S.; Chin, F.T.; et al. [F-18]-SuPAR: A radiofluorinated probe for noninvasive imaging of DNA damage-dependent poly(ADP-ribose) polymerase activity. Bioconjug. Chem. 2019, 30, 1331–1342. [Google Scholar] [CrossRef]
- Jiang, H.; Kim, J.H.; Frizzell, K.M.; Kraus, W.L.; Lin, H.N. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J. Am. Chem. Soc. 2010, 132, 9363–9372. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Xu, E.; Zhou, Q.; Chen, J.; Wei, W.; Liu, Y.; Liu, S. A label-free PFP-based photoelectrochemical biosensor for highly sensitive detection of PARP-1 activity. Biosens. Bioelectron. 2019, 138, 111308. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, P.; Wang, D.; Liu, Y.; Wei, W.; Zhang, Y.; Liu, S. Quartz crystal microbalance detection of poly(ADP-ribose) polymerase-1 based on gold nanorods signal amplification. Anal. Chem. 2019, 91, 11038–11044. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, E.; Wang, C.; Wei, W.; Liu, Y.; Liu, S. High specificity and efficiency electrochemical detection of poly(ADP-ribose) polymerase-1 activity based on versatile peptide-templated copper nanoparticles and detection array. Anal. Chim. Acta 2019, 1091, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, C.; Wang, Z.; Yang, H.; Wei, W.; Liu, Y.; Liu, S. Renewable electrochemical sensor for PARP-1 activity detection based on host-guest recognition. Biosens. Bioelectron. 2020, 148, 111810. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, J.; Shangguan, L.; Liu, Y.; Wei, Y.; Wei, W.; Liu, S. Ultrasensitive electrochemical detection of poly (ADP-ribose) polymerase-1 via polyaniline deposition. Talanta 2018, 180, 127–132. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, J.; Cao, Y.; Li, G. Gold nanoparticles based colorimetric assay of protein poly(ADP-ribosyl)ation. Analyst 2011, 136, 2044–2046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, K.; Wei, W.; Liu, S. Single-particle assay of poly(ADP-ribose) polymerase-1 activity with dark-field optical microscopy. ACS Sens. 2020, 5, 1198–1206. [Google Scholar] [CrossRef]
- Zhang, D.; Wei, W.; Liu, Y.; Pu, Y.; Liu, S. Dual imaging of poly(ADP-ribose) polymerase1 and endogenous H2O2 for the diagnosis of cancer cells using silver-coated gold nanorods. Anal. Chem. 2021, 93, 16248–16256. [Google Scholar] [CrossRef]
- Wang, C.; Liu, A.; Chen, J.; Liu, S.; Wei, W. Sensitive detection of PARP-1 activity by electrochemical impedance spectroscopy based on biomineralization. Anal. Chim. Acta 2023, 2023, 340937. [Google Scholar] [CrossRef]
- Yang, H.; Fu, F.; Li, W.; Wei, W.; Zhang, Y.; Liu, S. Telomerase and poly(ADP-ribose) polymerase-1 activity sensing based on the high fluorescence selectivity and sensitivity of TOTO-1 towards G bases in single-stranded DNA and poly(ADP-ribose). Chem. Sci. 2019, 10, 3706–3714. [Google Scholar] [CrossRef]
- Xu, E.; Yang, H.; Li, P.; Wang, Z.; Liu, Y.; Wei, W.; Liu, S. Dual-mode detection of PARP-1 by fluorescence andchemiluminescence. Sens. Actuators B Chem. 2021, 330, 129288. [Google Scholar] [CrossRef]
