Biomolecular Monitoring Tool Based on Lab-on-Chip for Virus Detection
Abstract
1. Introduction
2. Materials and Methods
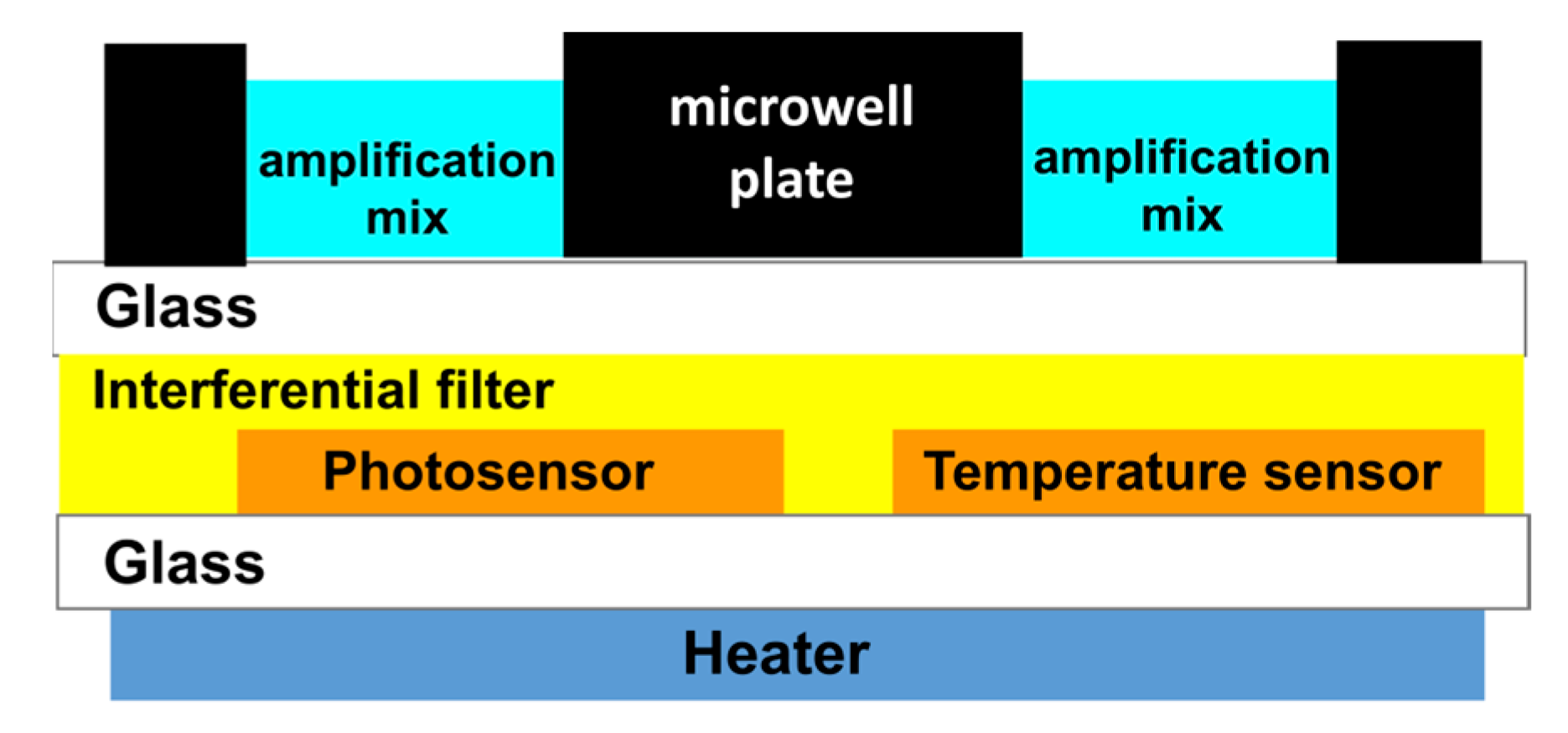
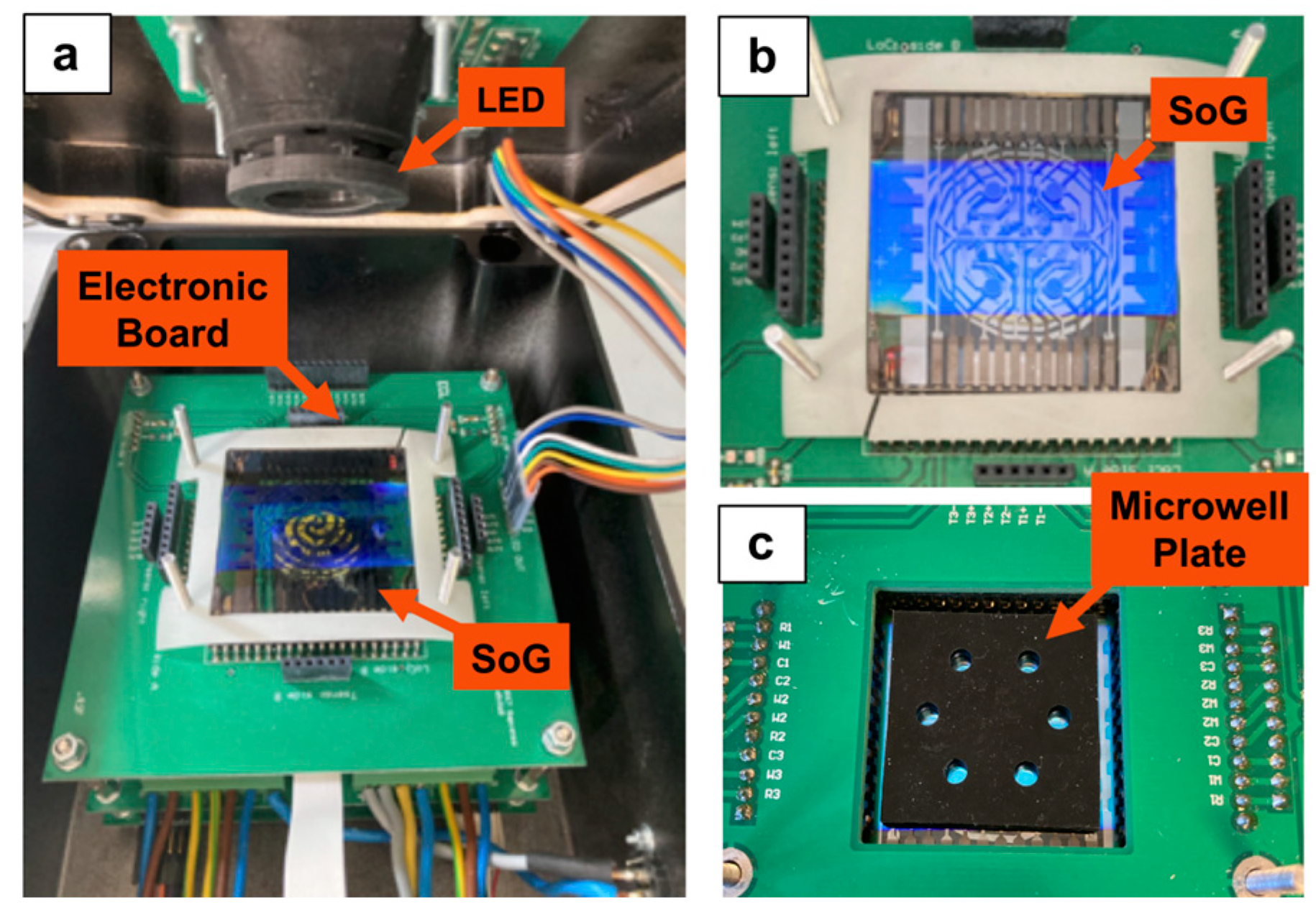
2.1. Fabrication of the System on Glass
- Immediately prior to use, glass slides of 2.8 × 3.2 cm2 were cleaned with ethanol and dried with a stream of nitrogen and subsequently irradiated for 10 min under UV/ozone (Diener Electronic, Ebhausen, Germany).
- A PDMS mixture containing curing agent SYLGARDTM 184 silicon elastomers in a ratio of 10:1 is placed in a vacuum chamber to eliminate the air bubbles.
- The PDMS (0.5 mL) mixture is spin-coated on a 5 × 5 cm2 glass slide having 1 mm thickness (previously cleaned with ethanol and dried with a stream of nitrogen) by using laurel WS-650-23 (Landsdale, PA, USA).
- The bottom side of the black PDMS microwell is placed on the PDMS spin-coated glass slide for a few minutes. Subsequently, it is removed and placed on the ultraclean 0.2 mm glass slides.
- The glass-PDMS microwell is placed in the vacuum chamber for at least 10 min to remove the gas bubbles which may form between glass and PDMS during the bonding. After degassing, the glass-PDMS microwell is heated at 100 °C for 30 min to finalize the bonding.
2.2. WMV Diagnosis
2.2.1. RNA Extraction
2.2.2. Real-Time RT-PCR by BIO-RAD CFX96
2.2.3. Real-Time RT-PCR-LoC System
2.3. SARS-CoV-2 Diagnosis
2.3.1. RNA Extraction
2.3.2. Real-Time RT-PCR by the BIO-RAD CFX96 and LoC System
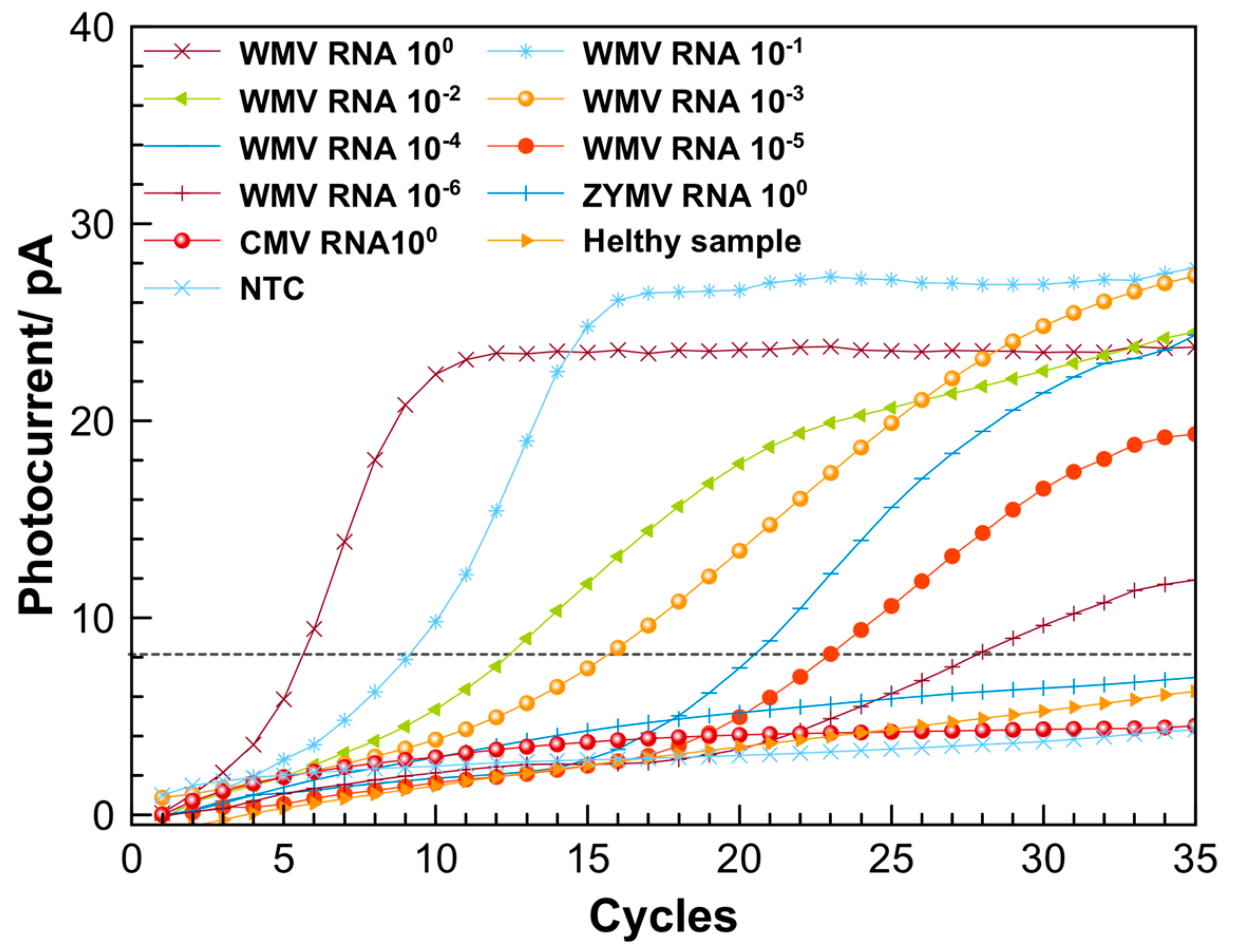
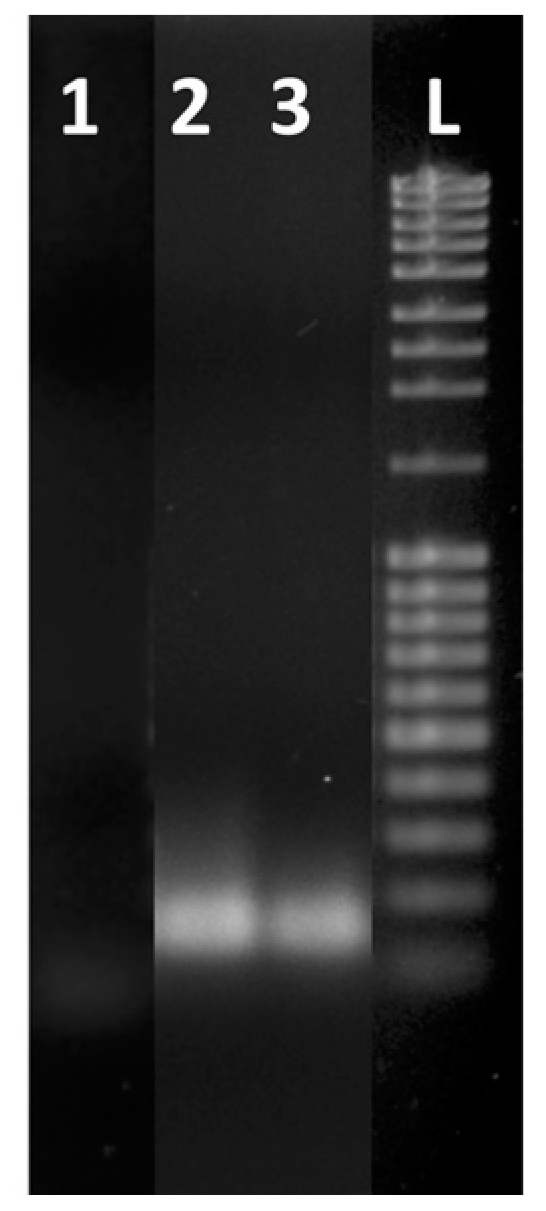
