Genetically Encoded Ratiometric pH Sensors for the Measurement of Intra- and Extracellular pH and Internalization Rates
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction of CoGFP-His6
2.2. Plasmid Construction of EGF-CoGFP-His6
2.3. Plasmid Construction of EGF-CoGFP-mTagBFP2-His6
2.4. Plasmid Construction of EGF-CoGFP-mCRISPRed-His6
2.5. Prokaryotic Expression and Protein Purification
2.6. NanoLC–ESI-MS/MS
2.7. Maturation Assay of EGF-CoGFP-mCRISPRed
2.8. 3D Fluorescence Spectroscopy
2.9. Live-Cell Imaging
2.10. Ratiometric Calibration
2.11. Flow Cytometry
2.12. Microfluidic Application
2.13. Fluorescence Ratio Analysis
3. Results
3.1. CoGFP_V0 Retains Dual-Excitation and Dual-Emission Maxima as C-Terminal Fusion Protein
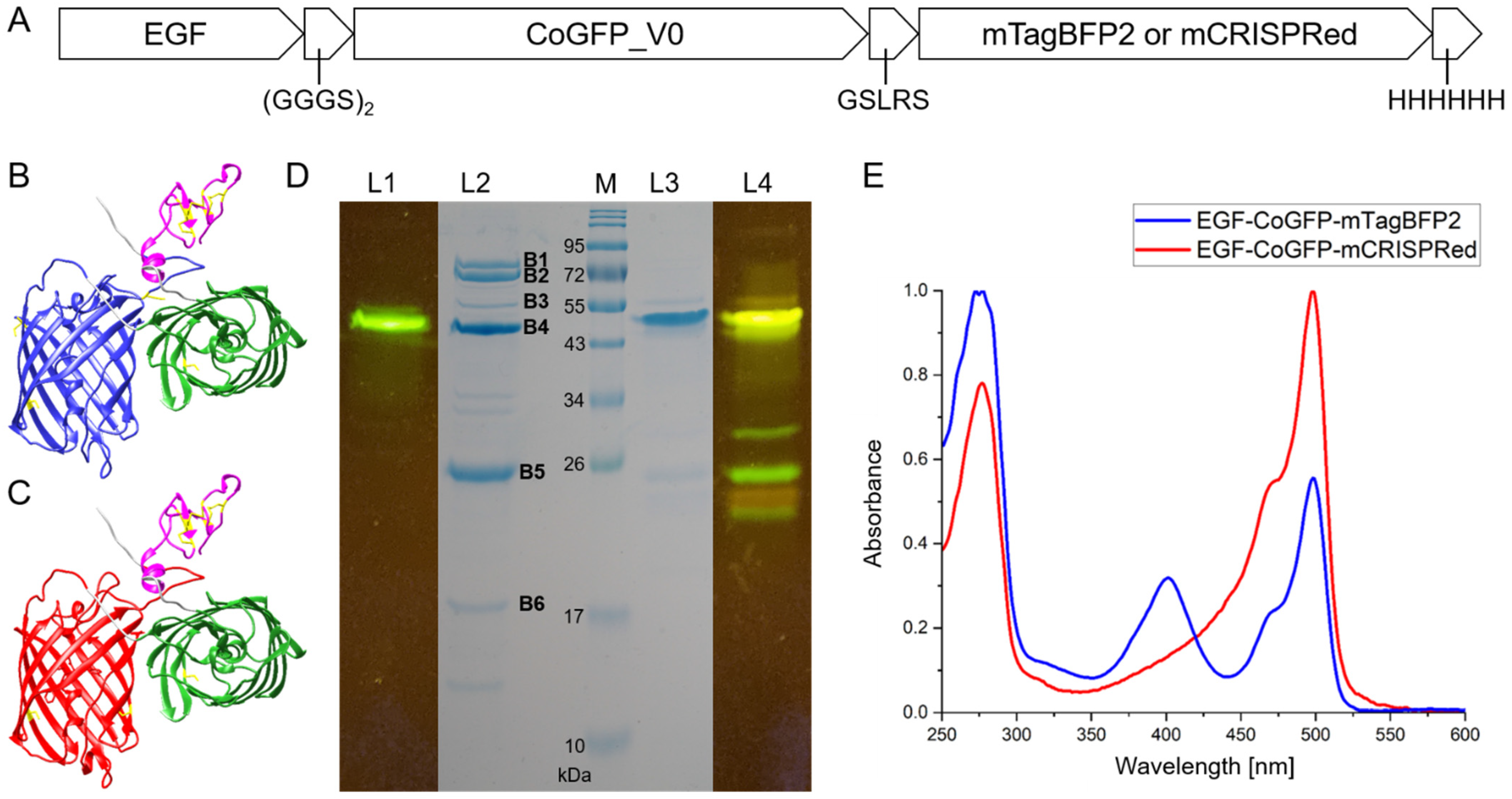
3.2. Design and Characterization of Tandem Fluorescent Protein Variants of EGF-CoGFP_V0
3.3. EGF-CoGFP-mTagBFP2 Reveals pH-Dependent FRET and EGF-CoGFP-mCRISPRed Shows Green/Red Fluorescence Ratio upon Single-Wavelength Excitation
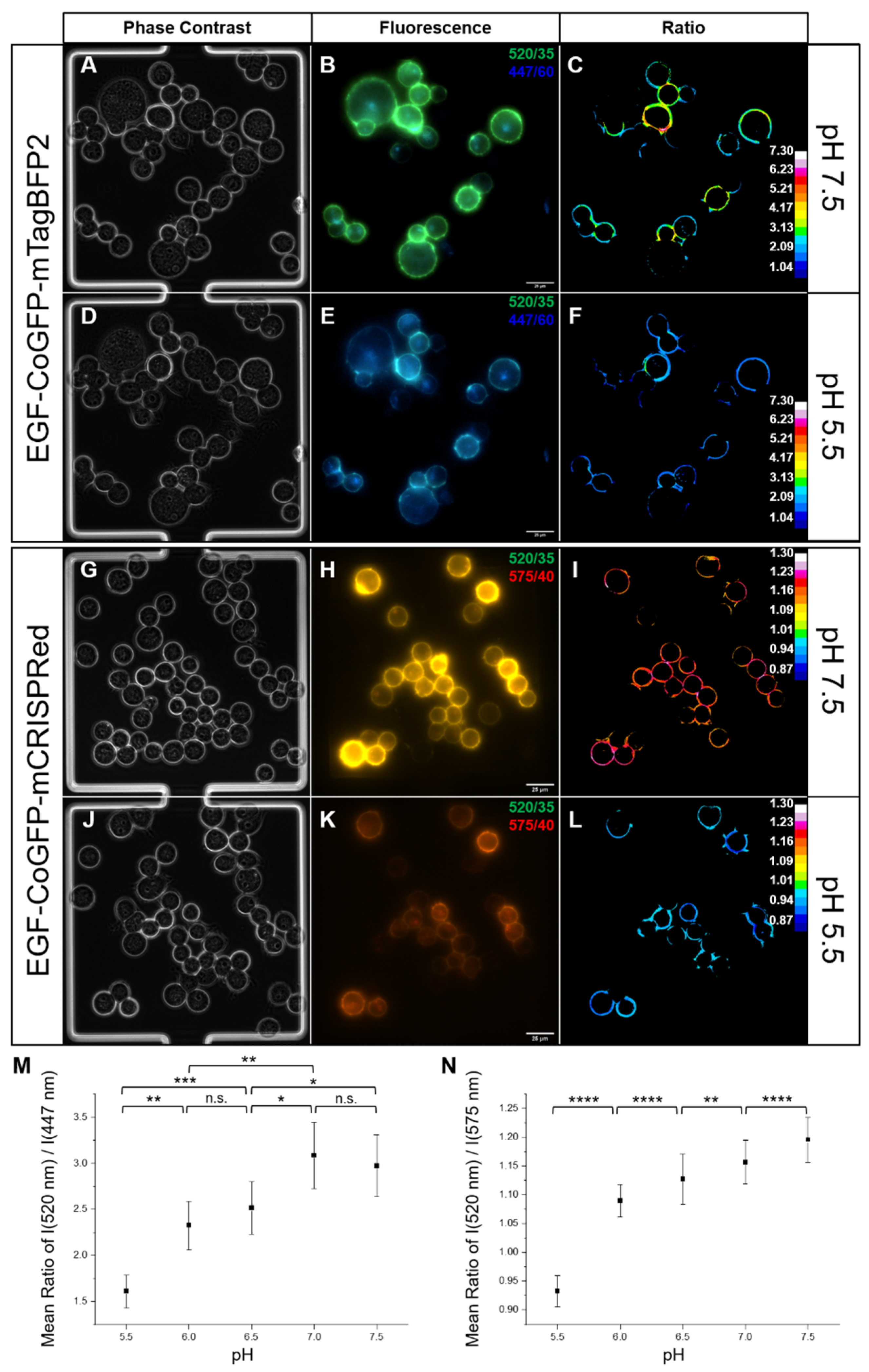
3.4. Intracellular pH Mapping Using pH Sensors
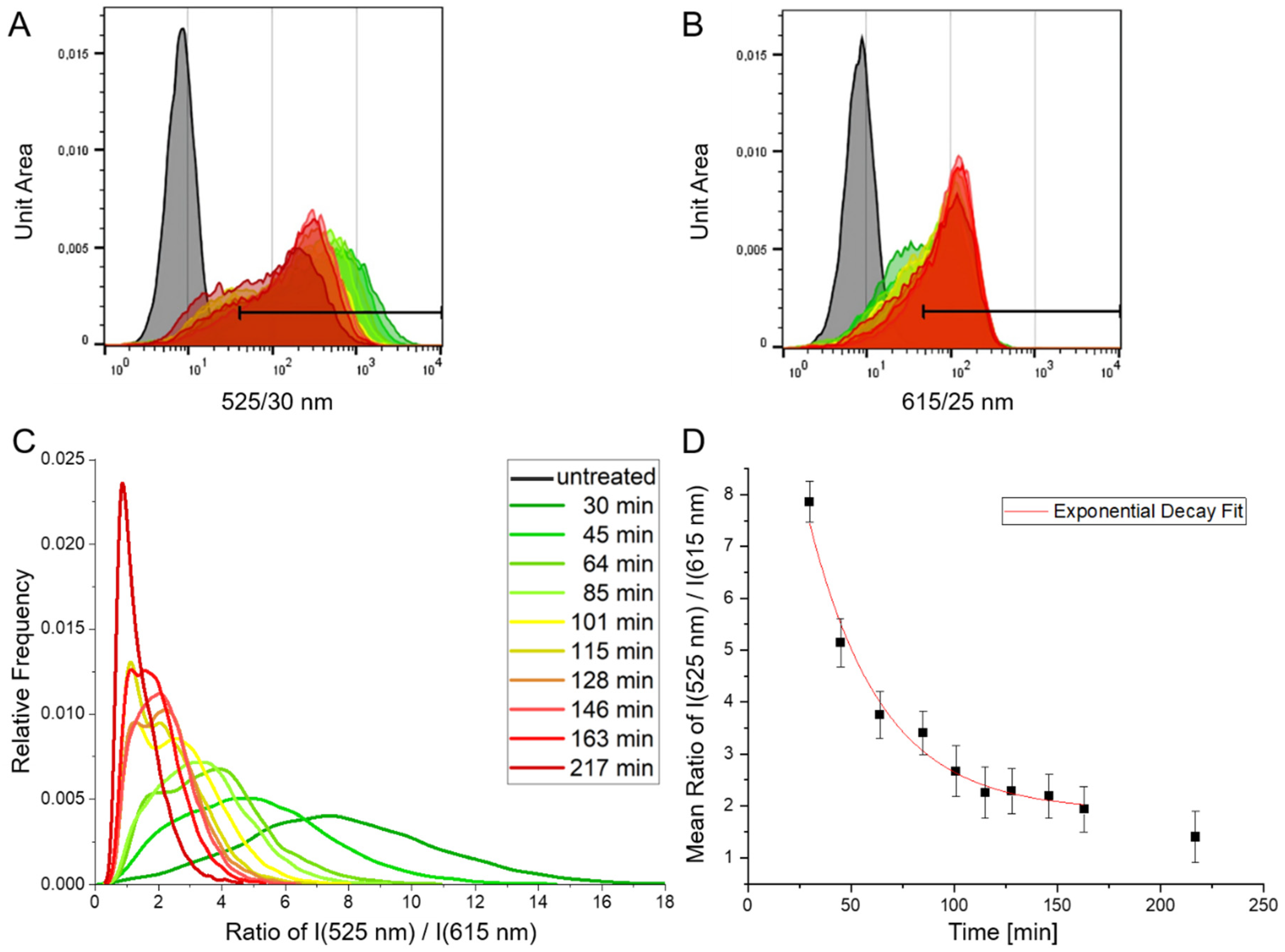
3.5. Time-Dependent Measurement of Cellular Uptake of the pH Sensors Using Flow Cytometry
3.6. Molecular pH Sensor as a Tool for Microfluidic Applications
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Dyke, R.W.; Belcher, J.D. Acidification of three types of liver endocytic vesicles: Similarities and differences. Am. J. Physiol.-Cell Physiol. 