The Unexpected Selectivity Switching from Mitochondria to Lysosome in a D-π-A Cyanine Dye
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedure for Synthesis
2.2. Cell Culture, Staining, and Fluorescence Confocal Microscopy Imaging
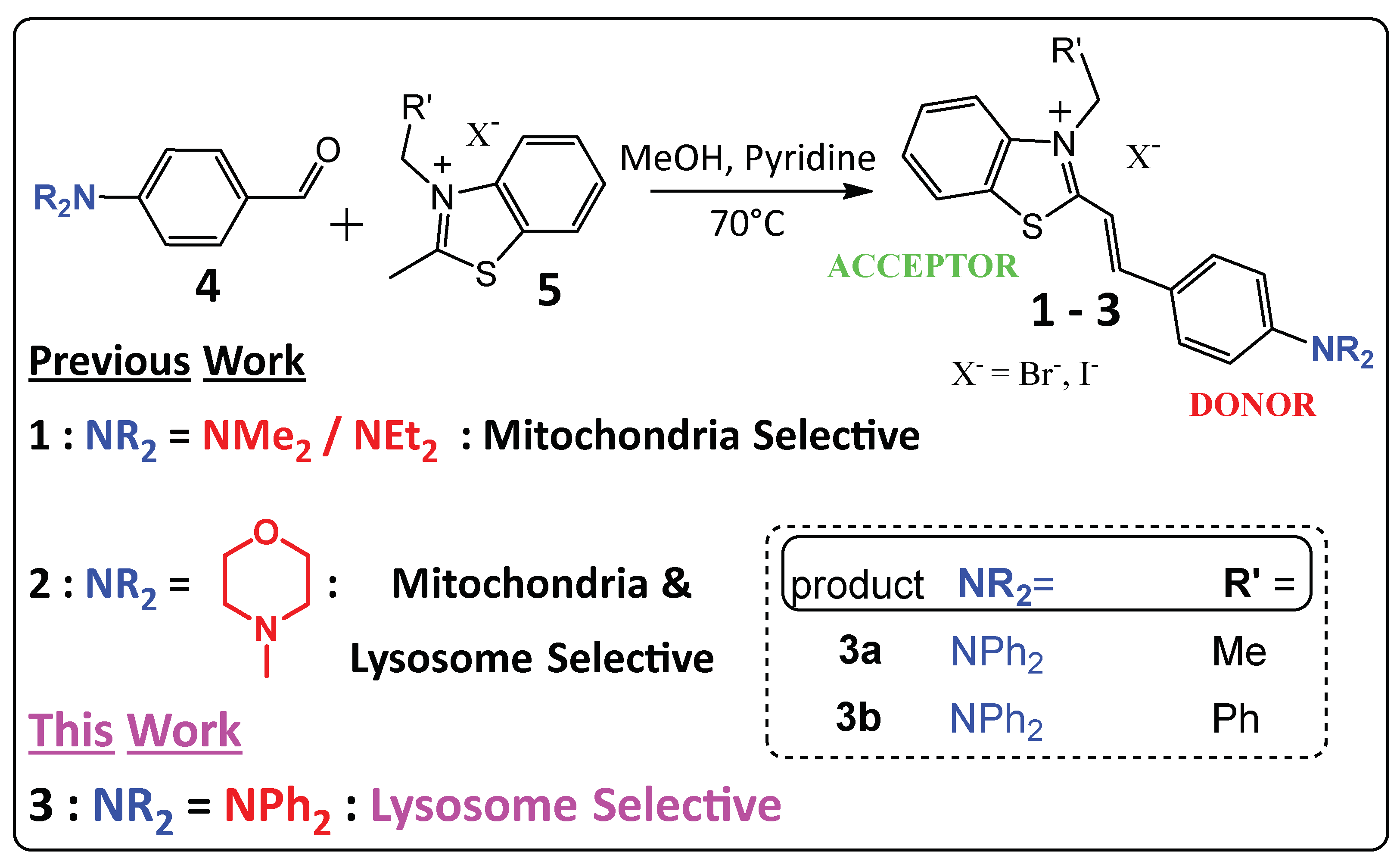
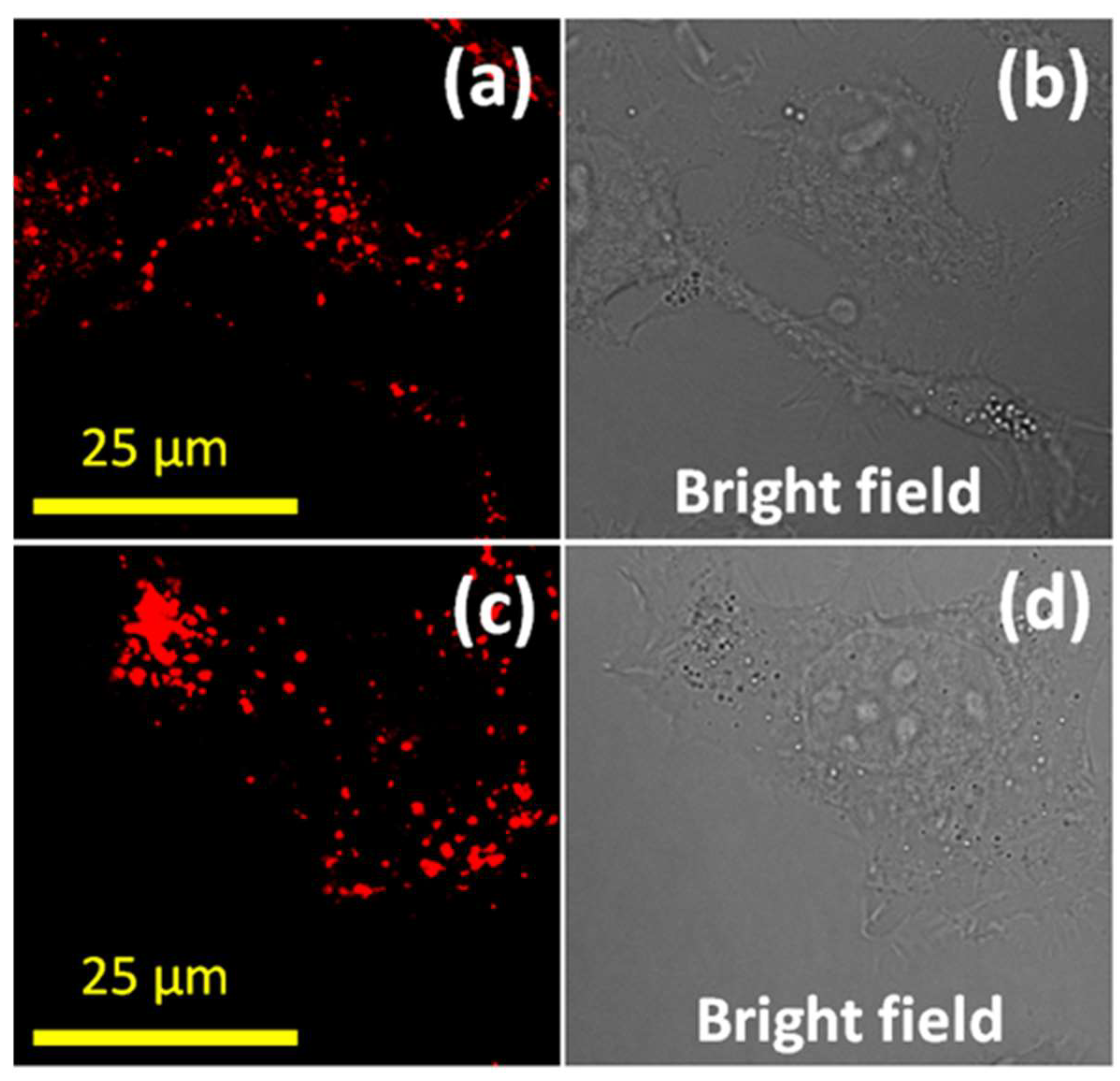
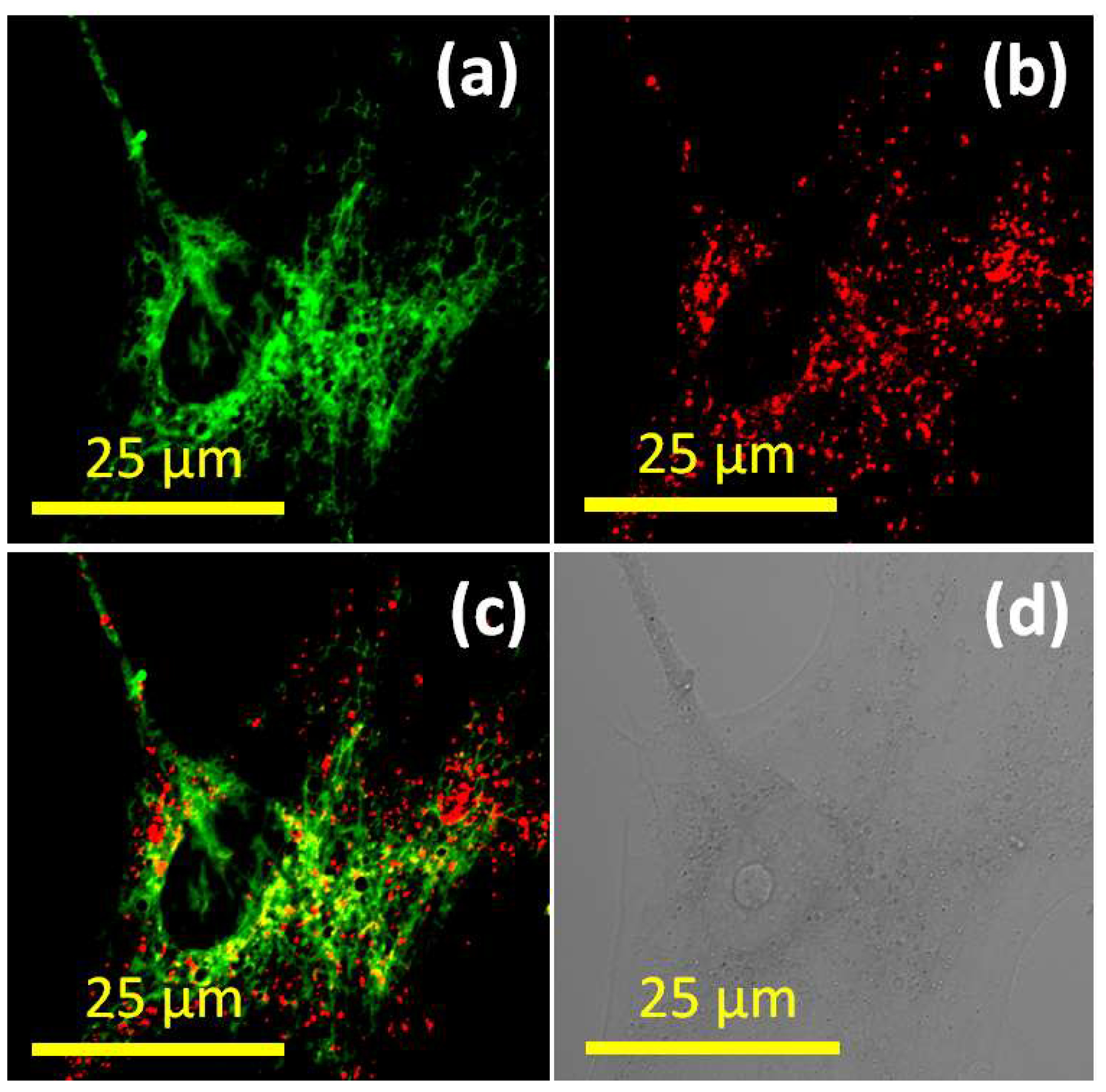
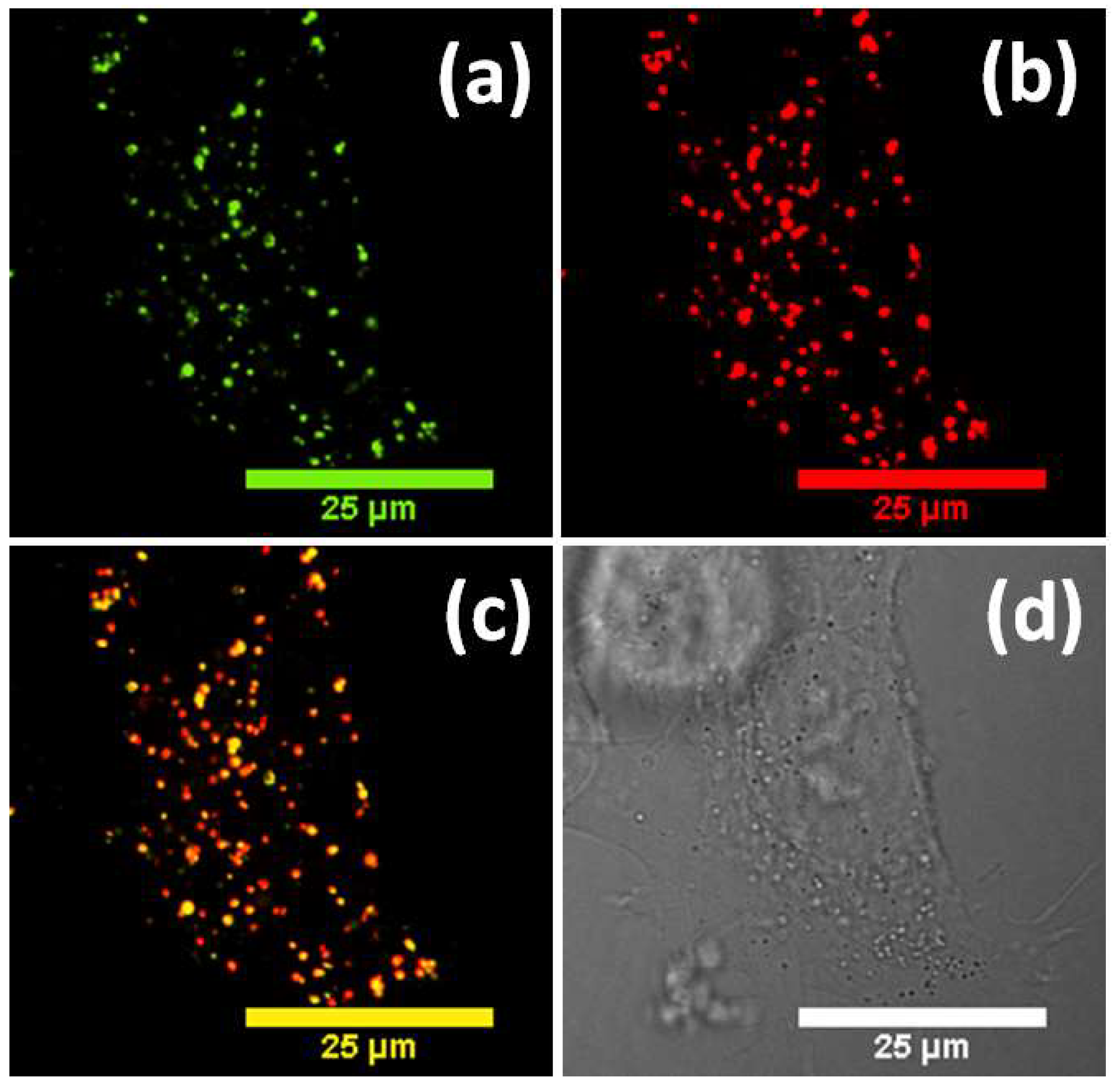
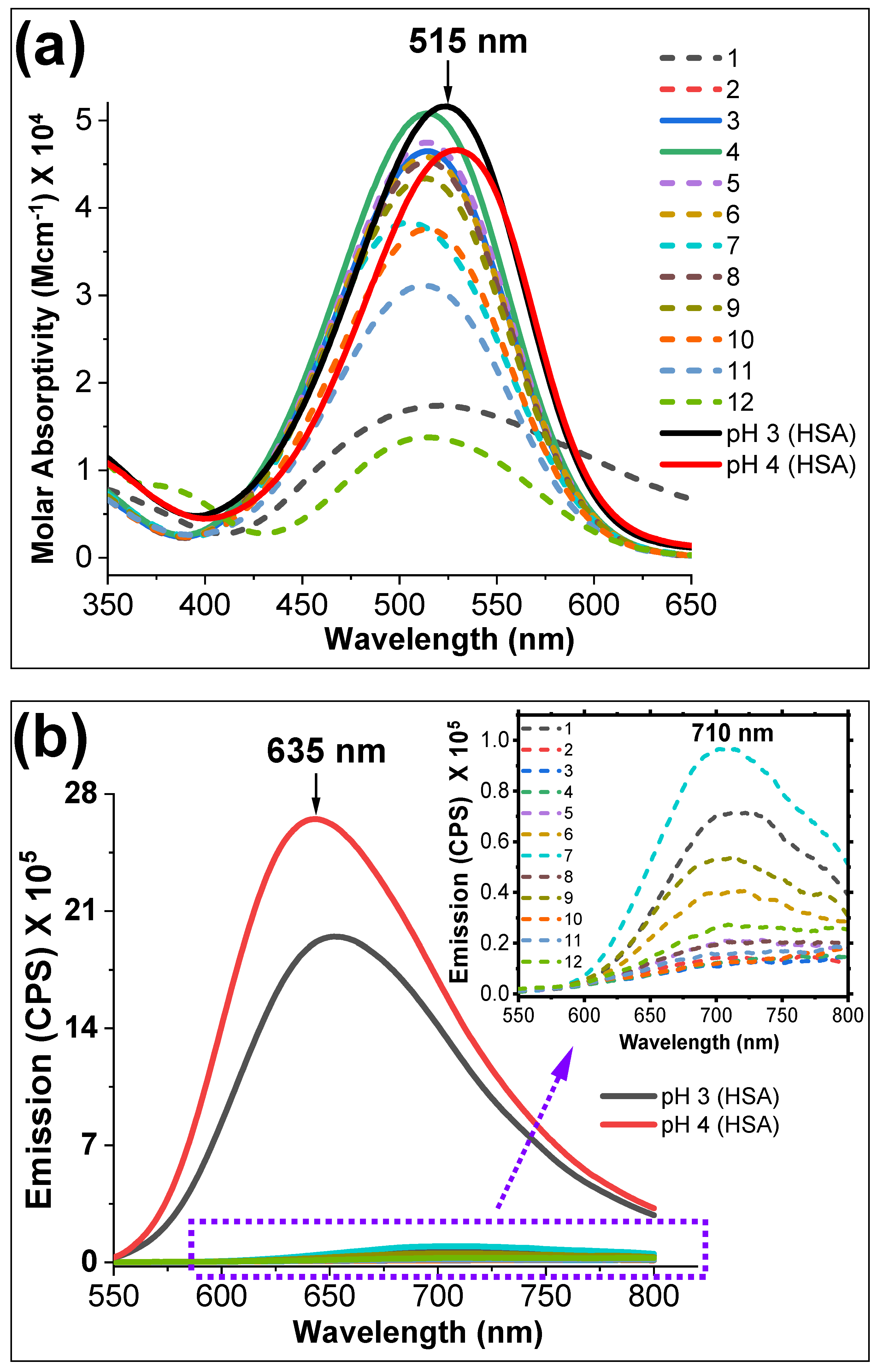
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shindy, H.A. Fundamentals in the Chemistry of Cyanine Dyes: A Review. Dye. Pigment. 2017, 145, 505–513. [Google Scholar] [CrossRef]
- Shi, C.; Wu, J.B.; Pan, D. Review on Near-Infrared Heptamethine Cyanine Dyes as Theranostic Agents for Tumor Imaging, Targeting, and Photodynamic Therapy. J. Biomed. Opt. 2016, 21, 050901. [Google Scholar] [CrossRef] [PubMed]
- Dave, R.; Terry, D.S.; Munro, J.B.; Blanchard, S.C. Mitigating Unwanted Photophysical Processes for Improved Single-Molecule Fluorescence Imaging. Biophys. J. 2009, 96, 2371–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Behera, R.K.; Behera, P.K.; Mishra, B.K.; Behera, G.B. Cyanines during the 1990s: A Review. Chem. Rev. 2000, 100, 1973–2012. [Google Scholar] [CrossRef] [PubMed]
