Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Probe Design and PCR Confirmation
2.3. GNP Synthesis and Surface Coating
2.4. Bacterial Culture
2.5. Biosensor Design and Optimization
2.6. Sensitivity and Specificity Testing
2.7. Biosensing from Large Food Samples
2.8. Statistical Analysis
3. Results and Discussion
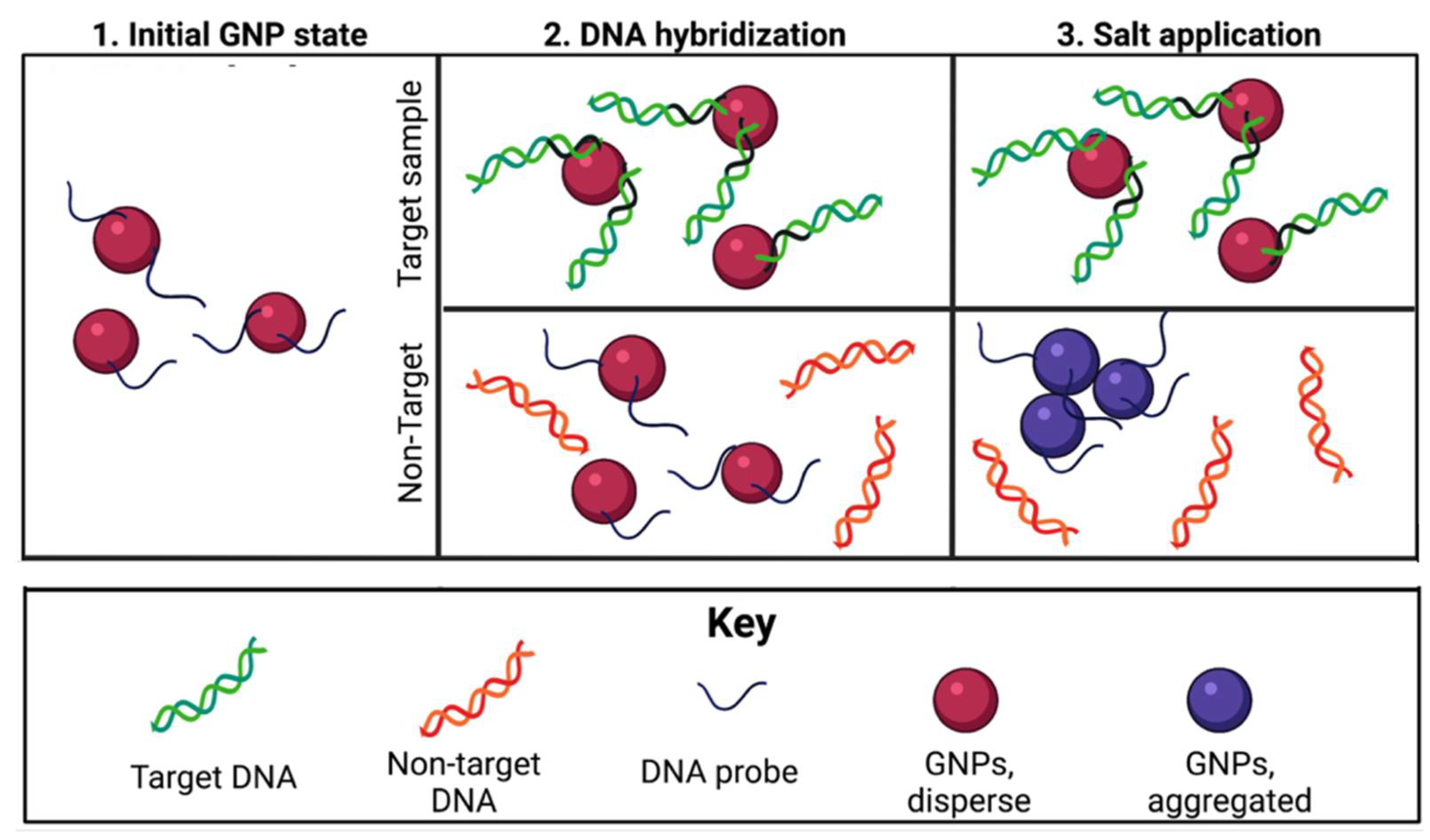
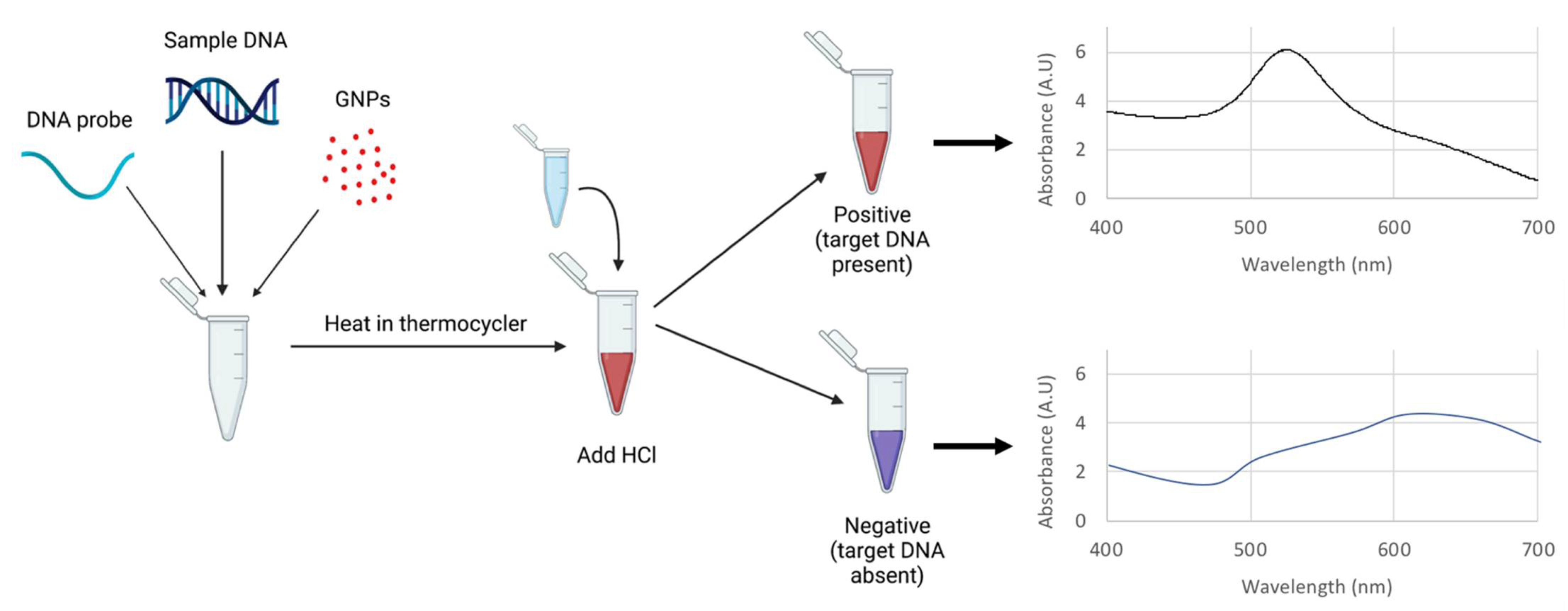
3.1. Principles of the E. coli O157 Nano-Biosensor
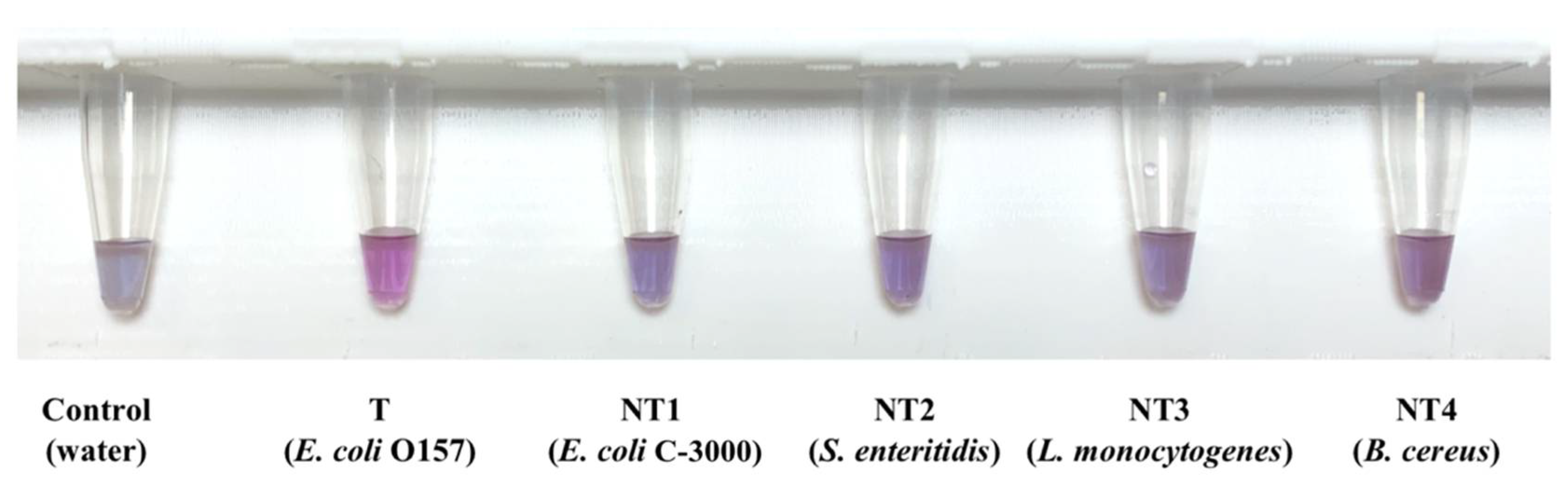
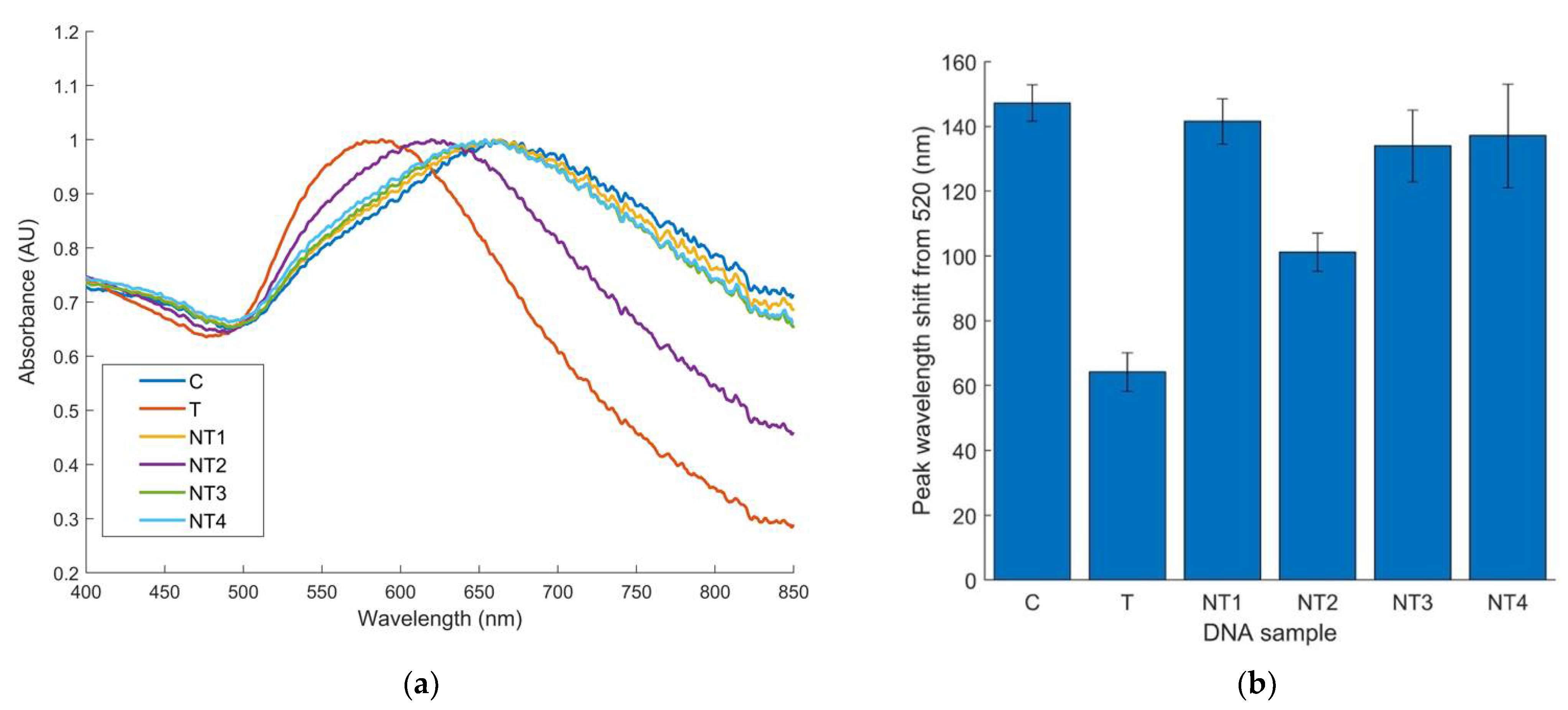
3.2. Optimization and Specificity Testing of Pure E. coli O157 Cultures
3.3. Sensitivity Testing of Pure E. coli O157 Cultures
3.4. Detection of DNA from Flour with E. coli O157 Biosensor
3.5. Improving Biosensor Sensitivity and Accessibility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. 2015. Available online: https://apps.who.int/iris/handle/10665/199350 (accessed on 12 November 2021).
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef] [PubMed]
- Tack, D.M.; Ray, L.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Rissman, T.; Jervis, R.; Lathrop, S.; Muse, A.; Duwell, M.; et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2016–2019. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Biswas, A.K.; Choi, K.; Pal, U.K. Methods for Rapid Detection of Foodborne Pathogens: An Overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, M.A.; Dawson, C.O. Economic impact of food safety outbreaks on food businesses. Foods 2013, 2, 585–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, H.; Shah, D.; Sayed, S.; Horton, S.; Schroeder, L.F. Availability of essential diagnostics in ten low-income and middle-income countries: Results from national health facility surveys. Lancet Glob. Health 2021, 9, e1553–e1560. [Google Scholar] [CrossRef]