1994, 266. [Google Scholar] [CrossRef] [PubMed]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Ligands for Clathrin-Mediated Endocytosis Are Differentially Sorted into Distinct Populations of Early Endosomes. Cell 2006, 124, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Serresi, M.; Bizzarri, R.; Cardarelli, F.; Beltram, F. Real-time measurement of endosomal acidification by a novel genetically encoded biosensor. Anal. Bioanal. Chem. 2009, 393, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-sensitive red fluorescent protein for imaging of exo- and endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef]
- Poburko, D.; Santo-Domingo, J.; Demaurex, N. Dynamic Regulation of the Mitochondrial Proton Gradient during Cytosolic Calcium Elevations. J. Biol. Chem. 2011, 286, 11672–11684. [Google Scholar] [CrossRef]
- Hanson, G.T.; McAnaney, T.B.; Park, E.S.; Rendell, M.E.P.; Yarbrough, D.K.; Chu, S.; Xi, L.; Boxer, S.G.; Montrose, M.H.; Remington, S.J. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry 2002, 41, 15477–15488. [Google Scholar] [CrossRef]
- Schmitt, F.-J.; Thaa, B.; Junghans, C.; Vitali, M.; Veit, M.; Friedrich, T. eGFP-pHsens as a highly sensitive fluorophore for cellular pH determination by fluorescence lifetime imaging microscopy (FLIM). Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 1581–1593. [Google Scholar] [CrossRef]
- Martynov, V.I.; Pakhomov, A.A.; Deyev, I.E.; Petrenko, A.G. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2924–2939. [Google Scholar] [CrossRef]
- Battisti, A.; Digman, M.A.; Gratton, E.; Storti, B.; Beltram, F.; Bizzarri, R. Intracellular pH measurements made simple by fluorescent protein probes and the phasor approach to fluorescence lifetime imaging. Chem. Commun. 2012, 48, 5127. [Google Scholar] [CrossRef]