- Abeywickrama, C.S.; Baumann, H.J.; Alexander, N.; Shriver, L.P.; Konopka, M.; Pang, Y. NIR-Emitting Benzothiazolium Cyanines with an Enhanced Stokes Shift for Mitochondria Imaging in Live Cells. Org. Biomol. Chem. 2018, 16, 3382–3388. [Google Scholar] [CrossRef]
- Onoe, S.; Temma, T.; Shimizu, Y.; Ono, M.; Saji, H. Investigation of Cyanine Dyes for in Vivo Optical Imaging of Altered Mitochondrial Membrane Potential in Tumors. Cancer Med. 2014, 3, 775–786. [Google Scholar] [CrossRef]
- Li, D.; Schreiber, C.L.; Smith, B.D. Sterically Shielded Heptamethine Cyanine Dyes for Bioconjugation and High Performance Near-infrared Fluorescence Imaging. Angew. Chem. 2020, 132, 12252–12259. [Google Scholar] [CrossRef]
- Zhao, N.; Zhang, C.; Zhao, Y.; Bai, B.; An, J.; Zhang, H.; Wu, J.B.; Shi, C. Optical Imaging of Gastric Cancer with Near-Infrared Heptamethine Carbocyanine Fluorescence Dyes. Oncotarget 2016, 7, 57277. [Google Scholar] [CrossRef] [Green Version]
- Lou, Z.; Li, P.; Song, P.; Han, K. Ratiometric Fluorescence Imaging of Cellular Hypochlorous Acid Based on Heptamethine Cyanine Dyes. Analyst 2013, 138, 6291–6295. [Google Scholar] [CrossRef]
- Ren, W.X.; Han, J.; Pradhan, T.; Lim, J.Y.; Lee, J.H.; Lee, J.; Kim, J.H.; Kim, J.S. A Fluorescent Probe to Detect Thiol-Containing Amino Acids in Solidtumors. Biomaterials 2014, 35, 4157–4167. [Google Scholar] [CrossRef]