- Fleming, K.A.; Horton, S.; Wilson, M.L.; Atun, R.; DeStigter, K.; Flanigan, J.; Sayed, S.; Adam, P.; Aguilar, B.; Andronikou, S.; et al. The Lancet Commission on diagnostics: Transforming access to diagnostics. Lancet 2021, 398, 1997–2050. [Google Scholar] [CrossRef]
- Park, S.H.; Aydin, M.; Khatiwara, A.; Dolan, M.C.; Gilmore, D.F.; Bouldin, J.L.; Ahn, S.; Ricke, S.C. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 2014, 38, 250–262. [Google Scholar] [CrossRef]
- Al-Awwal, N.; Masjedi, M.; El-Dweik, M.; Anderson, S.H.; Ansari, J. Nanoparticle immuno-fluorescent probes as a method for detection of viable E. coli O157:H7. J. Microbiol. Methods 2022, 193, 106403. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Ferreira, R.; Uria, N.; Abramova, N.; Gargallo, R.; Muñoz-Pascual, F.X.; Bratov, A. Novel impedimetric aptasensor for label-free detection of Escherichia coli O157:H7. Sens. Actuators B Chem. 2018, 255, 2988–2995. [Google Scholar] [CrossRef] [Green Version]
- You, S.M.; Luo, K.; Jung, J.Y.; Jeong, K.B.; Lee, E.S.; Oh, M.H.; Kim, Y.R. Gold Nanoparticle-Coated Starch Magnetic Beads for the Separation, Concentration, and SERS-Based Detection of E. coli O157:H7. ACS Appl. Mater. Interfaces 2020, 12, 18292–18300. [Google Scholar] [CrossRef] [PubMed]
- Zeinhom, M.M.A.; Wang, Y.; Song, Y.; Zhu, M.J.; Lin, Y.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157:H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef]
- Zhou, C.; Zou, H.; Li, M.; Sun, C.; Ren, D.; Li, Y. Fiber optic surface plasmon resonance sensor for detection of E. coli O157:H7 based on antimicrobial peptides and AgNPs-rGO. Biosens. Bioelectron. 