- Wu, S.; Chen, C.; Yang, H.; Wei, W.; Wei, M.; Zhang, Y.; Liu, S. A sensitive fluorescence “turn-off-on” biosensor for poly(ADP-ribose) polymerase-1 detection based on cationic conjugated polymer-MnO2 nanosheets. Sens. Actuators B Chem. 2018, 273, 1047–1053. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, E.; Xu, C.; Wei, W. Colorimetric method for PARP-1 detection based on preventing AuNRs from etching by molybdate. Sens. Actuators B Chem. 2020, 325, 128806. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, X.; Yang, H.; Xu, E.; Wu, S.; Wei, W.; Chen, J. Analysis of poly(ADP-ribose) polymerase-1 by enzyme-initiated auto-PARylation-controlled aggregation of hemin-graphene nanocomposites. Analyst 2018, 143, 2501–2507. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, L.; Wang, Z.; Dai, Z. Stable and reusable electrochemical biosensor for poly(ADP-ribose) polymerase and its inhibitor based on enzyme-initiated auto-PARylation. ACS Appl. Mater. Interfaces 2016, 8, 18669–18674. [Google Scholar] [CrossRef]
- Yang, L.M.; Yin, X.H.; An, B.; Li, F. Precise capture and direct quantification of tumor exosomes via highly efficient dual-aptamer recognition-assisted ratiometric immobilization-free electrochemical strategy. Anal. Chem. 2021, 93, 1709–1716. [Google Scholar] [CrossRef]
- Chang, J.F.; Wang, X.; Wang, J.; Li, H.Y.; Li, F. Nucleic acid functionalized MOFs-based homogeneous electrochemical biosensor for simultaneous detection of multiple tumor biomarkers. Anal. Chem. 2019, 91, 3604–3610. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Bürkle, A.; Hauser, K.; Mangerich, A. Real-time monitoring of PARP1-dependent PARylation by ATR-FTIR spectroscopy. Nat. Commun. 2020, 11, 2174. [Google Scholar] [CrossRef]
- Li, H.Y.; Yang, Q.T.; Wang, Z.X.; Li, F. Iridium complex with specific intercalation in G-quadruplex: A phosphorescence and electrochemiluminescence dual-mode homogeneous biosensor for enzyme-free and label-free detection of microRNA. ACS Sens. 2023, 8, 1529–1535. [Google Scholar] [CrossRef]
- Li, D.; Chen, Y.; Liu, Z. Boronate affinity materials for separation and molecular recognition: Structure, properties and applications. Chem. Soc. Rev. 2015, 44, 8097–8123. [Google Scholar] [CrossRef]
- Bull, S.D.; Davidson, M.G.; Van den Elsen, J.M.H.; Fossey, J.S.; Jenkins, A.T.A.; Jiang, Y.B.; Kubo, Y.; Marken, F.; Sakurai, K.; Zhao, J.; et al. Exploiting the reversible covalent bonding of boronic acids: Recognition, sensing, and assembly. Account. Chem. Res. 2013, 46, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, Z.; Shao, H.; Zhang, R.; Chen, L.; Yang, X. Boronate affinity material-based sensors for recognition and detection of glycoproteins. Analyst 2020, 145, 7511–7527. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Takahashi, S.; Du, X.; Anzai, J.-i. Electrochemical biosensors based on ferroceneboronic acid and its derivatives: A review. Biosensors 2014, 4, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Kedge, J.L.; Fossey, J.S. Molecular boronic acid-based saccharide sensors. ACS Sens. 2021, 6, 1508–1528. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Deng, D.; Zhang, L.; Yuan, B.; Jing, M.; Du, J.; Liu, L. Sandwich-type electrochemical biosensor for glycoproteins detection based on dual-amplification of boronic acid-gold nanoparticles and dopamine-gold nanoparticles. Biosens. Bioelectron. 2013, 43, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, N.; Liu, H.; Kang, X.; Liu, X.; Xue, C.; He, X. Highly sensitive and label-free electrochemical detection of microRNAs based on triple signal amplification of multifunctional gold nanoparticles, enzymes and redox-cycling reaction. Biosens. Bioelectron. 2014, 53, 399–405. [Google Scholar] [CrossRef]
