Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device
Abstract
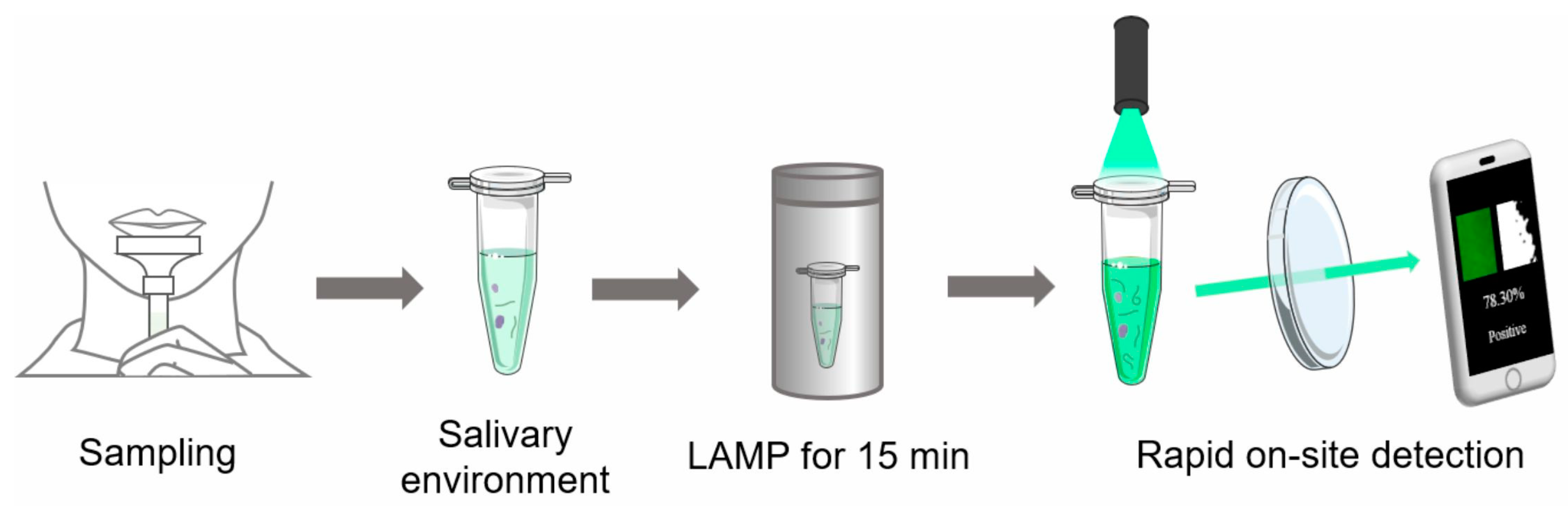
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Salivary Environment
2.3. Primer Design
2.4. RT-LAMP Assay
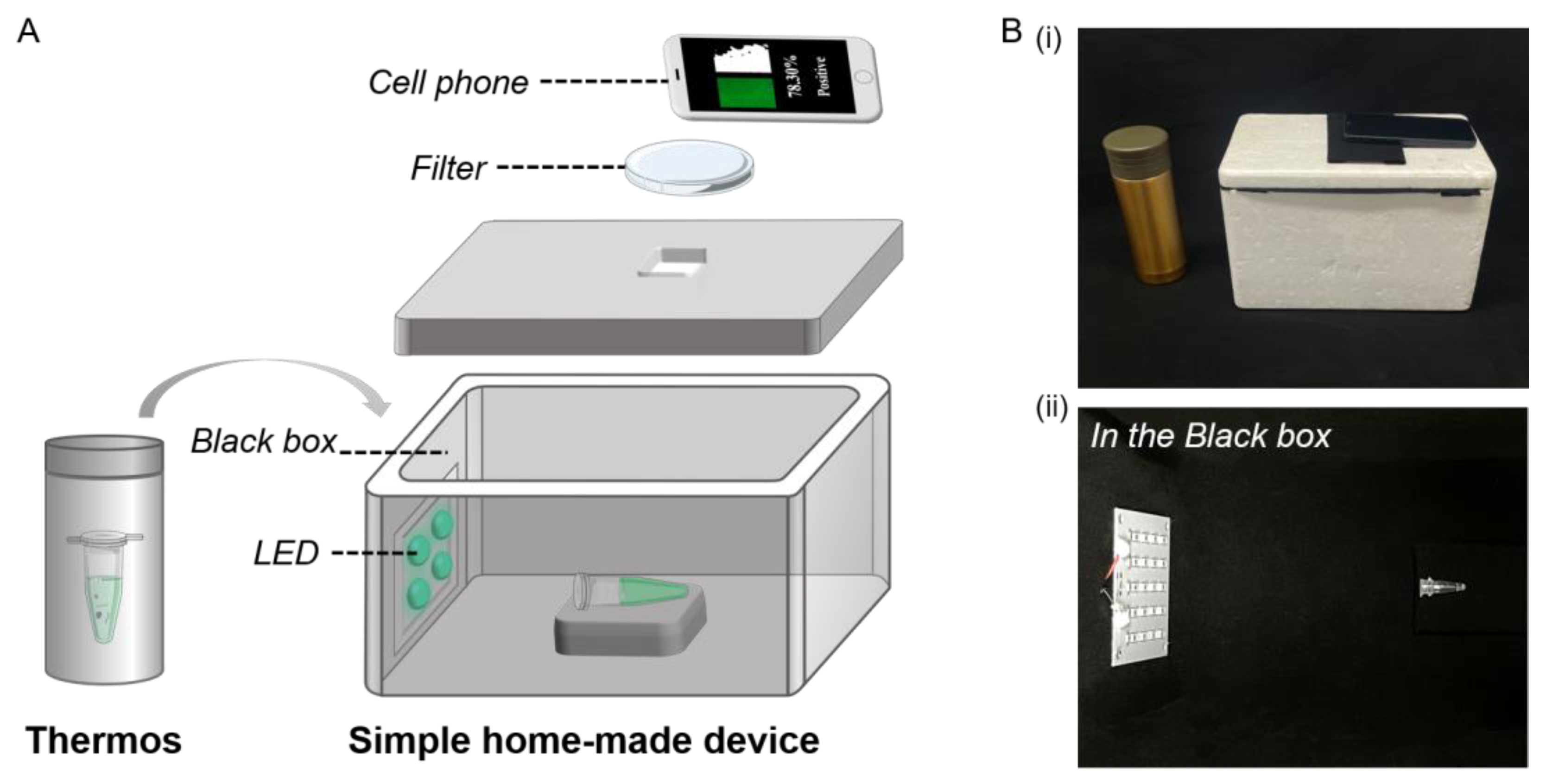
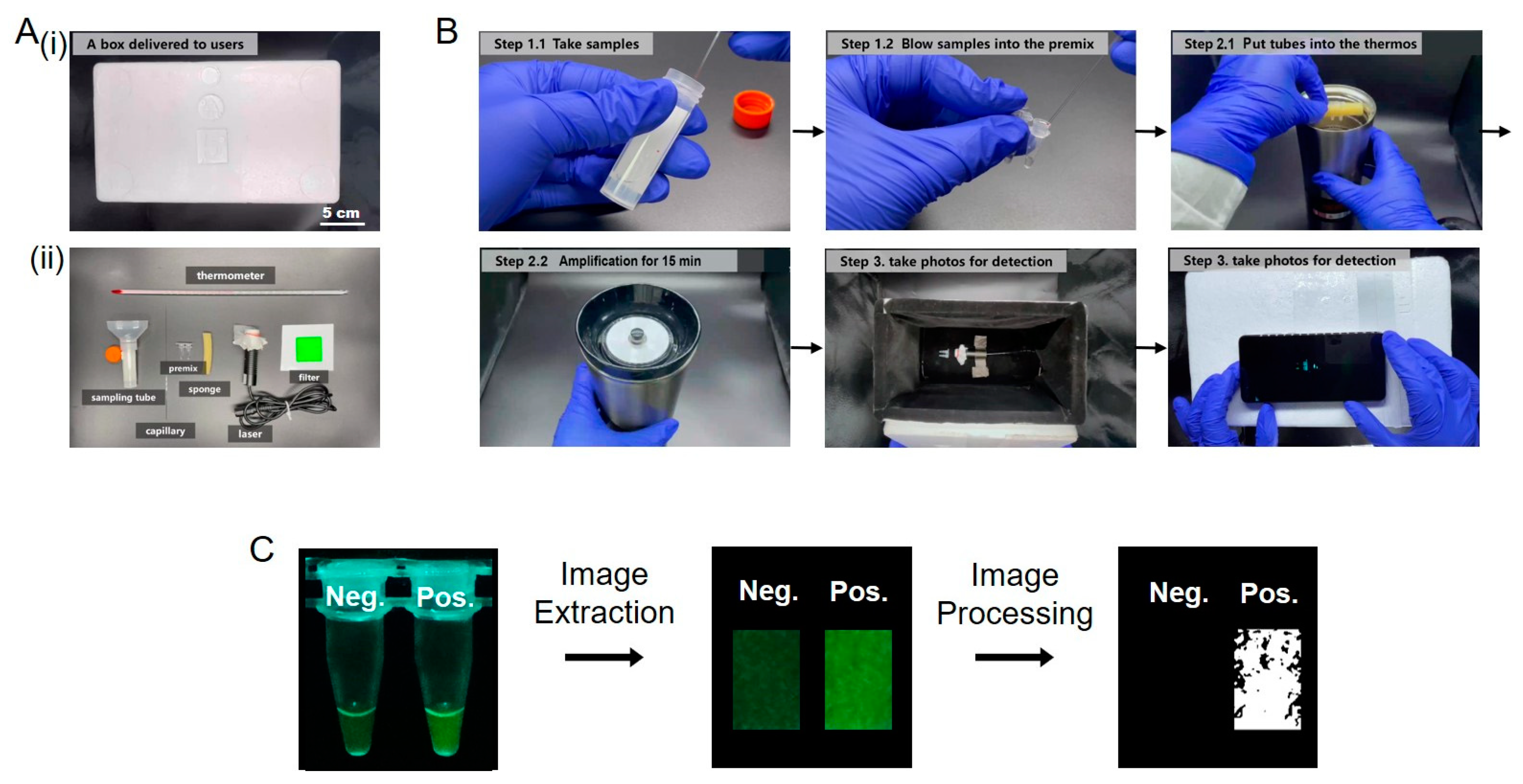
2.5. Detection Device
3. Results and Discussion
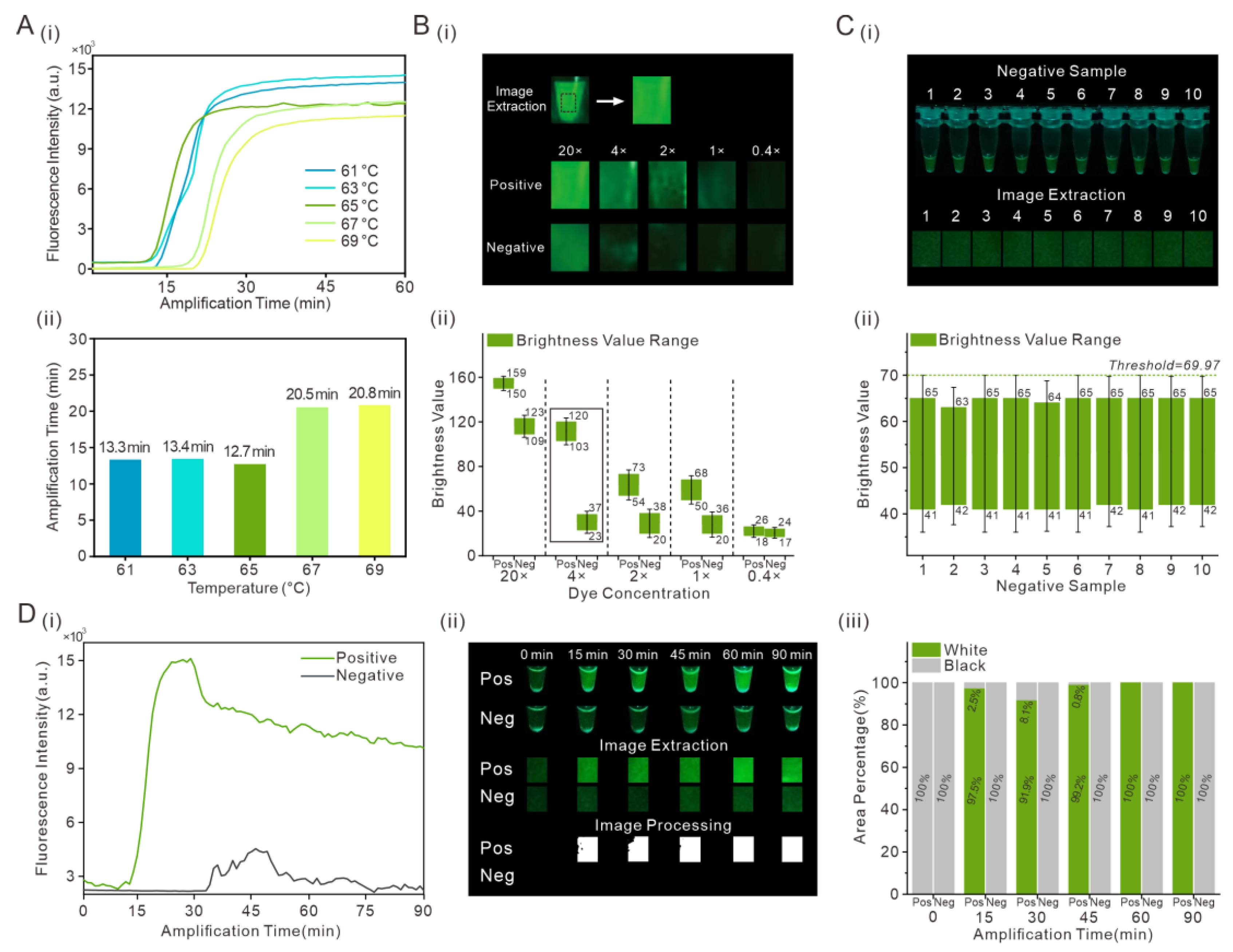
3.1. Optimization of RT-LAMP Assays and Determination of Brightness Threshold
3.2. Evaluation of the Detection of SARS-CoV-2 in the Portable Low-Cost Device
3.3. Optimization of the Portable Low-Cost Device
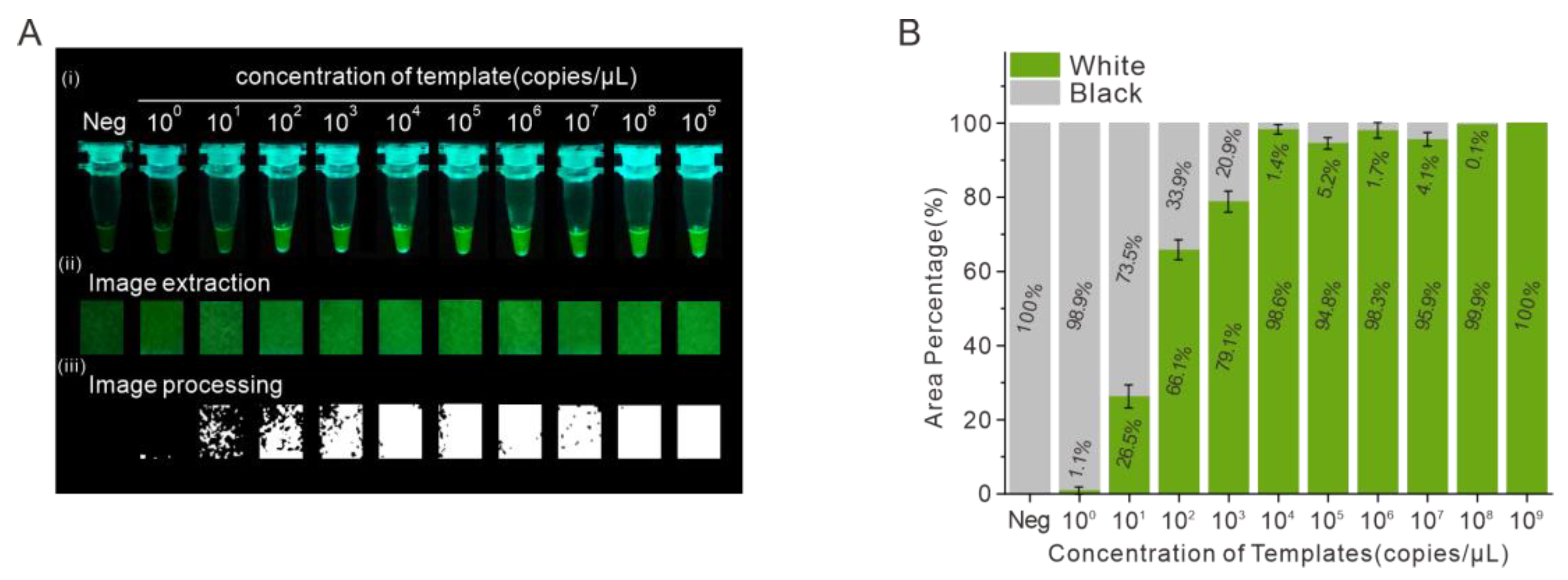
3.4. Detection of SARS-CoV-2 RNA Using the On-Site Detection Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, J.; Li, S.; Peng, Z.; Xiao, Z.; Wang, X.; Yan, R.; Luo, J. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel. Med. Infect. Dis. 2020, 37, 101673. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.P.; Peng, Y.S.; Huang, B.Y.; Ding, X.; Wang, X.Y.; Niu, P.H.; Meng, J.; Zhu, Z.Z.; Zhang, Z.; Wang, J.Y.; et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Lu, R.J.; Zhao, X.; Li, J.; Niu, P.H.; Yang, B.; Wu, H.L.; Wang, W.L.; Song, H.; Huang, B.Y.