2018, 117, 347–353. [Google Scholar] [CrossRef]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Song, X.C.; Mrázek, J.; Lamačová, J.; Scott Lynn, N.; Šedivák, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cho, H.Y.; Choi, H.K.; Lee, J.Y.; Choi, J.W. Application of Gold Nanoparticle to Plasmonic Biosensors. Int. J. Mol. Sci. 2018, 19, 2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, B.; Liu, H.; Zhang, X.; Tan, W. Aptamer-conjugated gold nanoparticles for bioanalysis. Nanomedicine 2013, 8, 983–993. [Google Scholar] [CrossRef]
- Yu, L.; Li, N. Noble Metal Nanoparticles-Based Colorimetric Biosensor for Visual Quantification: A Mini Review. Chemosensors 2019, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Ahirwar, R.; Nahar, P. Development of a label-free gold nanoparticle-based colorimetric aptasensor for detection of human estrogen receptor alpha. Anal. Bioanal. Chem. 2015, 408, 327–332. [Google Scholar] [CrossRef]
- Jiang, P.; Wang, Y.; Zhao, L.; Ji, C.; Chen, D.; Nie, L. Applications of Gold Nanoparticles in Non-Optical Biosensors. Nanomaterials 2018, 8, 877. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, M.; Goff, A.L.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [Green Version]
- Pingarrón, J.M.; Yáñez-Sedeño, P.; González-Cortés, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar] [CrossRef]
- Bakthavathsalam, P.; Rajendran, V.K.; Baquir Mohammed, J.A. A direct detection of Escherichia coli genomic DNA using gold nanoprobes. J. Nanobiotechnol. 2012, 10, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidari, Z.; Rezatofighi, S.E.; Rastegarzadeh, S. Development and comparison of cross-linking and non-crosslinking probe-gold nanoparticle hybridization assays for direct detection of unamplified bovine viral diarrhea virus-RNA. BMC Biotechnol. 2021, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Kuswandi, B.; Gani, A.A.; Kristiningrum, N.; Ahmad, M. Simple Colorimetric DNA Biosensor Based on Gold Nanoparticles for Pork Adulteration Detection in Processed Meats. Sens. Transducers 2017, 208, 7–13. [Google Scholar]