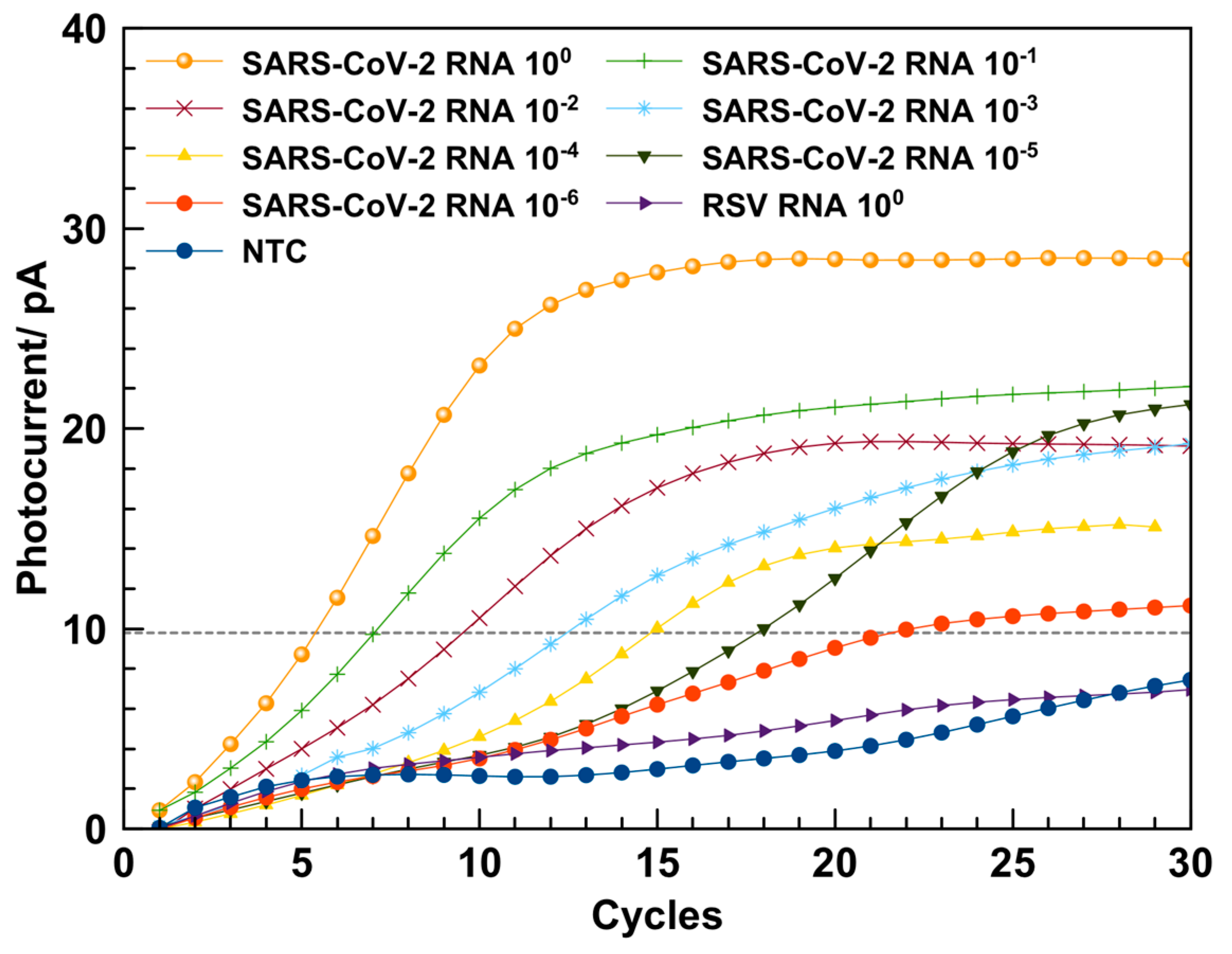
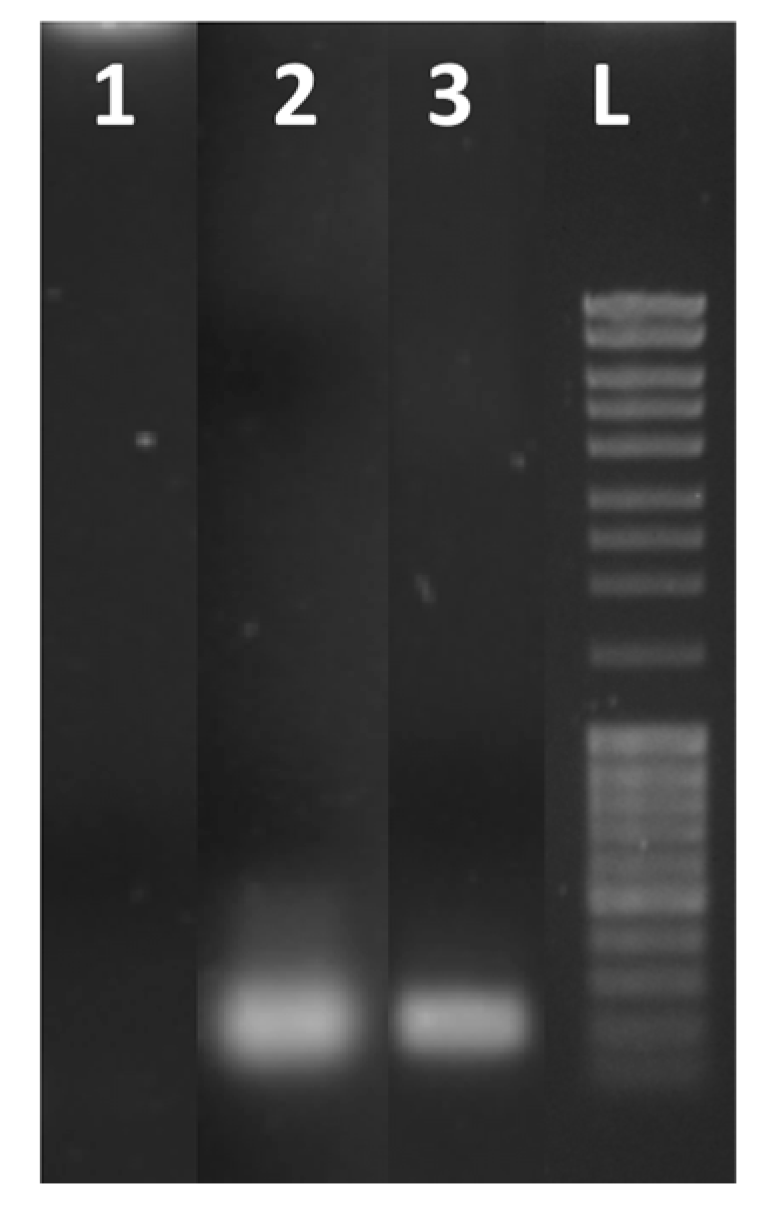
3. Results and Discussion
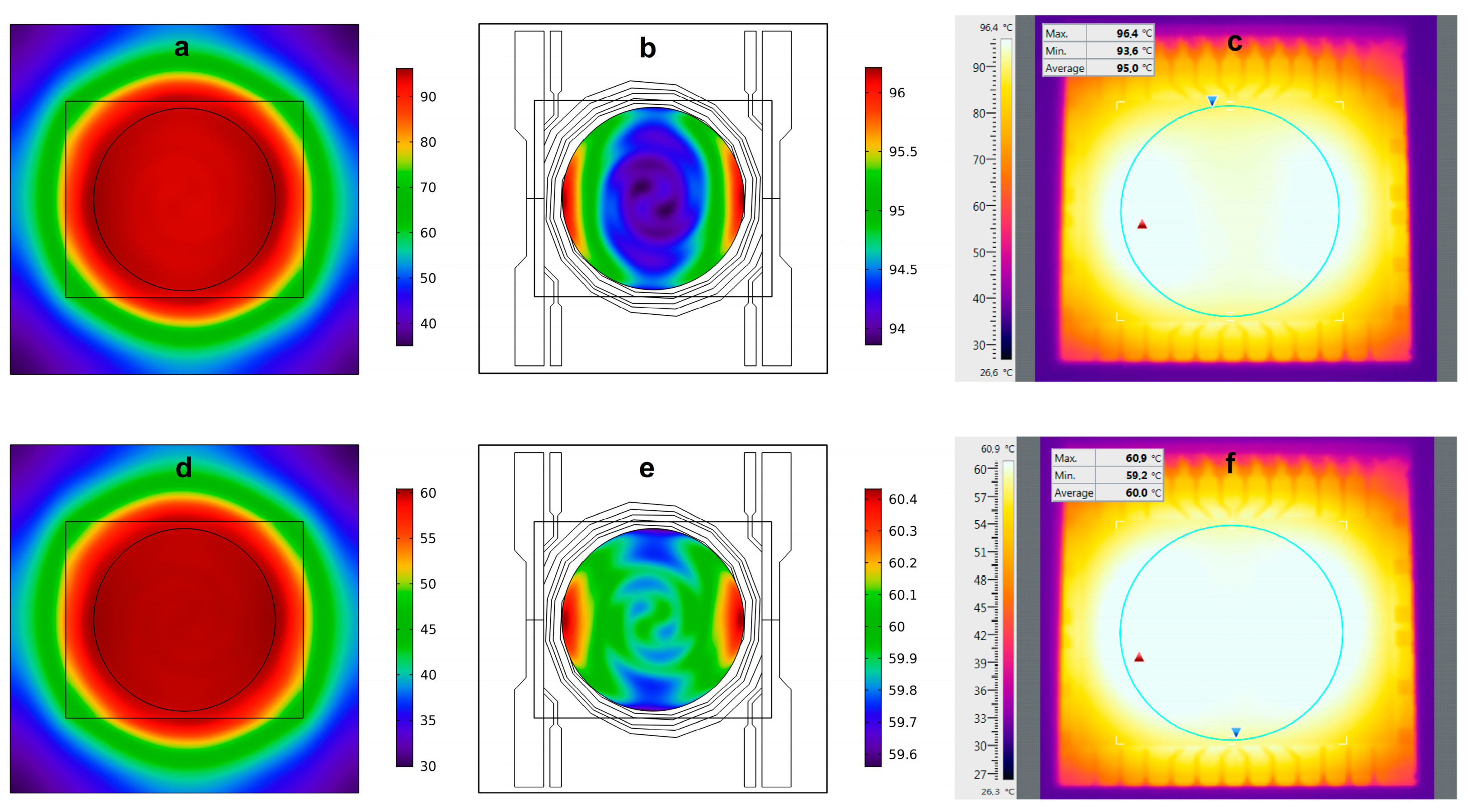
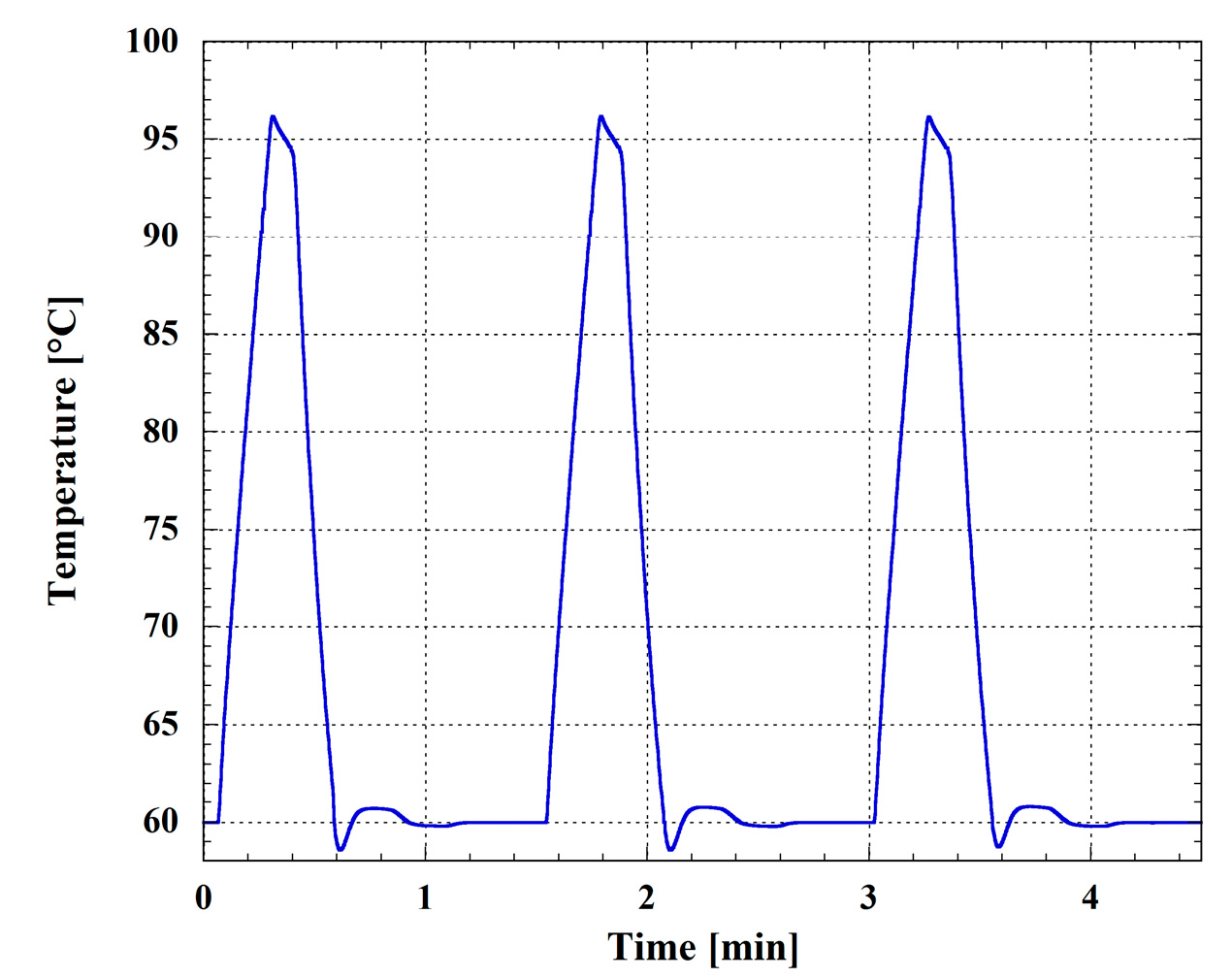
3.1. Temperature Generation and Control on the SoG
3.2. Optimization of Reaction Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- CHeid, A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real Time Quantitative PCR. Exp. Mol. Med. 2001, 33, 101–109. [Google Scholar]
- Tiberini, A.; Manglli, A.; Taglienti, A.; Vučurović, A.; Brodarič, J.; Ferretti, L.; Luigi, M.; Gentili, A.; Mehle, N. Development and Validation of a One-Step Reverse Transcription Real-Time PCR Assay for Simultaneous Detection and Identification of Tomato Mottle Mosaic Virus and Tomato Brown Rugose Fruit Virus. Plants 2022, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Safdar, M.; Abasıyanık, M.F. Simultaneous Identification of Pork and Poultry Origins in Pet Foods by a Quick Multiplex Real-Time PCR Assay Using Evagreen Florescence Dye. Appl. Biochem. Biotechnol. 2013, 171, 1855–1864. [Google Scholar] [CrossRef]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Estevez, M.C.; Alvarez, M.; Lechuga, L.M. Integrated Optical Devices for Lab-on-a-Chip Biosensing Applications. Laser Photonics Rev. 2012, 6, 463–487. [Google Scholar] [CrossRef]
- Dutse, S.W.; Yusof, N.A. Microfluidics-Based Lab-on-Chip Systems in DNA-Based Biosensing: An Overview. Sensors 2011, 11, 5754–5768. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Hashsham, S.A. Miniaturized Nucleic Acid Amplification Systems for Rapid and Point-of-Care Diagnostics: A Review. Anal. Chim. Acta 2012, 733, 1–15. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, D. Miniaturized PCR Chips for Nucleic Acid Amplification and Analysis: Latest Advances and Future Trends. Nucleic Acids Res. 2007, 35, 4223–4237. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, J.; Ma, W.; Zheng, W. PCR Microfluidic Devices for DNA Amplification. Biotechnol. Adv. 2006, 24, 243–284. [Google Scholar] [CrossRef]
- Gorgannezhad, L.; Sreejith, K.R.; Zhang, J.; Kijanka, G.; Christie, M.; Stratton, H.; Nguyen, N.T. Microfluidic Array Chip for Parallel Detection of Waterborne Bacteria. Micromachines 2019, 10, 883. [Google Scholar] [CrossRef]
- Zhang, Y.; Ozdemir, P. Microfluidic DNA Amplification—A Review. Anal. Chim. Acta 2009, 638, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Z.; Corstjens, P.L.A.M.; Mauk, M.G.; Bau, H.H. A Disposable Microfluidic Cassette for DNA Amplification and Detection. Lab Chip 2006, 6, 46–53. [Google Scholar] [CrossRef]
- Qiu, X.; Mauk, M.G.; Chen, D.; Liu, C.; Bau, H.H. A Large Volume, Portable, Real-Time PCR Reactor. Lab Chip 2010, 10, 3170–3177. [Google Scholar] [CrossRef] [PubMed]
- Velve Casquillas, G.; Fu, C.; Le Berre, M.; Cramer, J.; Meance, S.; Plecis, A.; Baigl, D.; Greffet, J.J.; Chen, Y.; Piel, M.; et al. Fast Microfluidic Temperature Control for High Resolution Live Cell Imaging. Lab Chip 2011, 11, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Moschou, D.; Vourdas, N.; Kokkoris, G.; Papadakis, G.; Parthenios, J.; Chatzandroulis, S.; Tserepi, A. All-Plastic, Low-Power, Disposable, Continuous-Flow PCR Chip with Integrated Microheaters for Rapid DNA Amplification. Sens. Actuators B Chem. 2014, 199, 470–478. [Google Scholar] [CrossRef]