- Hu, Q.; Wan, J.; Wang, H.; Cao, X.; Li, S.; Liang, Y.; Luo, Y.; Wang, W.; Niu, L. Boronate-affinity cross-linking-based ratiometric electrochemical detection of glycoconjugates. Anal. Chem. 2022, 94, 9481–9486. [Google Scholar] [CrossRef]
- Hu, Q.; Feng, W.; Liang, Y.; Liang, Z.; Cao, X.; Li, S.; Luo, Y.; Wan, J.; Ma, Y.; Han, D.; et al. Boronate affinity-amplified electrochemical aptasensing of lipopolysaccharide. Anal. Chem. 2022, 94, 17733–17738. [Google Scholar] [CrossRef]
- Hu, Q.; Hu, S.; Li, S.; Liu, S.; Liang, Y.; Cao, X.; Luo, Y.; Xu, W.; Wang, H.; Wan, J.; et al. Boronate affinity-based electrochemical aptasensor for point-of-care glycoprotein detection. Anal. Chem. 2022, 94, 10206–10212. [Google Scholar] [CrossRef]
- Hu, Q.; Wan, J.; Luo, Y.; Li, S.; Cao, X.; Feng, W.; Liang, Y.; Wang, W.; Niu, L. Electrochemical detection of femtomolar DNA via boronate affinity-mediated decoration of polysaccharides with electroactive tags. Anal. Chem. 2022, 94, 12860–12865. [Google Scholar] [CrossRef]
- Bai, C.-C.; Wang, D.; Liu, M.-X.; Ma, Y.-R.; Sun, Y.; Duan, R.; Dong, L.-Y.; Wang, X.H. Ultrasensitive and specific detection of glycoprotein with boronic acid-modified / fluorescein isothiocyanate-loaded graphene oxide as signal amplification matrix. Sens. Actuators B Chem. 2021, 344, 130327. [Google Scholar] [CrossRef]
- Chang, L.; He, X.; Chen, L.; Zhang, Y. Mercaptophenylboronic acid-capped Mn-doped ZnS quantum dots for highly selective and sensitive fluorescence detection of glycoproteins. Sens. Actuators B Chem. 2017, 243, 72–77. [Google Scholar] [CrossRef]
- Chen, J.; Hao, L.; Hu, J.; Zhu, K.; Li, Y.; Xiong, S.; Huang, X.; Xiong, Y.; Tang, B.Z. A universal boronate-affinity crosslinking-amplified dynamic light scattering immunoassay for point-of-care glycoprotein detection. Angew. Chem. Int. Ed. 2022, 61, e202112031. [Google Scholar]
- Chen, S.; Zhang, X.; Yu, Y.; Wang, J. Boronic acid-containing carbon dots array for sensitive identification of glycoproteins and cancer cells. Chin. Chem. Lett. 2021, 32, 3043–3047. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Wu, N.; Yang, T.; Wang, J.-H. Unusual selective response to glycoprotein over sugar facilitates ultrafast universal fluorescent immunoassay of biomarkers. Anal. Chem. 2020, 92, 5540–5545. [Google Scholar] [CrossRef]
- Kong, T.T.; Zhao, Z.; Li, Y.; Wu, F.; Jin, T.; Tang, B.Z. Detecting live bacteria instantly utilizing AIE strategies. J. Mater. Chem. B 2018, 6, 5986–5991. [Google Scholar] [CrossRef]
- Gao, X.; Yin, Y.; Wu, H.; Hao, Z.; Li, J.; Wang, S.; Liu, Y. Integrated SERS platform for reliable detection and photothermal elimination of bacteria in whole blood samples. Anal. Chem. 2021, 93, 1569–1577. [Google Scholar] [CrossRef]
- Lu, H.; Xu, S. Ultrasensitive turn on molecularly imprinted fluorescence sensor for glycoprotein detection based on nanoparticles signal amplification. Sens. Actuators B Chem. 2020, 306, 127566. [Google Scholar] [CrossRef]
- Son, S.E.; Gupta, P.K.; Hur, W.; Choi, H.; Lee, H.B.; Park, Y.; Seong, G.H. Determination of glycated albumin using a Prussian blue nanozymebased boronate affinity sandwich assay. Anal. Chim. Acta 2020, 1134, 41–49. [Google Scholar] [CrossRef]