- Hunter, R. Foundations of Colloid Science, 2nd ed.; Oxford University Press: Oxford, UK, 2001; ISBN 0198505027. [Google Scholar]
- Quintela, I.A.; de los Reyes, B.G.; Lin, C.-S.; Wu, V.C.H. Simultaneous Colorimetric Detection of a Variety of Salmonella spp. in Food and Environmental Samples by Optical Biosensing Using Oligonucleotide-Gold Nanoparticles. Front. Microbiol. 2019, 10, 1138. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Pan, W.; Liang, Q.; Song, X. Exonuclease I manipulating primer-modified gold nanoparticles for colorimetric telomerase activity assay. Biosens. Bioelectron. 2016, 77, 144–148. [Google Scholar] [CrossRef]
- Chen, Y.M.; Yu, C.J.; Cheng, T.L.; Tseng, W.L. Colorimetric detection of lysozyme based on electrostatic interaction with human serum albumin-modified gold nanoparticles. Langmuir 2008, 24, 3654–3660. [Google Scholar] [CrossRef]
- Annadhasan, M.; Muthukumarasamyvel, T.; Sankar Babu, V.R.; Rajendiran, N. Green synthesized silver and gold nanoparticles for colorimetric detection of Hg2+, Pb2+, and Mn2+ in aqueous medium. ACS Sustain. Chem. Eng. 2014, 2, 887–896. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, Y.; Shao, H.; Wang, Z.; Wang, X.; Jiang, X. Label-free colorimetric detection of cadmium ions in rice samples using gold nanoparticles. Anal. Chem. 2014, 86, 8530–8534. [Google Scholar] [CrossRef]