- Fuchiwaki, Y.; Nagai, H.; Saito, M.; Tamiya, E. Ultra-Rapid Flow-through Polymerase Chain Reaction Microfluidics Using Vapor Pressure. Biosens. Bioelectron. 2011, 27, 88–94. [Google Scholar] [CrossRef]
- Wheeler, E.K.; Hara, C.A.; Frank, J.; Deotte, J.; Hall, S.B.; Benett, W.; Spadaccini, C.; Beer, N.R. Under-Three Minute PCR: Probing the Limits of Fast Amplification. Analyst 2011, 136, 3707–3712. [Google Scholar] [CrossRef]
- Wang, Q.; Tan, Y.; Gong, H. An Integrated System for Real-Time Pcr Analysis Based on Microfluidic Biochip. Int. J. Comput. Eng. Sci. 2003, 4, 285–288. [Google Scholar] [CrossRef]
- Houssin, T.; Cramer, J.; Grojsman, R.; Bellahsene, L.; Colas, G.; Moulet, H.; Minnella, W.; Pannetier, C.; Leberre, M.; Plecis, A.; et al. Ultrafast, Sensitive and Large-Volume on-Chip Real-Time PCR for the Molecular Diagnosis of Bacterial and Viral Infections. Lab Chip 2016, 16, 1401–1411. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Manz, A.; Chung, B.G. Polymerase Chain Reaction in Microfluidic Devices. Lab Chip 2016, 16, 3866–3884. [Google Scholar] [CrossRef]
- Shi, B.; Li, Y.; Wu, D.; Wu, W. A Handheld Continuous-Flow Real-Time Fluorescence QPCR System with a PVC Microreactor. Analyst 2020, 145, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, N.; Rui, Z.; Liu, H.B.; Dai, C.C.; Kaushik, R.; Ratnaharika, B.; Gong, H.Q. Real-Time PCR-Based Microfluidic Array Chip for Simultaneous Detection of Multiple Waterborne Pathogens. Sens. Actuators B Chem. 2010, 145, 543–552. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Ilic, B.R.; Manz, A.; Neužil, P. Handheld Real-Time PCR Device. Lab Chip 2016, 16, 586–592. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Jiang, Y.; Shi, B.; Wu, D.; Wu, W. Low-Cost Battery-Powered and User-Friendly Real-Time Quantitative PCR System for the Detection of Multigene. Micromachines 2020, 11, 435. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, D.; Li, Y. Micropumps, Microvalves, and Micromixers within PCR Microfluidic Chips: Advances and Trends. Biotechnol. Adv. 2007, 25, 483–514. [Google Scholar] [CrossRef]
- Xiang, Q.; Xu, B.; Fu, R.; Li, D. Real Time PCR on Disposable PDMS Chip with a Miniaturized Thermal Cycler. Biomed. Microdevices 2005, 7, 273–279. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, J.; Wu, X.; Zhu, C.; Liu, Y.; Wang, A.; Deng, G.; Zhu, L. A Rapid Microfluidic Platform with Real-Time Fluorescence Detection System for Molecular Diagnosis. Biotechnol. Biotechnol. Equip. 2019, 33, 223–230. [Google Scholar] [CrossRef]
- Dong, X.; Liu, L.; Tu, Y.; Zhang, J.; Miao, G.; Zhang, L.; Ge, S.; Xia, N.; Yu, D.; Qiu, X. Rapid PCR Powered by Microfluidics: A Quick Review under the Background of COVID-19 Pandemic. TrAC-Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef]
- Lovecchio, N.; Costantini, F.; Nascetti, A.; de Cesare, G.; Caputo, D. Thin-Film-Based Multifunctional System for Optical Detection and Thermal Treatment of Biological Samples. Biosensors 2022, 12, 969. [Google Scholar] [CrossRef]