- Niko, Y.; Didier, P.; Mely, Y.; Konishi, G.I.; Klymchenko, A.S. Bright and Photostable Push-Pull Pyrene Dye Visualizes Lipid Order Variation between Plasma and Intracellular Membranes. Sci. Rep. 2016, 6, 18870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeywickrama, C.S.; Wijesinghe, K.J.; Stahelin, R.V.; Pang, Y. Red-Emitting Pyrene–Benzothiazolium: Unexpected Selectivity to Lysosomes for Real-Time Cell Imaging without Alkalinizing Effect. Chem. Commun. 2019, 55, 3469–3472. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Sun, X.; Guo, Z.; Tang, J.; Shen, Y.; James, T.D.; Tian, H.; Zhu, W. In Vivo and in Situ Tracking Cancer Chemotherapy by Highly Photostable NIR Fluorescent Theranostic Prodrug. J. Am. Chem. Soc. 2014, 136, 3579–3588. [Google Scholar] [CrossRef] [PubMed]
- Araneda, J.F.; Piers, W.E.; Heyne, B.; Parvez, M.; McDonald, R. High Stokes Shift Anilido-pyridine Boron Difluoride Dyes. Angew. Chem. Int. Ed. 2011, 50, 12214–12217. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Hink, M.A.; Joosen, L.; Gadella, T.W.J.; Verkhusha, V.V. An Orange Fluorescent Protein with a Large Stokes Shift for Single-Excitation Multicolor FCCS and FRET Imaging. J. Am. Chem. Soc. 2012, 134, 7913–7923. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Sarkar, N.; Ghosh, A.K.; Majumdar, D.; Nath, D.N.; Bhattacharyya, K. Excited-State Intramolecular Proton Transfer in 2-(2′-Hydroxyphenyl)Benzimidazole and -Benzoxazole: Effect of Rotamerism and Hydrogen Bonding. J. Phys. Chem. 1994, 98, 9126–9132. [Google Scholar] [CrossRef]
- Abeywickrama, C.S.; Pang, Y. Synthesis of Fused 2-(2′-Hydroxyphenyl) Benzoxazole Derivatives: The Impact of Meta-/Para-Substitution on Fluorescence and Zinc Binding. Tetrahedron Lett. 2016, 57, 3518–3522. [Google Scholar] [CrossRef]
- Jin, X.; Sun, X.; Di, X.; Zhang, X.; Huang, H.; Liu, J.; Ji, P.; Zhu, H. Novel Fluorescent ESIPT Probe Based on Flavone for Nitroxyl in Aqueous Solution and Serum. Sens. Actuators B Chem. 2016, 224, 209–216. [Google Scholar] [CrossRef]
- Li, Y.; Dahal, D.; Abeywickrama, C.S.; Pang, Y. Progress in Tuning Emission of the Excited-State Intramolecular Proton Transfer (ESIPT)-Based Fluorescent Probes. ACS Omega 2021, 6, 6547–6553. [Google Scholar] [CrossRef]
- Bi, X.; Liu, B.; Mcdonald, L.; Pang, Y. Excited-State Intramolecular Proton Transfer (ESIPT) of Fluorescent Flavonoid Dyes: A Close Look by Low Temperature Fluorescence. J. Phys. Chem. B 2017, 121, 4981–4986. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Bi, X.; Liu, D.; Pan, C.; Wang, L.; Pang, Y. A Novel Flavonoid-Based Bioprobe for Intracellular Recognition of Cu2+ and Its Complex with Cu2+ for Secondary Sensing of Pyrophosphate. Sens. Actuators B Chem. 2016, 229, 131–137. [Google Scholar] [CrossRef]
- Bertman, K.A.; Abeywickrama, C.S.; Pang, Y. A NIR Emitting Cyanine with Large Stokes’ Shift for Mitochondria and Identification of Their Membrane Potential Disruption. ChemBioChem 2022, 23, e202100516. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, Z.R.; Rotkiewicz, K.; Rettig, W. Structural Changes Accompanying Intramolecular Electron Transfer: Focus on Twisted Intramolecular Charge-Transfer States and Structures. Chem. Rev. 2003, 103, 3899–4032. [Google Scholar] [CrossRef] [PubMed]
- Haidekker, M.A.; Brady, T.P.; Lichlyter, D.; Theodorakis, E.A. A Ratiometric Fluorescent Viscosity Sensor. J. Am. Chem. Soc. 2006, 128, 398–399. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Geddes, C.D. (Eds.) Topics in Fluorescence Spectroscopy; Plenum Press: New York, NY, USA, 1991; ISBN 978-0-306-43874-5. [Google Scholar]
- Barman, S.; Mukhopadhyay, S.K.; Gangopadhyay, M.; Biswas, S.; Dey, S.; Singh, N.D.P. Coumarin–Benzothiazole–Chlorambucil (Cou–Benz–Cbl) Conjugate: An ESIPT Based PH Sensitive Photoresponsive Drug Delivery System. J. Mater. Chem. B 2015, 3, 3490–3497. [Google Scholar] [CrossRef]