- Bizzarri, R.; Arcangeli, C.; Arosio, D.; Ricci, F.; Faraci, P.; Cardarelli, F.; Beltram, F. Development of a novel GFP-based ratiometric excitation and emission pH indicator for intracellular studies. Biophys. J. 2006, 90, 3300–3314. [Google Scholar] [CrossRef] [PubMed]
- Ogoh, K.; Kinebuchi, T.; Murai, M.; Takahashi, T.; Ohmiya, Y.; Suzuki, H. Dual-color-emitting green fluorescent protein from the sea cactus Cavernularia obesa and its use as a pH indicator for fluorescence microscopy. Luminescence 2013, 28, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Goryashchenko, A.S.; Pakhomov, A.A.; Ryabova, A.V.; Romanishkin, I.D.; Maksimov, E.G.; Orsa, A.N.; Serova, O.V.; Mozhaev, A.A.; Maksimova, M.A.; Martynov, V.I.; et al. FLIM-Based Intracellular and Extracellular pH Measurements Using Genetically Encoded pH Sensor. Biosensors 2021, 11, 340. [Google Scholar] [CrossRef] [PubMed]
- Disbrow, G.L.; Hanover, J.A.; Schlegel, R. Endoplasmic Reticulum-Localized Human Papillomavirus Type 16 E5 Protein Alters Endosomal pH but Not trans -Golgi pH. J. Virol. 2005, 79, 5839–5846. [Google Scholar] [CrossRef] [PubMed]
- Arosio, D.; Ricci, F.; Marchetti, L.; Gualdani, R.; Albertazzi, L.; Beltram, F. Simultaneous intracellular chloride and pH measurements using a GFP-based sensor. Nat. Methods 2010, 7, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Gjetting, S.K.; Ytting, C.K.; Schulz, A.; Fuglsang, A.T. Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 2012, 63, 3207–3218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhong, W.; Zhou, J.; Sheng, F.; Fang, Z.; Wei, Y.; Chen, Y.; Deng, X.; Xia, B.; Lin, J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012, 8, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Harada, H.; Hiraoka, M. A novel method to visually determine the intracellular pH of xenografted tumor in vivo by utilizing fluorescent protein as an indicator. Biochem. Biophys. Res. Commun. 2015, 464, 1151–1156. [Google Scholar] [CrossRef]
- Chin, M.Y.; Patwardhan, A.R.; Ang, K.-H.; Wang, A.L.; Alquezar, C.; Welch, M.; Nguyen, P.T.; Grabe, M.; Molofsky, A.V.; Arkin, M.R.; et al. Genetically Encoded, pH-Sensitive mTFP1 Biosensor for Probing Lysosomal pH. ACS Sens. 2021, 6, 2168–2180. [Google Scholar] [CrossRef]
- Awaji, T.; Hirasawa, A.; Shirakawa, H.; Tsujimoto, G.; Miyazaki, S. Novel Green Fluorescent Protein-Based Ratiometric Indicators for Monitoring pH in Defined Intracellular Microdomains. Biochem. Biophys. Res. Commun. 2001, 289, 457–462. [Google Scholar] [CrossRef]
- Esposito, A.; Gralle, M.; Dani, M.A.C.; Lange, D.; Wouters, F.S. pHlameleons: A Family of FRET-Based Protein Sensors for Quantitative pH Imaging. Biochemistry 2008, 47, 13115–13126. [Google Scholar] [CrossRef] [PubMed]
- Shimozono, S.; Hosoi, H.; Mizuno, H.; Fukano, T.; Tahara, T.; Miyawaki, A. Concatenation of Cyan and Yellow Fluorescent Proteins for Efficient Resonance Energy Transfer. Biochemistry 2006, 45, 6267–6271. [Google Scholar] [CrossRef] [PubMed]