- Petrucci, G.; Caputo, D.; Lovecchio, N.; Costantini, F.; Legnini, I.; Bozzoni, I.; Nascetti, A.; de Cesare, G. Multifunctional System-on-Glass for Lab-on-Chip Applications. Biosens. Bioelectron. 2017, 93, 315–321. [Google Scholar] [CrossRef]
- Bruijns, B.B.; Costantini, F.; Lovecchio, N.; Tiggelaar, R.M.; Di Timoteo, G.; Nascetti, A.; de Cesare, G.; Gardeniers, J.G.E.; Caputo, D. On-Chip Real-Time Monitoring of Multiple Displacement Amplification of DNA. Sens. Actuators B Chem. 2019, 293, 16–22. [Google Scholar] [CrossRef]
- Mirasoli, M.; Bonvicini, F.; Lovecchio, N.; Petrucci, G.; Zangheri, M.; Calabria, D.; Costantini, F.; Roda, A.; Gallinella, G.; Caputo, D.; et al. On-Chip LAMP-BART Reaction for Viral DNA Real-Time Bioluminescence Detection. Sens. Actuators B Chem. 2018, 262, 1024–1033. [Google Scholar] [CrossRef]
- Costantini, F.; Tiggelaar, R.M.; Salvio, R.; Nardecchia, M.; Schlautmann, S.; Manetti, C.; Gardeniers, H.J.G.E.; de Cesare, G.; Caputo, D.; Nascetti, A. An All-Glass Microfluidic Network with Integrated Amorphous Silicon Photosensors for on-Chip Monitoring of Enzymatic Biochemical Assay. Biosensors 2017, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Lovecchio, N.; Costantini, F.; Parisi, E.; Nardecchia, M.; Tucci, M.; Nascetti, A.; De Cesare, G.; Caputo, D. Integrated Optoelectronic Device for Detection of Fluorescent Molecules. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Lovecchio, N.; Caputo, D.; Costantini, F.; Meo, V.D.; Nascetti, A.; de Cesare, G. On the Stability of Amorphous Silicon Temperature Sensors. IEEE Trans. Electron Devices 2020, 67, 3348–3354. [Google Scholar] [CrossRef]