- Li, W.; Chen, D.; Wang, H.; Luo, S.; Dong, L.; Zhang, Y.; Shi, J.; Tong, B.; Dong, Y. Quantitation of Albumin in Serum Using “Turn-on” Fluorescent Probe with Aggregation-Enhanced Emission Characteristics. ACS Appl. Mater. Interfaces 2015, 7, 26094–26100. [Google Scholar] [CrossRef]
- Boonkitpatarakul, K.; Wang, J.; Niamnont, N.; Liu, B.; Mcdonald, L.; Pang, Y.; Sukwattanasinitt, M. Novel Turn-on Fluorescent Sensors with Mega Stokes Shifts for Dual Detection of Al3+ and Zn2+. ACS Sens. 2015, 1, 144–150. [Google Scholar] [CrossRef]
- Niko, Y.; Moritomo, H.; Sugihara, H.; Suzuki, Y.; Kawamata, J.; Konishi, G. A Novel Pyrene-Based Two-Photon Active Fluorescent Dye Efficiently Excited and Emitting in the ‘Tissue Optical Window (650–1100 Nm)’. J. Mater. Chem. B 2015, 3, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Inoue, K.; Kawakami, R.; Murakami, M.; Nakayama, T.; Yamamoto, S.; Inoue, K.; Tsuda, T.; Sayama, K.; Imamura, T.; Kaneno, D.; et al. Synthesis and Photophysical Properties of a New Push–Pull Pyrene Dye with Green-to-Far-Red Emission and Its Application to Human Cellular and Skin Tissue Imaging. J. Mater. Chem. B 2022, 10, 1641–1649. [Google Scholar] [CrossRef]