- Jazayeri, M.H.; Aghaie, T.; Avan, A.; Vatankhah, A.; Ghaffari, M.R.S. Colorimetric detection based on gold nano particles (GNPs): An easy, fast, inexpensive, low-cost and short time method in detection of analytes (protein, DNA, and ion). Sens. Bio-Sens. Res. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Ahmadi, S.; Kamaladini, H.; Haddadi, F.; Sharifmoghadam, M.R. Thiol-Capped Gold Nanoparticle Biosensors for Rapid and Sensitive Visual Colorimetric Detection of Klebsiella pneumoniae. J. Fluoresc. 2018, 28, 987–998. [Google Scholar] [CrossRef] [PubMed]
- ThermoFisher Scientific. NanoDrop One User Guide; ThermoFisher Scientific: Waltham, MA, USA, 2017. [Google Scholar]
- ThermoFisher Scientific NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer. Available online: https://www.thermofisher.com/order/catalog/product/ND-ONEC-W (accessed on 15 April 2022).
- National Center for Biotechnology Information. Escherichia coli (ID 167). Available online: https://www-ncbi-nlm-nih-gov.proxy1.cl.msu.edu/genome/?term=e.coliO157 (accessed on 30 March 2022).
- Feng, P.; Weagant, S.; Jinneman, K. BAM Chapter 4A: Diarrheagenic Escherichia coli|FDA. In Bacteriological Analytical Manual; Association of Official Analytical Chemists: Washington, DC, USA, 2020. [Google Scholar]
- Surendraraj, A.; Thampuran, N.; Joseph, T.C. Molecular Screening, Isolation, and Characterization of Enterohemorrhagic Escherichia coli O157:H7 from Retail Shrimp. J. Food Prot. 2010, 73, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Yrad, F.M.; Castañares, J.M.; Alocilja, E.C. Visual Detection of Dengue-1 RNA Using Gold Nanoparticle-Based Lateral Flow Biosensor. Diagnostics 2019, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Ott, L.R.; Longnecker, M. Multiple Comparisons. In An Introduction to Statistical Methods and Data Analysis; Cole, B., Ed.; Cengage Learning: Boston, MA, USA, 2010; pp. 451–498. ISBN 9780495017585. [Google Scholar]
- Pennington, H. Escherichia coli O157. Lancet 2010, 376, 1428–1435. [Google Scholar] [CrossRef]