- Nandimandalam, M.; Costantini, F.; Lovecchio, N.; Iannascoli, L.; Nascetti, A.; de Cesare, G.; Caputo, D.; Manetti, C. Split Aptamers Immobilized on Polymer Brushes Integrated in a Lab-on-Chip System Based on an Array of Amorphous Silicon Photosensors: A Novel Sensor Assay. Materials 2021, 14, 7210. [Google Scholar] [CrossRef]
- Manglli, A.; Bertin, S.; Tomassoli, L. Preliminary analysis of ZYMV and WMV interaction in mixed infection by ΔΔCt Rt-qPCR. In Proceedings of the International Advances in Plant Virology, Rome, Italy, 29 October 2019. [Google Scholar]
- Jiang, Y.; Fang, X.; Bai, C. Signaling Aptamer/Protein Binding by a Molecular Light Switch Complex. Anal. Chem. 2004, 76, 5230–5235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Deng, F.; Yan, Z.; Xia, Y.; Wang, Z.; Ye, J.; Deng, Y.; Zhang, Z.; Qiao, M.; et al. TqPCR: A Touchdown QPCR Assay with Significantly Improved Detection Sensitivity and Amplification Efficiency of SYBR Green QPCR. PLoS ONE 2015, 10, e0132666. [Google Scholar] [CrossRef]
- Giangaspero, A.; Marangi, M.; Latrofa, M.S.; Annoscia, G.; Putignani, L.; Capelli, G.; Bonassisa, L.; Normanno, G.; Otranto, D.; Cereda, M.; et al. Efficiency of the Q3 Lab-on-Chip Real Time-PCR Platform for Detecting Protozoan Pathogens in Bivalve Mollusks. J. Food Sci. Technol. 2019, 56, 5000–5008. [Google Scholar] [CrossRef]