- Abeywickrama, C.S.; Baumann, H.J.; Pang, Y. Simultaneous Visualization of Mitochondria and Lysosome by a Single Cyanine Dye: The Impact of the Donor Group (-NR2) Towards Organelle Selectivity. J. Fluoresc. 2021, 31, 1227–1234. [Google Scholar] [CrossRef]
- Pierzyńska-Mach, A.; Janowski, P.A.; Dobrucki, J.W. Evaluation of Acridine Orange, LysoTracker Red, and Quinacrine as Fluorescent Probes for Long-Term Tracking of Acidic Vesicles. Cytom. Part A 2014, 85, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Wiederschain, G.Y. The Molecular Probes Handbook. A Guide to Fluorescent Probes and Labeling Technologies. Biochemistry 2011, 76, 1276. [Google Scholar] [CrossRef] [Green Version]
- Yapici, N.B.; Bi, Y.; Li, P.; Chen, X.; Yan, X.; Mandalapu, S.R.; Faucett, M.; Jockusch, S.; Ju, J.; Gibson, K.M.; et al. Highly Stable and Sensitive Fluorescent Probes (LysoProbes) for Lysosomal Labeling and Tracking. Sci. Rep. 2015, 5, srep08576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeywickrama, C.S.; Li, Y.; Ramanah, A.; Owitipana, D.N.; Wijesinghe, K.J.; Pang, Y. Albumin-Induced Large Fluorescence Turn ON in 4-(Diphenylamino)Benzothiazolium Dyes for Clinical Applications in Protein Detection. Sens. Actuators B Chem. 2022, 368, 132199. [Google Scholar] [CrossRef]

| Solvent | 3a | 3b | ||||
|---|---|---|---|---|---|---|