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Ma, Y.D.; Chen, Y.S.; Lee, G.B. An integrated self-driven microfluidic device for rapid detection of the influenza A (H1N1) virus by reverse transcription loop-mediated isothermal amplification. Sens. Actuators B Chem. 2019, 296, 126647. [Google Scholar] [CrossRef]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the road for rapid detection and point-of care diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Patrice, F.; Manuela, T.; Jonathan, H.; Eve-Julie, B.; Boehme, C.C.; Tsugunori, N.; Mark, D.P.; Jacques, S. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef]
- Yu, L.; Wu, S.S.; Hao, X.W.; Dong, X.; Mao, L.L.; Pelechano, V.; Chen, W.H.; Yin, X.S. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020, 7, 975–977. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef]
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Jin, X.; Zhang, C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 2826. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xu, G.; Reboud, J.; Kasprzyk-Hordern, B.; Cooper, J.M. Monitoring genetic population biomarkers for wastewater-based epidemiology. Anal. Chem. 2017, 89, 9941–9945. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Mauk, M.G.; Hackett, B.A.; Cherry, S.; Bau, H.H.; Liu, C. Instrument-free point-of-care molecular detection of Zika virus. Anal. Chem. 2016, 88, 7289–7294. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Ganguli, A.; Nguyen, J.; Brisbin, R.; Shanmugam, K.; Hirschberg, D.L.; Wheeler, M.B.; Bashir, R.; Nash, D.M.; Cunningham, B.T. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip 2020, 20, 1621. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; Williamson, D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00720–e00776. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Geng, B.; Muenker, M.C.; Moore, A.J.; Vogels, C.B.F.; et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2 Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Ning, B.; Yu, T.; Zhang, S.W.; Huang, Z.; Tian, D.; Lin, Z.; Niu, A.; Golden, N.; Hensley, K.; Threeton, B.; et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2021, 7, eabe3703. [Google Scholar] [CrossRef]
- To, K.K.W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.Y.; Cai, J.P.; Chan, J.M.C.; Chik, T.S.H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Choi, J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A. Molecular diagnostic technologies for COVID-19: Limitations and challenges. J. Adv. Res. 2020, 26, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.J.; Feng, J.P.; Guo, M.; Hu, W.J.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.H.; Wang, X. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2021, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Dorlass, E.G.; Monteiro, C.O.; Viana, A.O.; Soares, C.P.; Guaragna, M.R.R.; Thomazelli, L.M.; Araujo, D.B.; Leal, F.B.; Candido, E.D.; Telezynski, B.L.; et al. Lower cost alternatives for molecular diagnosis of COVID-19: Conventional RT-PCR and SYBR Green-based RT-qPCR. Braz. J. Microbiol. 2020, 51, 1117–1123. [Google Scholar] [CrossRef]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.J.; Jia, H.D.; Wang, Y.; Zeng, Y.D.; Ji, M.M. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.; Pang, X.; Su, Z.; Yang, Y.; Liu, Y.; Zhang, Z.; Fu, Y.; Wang, J.; Zhou, J. Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device. Biosensors 2023, 13, 724. https://doi.org/10.3390/bios13070724
Fu Q, Pang X, Su Z, Yang Y, Liu Y, Zhang Z, Fu Y, Wang J, Zhou J. Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device. Biosensors. 2023; 13(7):724. https://doi.org/10.3390/bios13070724
Chicago/Turabian StyleFu, Quanying, Xueyuan Pang, Zhenning Su, Yuxiao Yang, Yiren Liu, Ziyue Zhang, Yuqiu Fu, Jiasi Wang, and Jianhua Zhou. 2023. "Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device" Biosensors 13, no. 7: 724. https://doi.org/10.3390/bios13070724
APA StyleFu, Q., Pang, X., Su, Z., Yang, Y., Liu, Y., Zhang, Z., Fu, Y., Wang, J., & Zhou, J. (2023). Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device. Biosensors, 13(7), 724. https://doi.org/10.3390/bios13070724