- Battaglia, S.; Petralia, S.; Vicario, N.; Cirillo, D.; Conoci, S. An Innovative Silicon-Chip for Sensitive Real Time PCR Improvement in Pathogen Detection. Analyst 2019, 144, 2353–2358. [Google Scholar] [CrossRef]
- Petralia, S.; Conoci, S. PCR Technologies for Point of Care Testing: Progress and Perspectives. ACS Sens. 2017, 2, 876–891. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costantini, F.; Lovecchio, N.; Nandimandalam, M.; Manglli, A.; Faggioli, F.; Biasin, M.; Manetti, C.; Roversi, P.F.; Nascetti, A.; de Cesare, G.; et al. Biomolecular Monitoring Tool Based on Lab-on-Chip for Virus Detection. Biosensors 2023, 13, 544. https://doi.org/10.3390/bios13050544
Costantini F, Lovecchio N, Nandimandalam M, Manglli A, Faggioli F, Biasin M, Manetti C, Roversi PF, Nascetti A, de Cesare G, et al. Biomolecular Monitoring Tool Based on Lab-on-Chip for Virus Detection. Biosensors. 2023; 13(5):544. https://doi.org/10.3390/bios13050544
Chicago/Turabian StyleCostantini, Francesca, Nicola Lovecchio, Manasa Nandimandalam, Ariana Manglli, Francesco Faggioli, Mara Biasin, Cesare Manetti, Pio Federico Roversi, Augusto Nascetti, Giampiero de Cesare, and et al. 2023. "Biomolecular Monitoring Tool Based on Lab-on-Chip for Virus Detection" Biosensors 13, no. 5: 544. https://doi.org/10.3390/bios13050544
APA StyleCostantini, F., Lovecchio, N., Nandimandalam, M., Manglli, A., Faggioli, F., Biasin, M., Manetti, C., Roversi, P. F., Nascetti, A., de Cesare, G., & Caputo, D. (2023). Biomolecular Monitoring Tool Based on Lab-on-Chip for Virus Detection. Biosensors, 13(5), 544. https://doi.org/10.3390/bios13050544