- Gyles, C.L. Shiga toxin-producing Escherichia coli: An overview. J. Anim. Sci. 2007, 85, E45–E62. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.M.S.; Filippini, D.; Nielen, M.W.F.; Salentijn, G.I.J. Unraveling the Hook Effect: A Comprehensive Study of High Antigen Concentration Effects in Sandwich Lateral Flow Immunoassays. Anal. Chem. 2020, 92, 15587–15595. [Google Scholar] [CrossRef] [PubMed]
- Vetrone, S.A.; Huarng, M.C.; Alocilja, E.C. Detection of Non-PCR Amplified S. enteritidis Genomic DNA from Food Matrices Using a Gold-Nanoparticle DNA Biosensor: A Proof-of-Concept Study. Sensors 2012, 12, 10487–10499. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.E.; Carrique-Mas, J.J.; McLaren, I.; Davies, R.H. A comparison of pooled and individual bird sampling for detection of Salmonella in commercial egg laying flocks. Prev. Vet. Med. 2011, 99, 176–184. [Google Scholar] [CrossRef]
- Wang, Y.; Salazar, J.K. Culture-Independent Rapid Detection Methods for Bacterial Pathogens and Toxins in Food Matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- He, S.; Huang, Y.; Ma, Y.; Yu, H.; Pang, B.; Liu, X.; Yin, C.; Wang, X.; Wei, Y.; Tian, Y.; et al. Detection of four foodborne pathogens based on magnetic separation multiplex PCR and capillary electrophoresis. Biotechnol. J. 2022, 17, e2100335. [Google Scholar] [CrossRef]
- Zheng, L.; Cai, G.; Wang, S.; Liao, M.; Li, Y.; Lin, J. A microfluidic colorimetric biosensor for rapid detection of Escherichia coli O157:H7 using gold nanoparticle aggregation and smart phone imaging. Biosens. Bioelectron. 2019, 124–125, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, J.; Shen, Z.; Liu, Y.; Song, Y.; Liang, Y.; Li, Z.; Nie, L.; Fang, Y.; Zhao, Y. A newly developed paper embedded microchip based on LAMP for rapid multiple detections of foodborne pathogens. BMC Microbiol. 2021, 21, 197. [Google Scholar] [CrossRef] [PubMed]