| λabs (nm) | λem (nm) | ϕfl | λabs (nm) | λem (nm) | ϕfl | |
| DCM | 549 | 734 | 0.003 | 558 | 747 | 0.001 |
| ACN | 507 | 720 | 0.0005 | 520 | 750 | 0.0003 |
| DMSO | 506 | 728 | 0.002 | 519 | 745 | 0.002 |
| EtOH | 518 | 708 | 0.003 | 532 | 711 | 0.002 |
| Water | 501 | 720 | 0.0001 | 517 | 731 | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abeywickrama, C.S.; Baumann, H.J.; Bertman, K.A.; Corbin, B.; Pang, Y. The Unexpected Selectivity Switching from Mitochondria to Lysosome in a D-π-A Cyanine Dye. Biosensors 2022, 12, 504. https://doi.org/10.3390/bios12070504
Abeywickrama CS, Baumann HJ, Bertman KA, Corbin B, Pang Y. The Unexpected Selectivity Switching from Mitochondria to Lysosome in a D-π-A Cyanine Dye. Biosensors. 2022; 12(7):504. https://doi.org/10.3390/bios12070504
Chicago/Turabian StyleAbeywickrama, Chathura S., Hannah J. Baumann, Keti A. Bertman, Brian Corbin, and Yi Pang. 2022. "The Unexpected Selectivity Switching from Mitochondria to Lysosome in a D-π-A Cyanine Dye" Biosensors 12, no. 7: 504. https://doi.org/10.3390/bios12070504
APA StyleAbeywickrama, C. S., Baumann, H. J., Bertman, K. A., Corbin, B., & Pang, Y. (2022). The Unexpected Selectivity Switching from Mitochondria to Lysosome in a D-π-A Cyanine Dye. Biosensors, 12(7), 504. https://doi.org/10.3390/bios12070504