- Imbriano, A.; Tricase, A.; Macchia, E.; Torsi, L.; Bollella, P. Self-powered logically operated fluorescent detection of hepatitis B virus (HBV). Anal. Chim. Acta 2023, 1252, 341037. [Google Scholar] [CrossRef]
- Mikagi, A.; Tsurufusa, R.; Tsuchido, Y.; Hashimoto, T.; Hayashita, T. Fast and sensitive bacteria detection by boronic acid modified fluorescent dendrimer. Sensors 2021, 21, 3115. [Google Scholar] [CrossRef]
- Sukla, S.; Mondal, P.; Biswas, S.; Ghosh, S. A rapid and easy-to-perform method of nucleic-acid based dengue virus diagnosis using fluorescence-based molecular beacons. Biosensors 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Oh, Y.-H.; Ko, D.-H.; Sung, H.; Oh, H.-B.; Hwang, S.-H. Nanoparticle-based visual detection of amplified DNA for diagnosis of hepatitis C virus. Biosensors 2022, 12, 744. [Google Scholar] [CrossRef]
- Heinisch, T.; Ward, T.R. Artificial metalloenzymes based on the biotin−streptavidin technology: Challenges and opportunities. Account. Chem. Res. 2016, 49, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Dundas, C.M.; Demonte, D.; Park, S. Streptavidin–biotin technology: Improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 2013, 97, 9343–9353. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Liu, L.; Harrington, M.G.; Wang, J.; Zhou, F. Regenerable and simultaneous surface plasmon resonance detection of Ab(1-40) and Ab(1-42) peptides in cerebrospinal fluids with signal amplification by streptavidin conjugated to an N-terminus-specific antibody. Anal. Chem. 2010, 82, 10151–10157. [Google Scholar] [CrossRef]
- Bearden, S.; Wang, F.; Hall, A.R. Simple and efficient room-temperature release of biotinylated nucleic acids from streptavidin and its application to selective molecular detection. Anal. Chem. 2019, 91, 7996–8001. [Google Scholar] [CrossRef]
- Xia, N.; Wu, D.; Sun, T.; Wang, Y.; Ren, X.; Zhao, F.; Liu, L.; Yi, X. Magnetic bead-based electrochemical and colorimetric methods for the detection of poly(ADP-ribose) polymerase-1 with boronic acid derivatives as the signal probes. Sens. Actuators B Chem. 2021, 327, 128913. [Google Scholar] [CrossRef]
- Deng, D.; Hao, Y.; Yang, P.; Xia, N.; Yu, W.; Liu, X.; Liu, L. Single-labeled peptide substrates for detection of protease activity based on the inherent fluorescence quenching ability of Cu2+. Anal. Methods 2019, 11, 1248–1253. [Google Scholar] [CrossRef]
- Zhu, L.; Chang, Y.; Li, Y.; Qiao, M.; Liu, L. Biosensors based on the binding events of nitrilotriacetic acid–metal complexes. Biosensors 2023, 13, 507. [Google Scholar] [CrossRef]
- Xia, N.; Wu, D.; Yu, H.; Sun, W.; Yi, X.; Liu, L. Magnetic bead-based electrochemical and colorimetric assays of circulating tumor cells with boronic acid derivatives as the recognition elements and signal probes. Talanta 2021, 221, 121640. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cheng, C.; Chang, Y.; Ma, H.; Hao, Y. Two sensitive electrochemical strategies for the detection of protein kinase activity based on the 4-mercaptophenylboronic acid-induced in situ assembly of silver nanoparticles. Sens. Actuators B Chem. 2017, 248, 178–186. [Google Scholar] [CrossRef]