- Burgstaller, S.; Bischof, H.; Gensch, T.; Stryeck, S.; Gottschalk, B.; Ramadani-Muja, J.; Eroglu, E.; Rost, R.; Balfanz, S.; Baumann, A.; et al. pH-Lemon, a Fluorescent Protein-Based pH Reporter for Acidic Compartments. ACS Sens. 2019, 4, 883–891. [Google Scholar] [CrossRef]
- Liu, J.; Abdullah, M.A.A.; Yang, L.; Wang, J. Fast Affinity Induced Reaction Sensor Based on a Fluorogenic Click Reaction for Quick Detection of Protein Biomarkers. Anal. Chem. 2020, 92, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Song, B.; Ji, X.; Su, Y.; Wang, H.; He, Y. Fluorescent Silicon Nanorods-Based Ratiometric Sensors for Long-Term and Real-Time Measurements of Intracellular pH in Live Cells. Anal. Chem. 2017, 89, 12152–12159. [Google Scholar] [CrossRef]
- Janson, N.; Krüger, T.; Karsten, L.; Boschanski, M.; Dierks, T.; Müller, K.M.; Sewald, N. Bifunctional Reagents for Formylglycine Conjugation: Pitfalls and Breakthroughs. Chembiochem 2020, 21, 3580–3593. [Google Scholar] [CrossRef]
- Boschanski, M.; Krüger, T.; Karsten, L.; Falck, G.; Alam, S.; Gerlach, M.; Müller, B.; Müller, K.M.; Sewald, N.; Dierks, T. Site-Specific Conjugation Strategy for Dual Antibody–Drug Conjugates Using Aerobic Formylglycine-Generating Enzymes. Bioconjug. Chem. 2021, 32, 1167–1174. [Google Scholar] [CrossRef]
- Feiner, R.C.; Teschner, J.; Teschner, K.E.; Radukic, M.T.; Baumann, T.; Hagen, S.; Hannappel, Y.; Biere, N.; Anselmetti, D.; Arndt, K.M.; et al. rAAV Engineering for Capsid-Protein Enzyme Insertions and Mosaicism Reveals Resilience to Mutational, Structural and Thermal Perturbations. Int. J. Mol. Sci. 2019, 20, 5702. [Google Scholar] [CrossRef]