| Biosensor | Analyte, Capturing Molecule | Sample Preparation | Samples | Assay Time | LOD (CFU/mL) | Source |
|---|---|---|---|---|---|---|
| Immuno-fluorescent probes * | Antigens, antibodies | 6 h incubation | Water sample | 5 min | 103 | [10] |
| Impedimetric aptasensor | Outer membrane proteins, aptamer | 10 min centrifugation | Pure culture | 30 min | 102 | [11] |
| SERS * | Antigens, antibodies | N/A | Pure culture | 1 h | 100 | [12] |
| Smartphone-based fluorescence device | Antigens, antibodies | Suspension in PBS, sandwich immunoassay | Yogurt, Egg | <2 h | 1–10 | [13] |
| Fiber optic SPR * | Lipopolysaccharides, antimicrobial peptides | 24-h bacterial inactivation | Water, juice | ~1 h | 5 × 102 | [14] |
| SPR * | Antigens, antibodies | Homogenization and sedimentation (<10 min) | Hamburger, cucumber | <80 min | <50 | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dester, E.; Kao, K.; Alocilja, E.C. Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor. Biosensors 2022, 12, 274. https://doi.org/10.3390/bios12050274
Dester E, Kao K, Alocilja EC. Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor. Biosensors. 2022; 12(5):274. https://doi.org/10.3390/bios12050274
Chicago/Turabian StyleDester, Emma, Kaily Kao, and Evangelyn C. Alocilja. 2022. "Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor" Biosensors 12, no. 5: 274. https://doi.org/10.3390/bios12050274
APA StyleDester, E., Kao, K., & Alocilja, E. C. (2022). Detection of Unamplified E. coli O157 DNA Extracted from Large Food Samples Using a Gold Nanoparticle Colorimetric Biosensor. Biosensors, 12(5), 274. https://doi.org/10.3390/bios12050274