- Wu, S.; Wei, M.; Yang, H.; Fan, J.; Wei, W.; Zhang, Y.; Liu, S. Counter ions-mediated gold nanorods-based sensor for label-free detection of poly(ADP-ribose) polymerase-1 activity and its inhibitor. Sens. Actuators B Chem. 2018, 259, 565–572. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, J.; Yang, H.; Xu, E.; Wei, W.; Zhang, Y.; Liu, S. Detection of PARP-1 activity based on hyperbranched-poly (ADP-ribose) polymers responsive current in artificial nanochannels. Biosens. Bioelectron. 2018, 113, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Yang, H.; Wu, L.; Chen, J.; Wei, W.; Liu, Y.; Liu, S. Label-free poly(ADP-ribose) polymerase-1 activity assay based on perpendicular orientated mesoporous silica films. Sens. Actuators B Chem. 2019, 294, 185–191. [Google Scholar] [CrossRef]
- Desroches, A.; Denault, J.B. Caspase-7 uses RNA to enhance proteolysis of poly(ADP-ribose) polymerase 1 and other RNA-binding proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 21521–21528. [Google Scholar] [CrossRef]
- Koşar, P.A.; Tuncer, H.; Uğuz, A.C.; Palma, J.E.; Darıcı, H.; Onaran, I.; Çiğ, B.; Koşar, A.; Moratinos, A.B.R. The efficiency of poly(ADP-ribose) polymerase (PARP) cleavage on detection of apoptosis in an experimental model of testicular torsion. Int. J. Exp. Pathol. 2015, 96, 294–300. [Google Scholar] [CrossRef]


| Method | Signal Reporter | Linear Range | Detection Limit | Ref. |
|---|---|---|---|---|
| photoelectrochemistry | PFP | 0.01~2 U | 0.007 U | [11] |
| QCM | CTAB-GNRs | 0.06~1.2 nM | 0.04 nM | [12] |
| colorimetry | NAD-AuNPs | 0.43~1.74 nM | 0.32 nM | [16] |
| colorimetry | CTAB-GNRs | 0.05~1.0 U | 0.006 U | [63] |
| colorimetry | hemin-graphene | 0.05~1.0 U | 0.003 U | [24] |
| chemiluminescence | AuNCs | 0.01~1.0 U | 0.009 U | [21] |
| electrochemistry | MBs/FcBA | 0.1~50 U | 0.1 U | [58] |
| electrochemistry | [Ru(NH3)6]3+ | 0.01~1 U | 0.003 U | [25] |
| electrochemistry | Polyaniline | 0.005~1.0 U | 0.002 U | [15] |
| electrochemistry | Artificial nanochannels | 0.05~1.5 U | 0.006 U | [64] |
| electrochemistry | NH2-MSFs | 0.01~1.2 U | 0.005 U | [65] |
| electrochemistry | P-CuNPs | 0.01~1U | 0.004 U | [13] |
| electrochemistry | PMo12O403− | 0.01~1.0 U | 0.008 U | [14] |
| fluorescence | PFP/MnO2 | 0.024~1.2 nM | 0.003 nM | [22] |
| fluorescence | TOTO-1 | 0.02~1.5 U | 0.02 U | [20] |
| fluorescence | FITC-PBA | 0.01~20 U | 0.01 U | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, F.; Liu, G.; Qiao, Y.; Dong, X.; Liu, L. Streptavidin-Conjugated DNA for the Boronate Affinity-Based Detection of Poly(ADP-Ribose) Polymerase-1 with Improved Sensitivity. Biosensors 2023, 13, 723. https://doi.org/10.3390/bios13070723
Gao F, Liu G, Qiao Y, Dong X, Liu L. Streptavidin-Conjugated DNA for the Boronate Affinity-Based Detection of Poly(ADP-Ribose) Polymerase-1 with Improved Sensitivity. Biosensors. 2023; 13(7):723. https://doi.org/10.3390/bios13070723
Chicago/Turabian StyleGao, Fengli, Gang Liu, Yishu Qiao, Xiuwen Dong, and Lin Liu. 2023. "Streptavidin-Conjugated DNA for the Boronate Affinity-Based Detection of Poly(ADP-Ribose) Polymerase-1 with Improved Sensitivity" Biosensors 13, no. 7: 723. https://doi.org/10.3390/bios13070723
APA StyleGao, F., Liu, G., Qiao, Y., Dong, X., & Liu, L. (2023). Streptavidin-Conjugated DNA for the Boronate Affinity-Based Detection of Poly(ADP-Ribose) Polymerase-1 with Improved Sensitivity. Biosensors, 13(7), 723. https://doi.org/10.3390/bios13070723