- Feiner, R.C.; Kemker, I.; Krutzke, L.; Allmendinger, E.; Mandell, D.J.; Sewald, N.; Kochanek, S.; Müller, K.M. EGFR-Binding Peptides: From Computational Design towards Tumor-Targeting of Adeno-Associated Virus Capsids. Int. J. Mol. Sci. 2020, 21, 9535. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, J.; Shi, B.Z.; Xu, Y.H.; Li, Z.H.; Gu, J.R. Application of EGFP-EGF fusions to explore mechanism of endocytosis of epidermal growth factor. Acta Pharmacol. Sin. 2007, 28, 111–117. [Google Scholar] [CrossRef]
- Nicholson, R.I.; Gee, J.M.W.; Harper, M.E. EGFR and cancer prognosis. Eur. J. Cancer 2001, 37, S9–S15. [Google Scholar] [CrossRef]
- Karsten, L.; Janson, N.; Le Joncour, V.; Alam, S.; Müller, B.; Tanjore Ramanathan, J.; Laakkonen, P.; Sewald, N.; Müller, K.M. Bivalent EGFR-Targeting DARPin-MMAE Conjugates. Int. J. Mol. Sci. 2022, 23, 2468. [Google Scholar] [CrossRef] [PubMed]
- Falck, G.; Müller, K. Enzyme-Based Labeling Strategies for Antibody–Drug Conjugates and Antibody Mimetics. Antibodies 2018, 7, 4. [Google Scholar] [CrossRef]
- Krüger, T.; Weiland, S.; Falck, G.; Gerlach, M.; Boschanski, M.; Alam, S.; Müller, K.M.; Dierks, T.; Sewald, N. Two-fold Bioorthogonal Derivatization by Different Formylglycine-Generating Enzymes. Angew. Chemie Int. Ed. 2018, 57, 7245–7249. [Google Scholar] [CrossRef] [PubMed]
- Krüger, T.; Weiland, S.; Boschanski, M.; Sinha, P.K.; Falck, G.; Müller, K.M.; Dierks, T.; Sewald, N. Conversion of Serine-Type Aldehyde Tags by the Radical SAM Protein AtsB from Methanosarcina mazei. ChemBioChem 2019, 20, 2074–2078. [Google Scholar] [CrossRef]
- Feiner, R.C.; Müller, K.M. Recent progress in protein-protein interaction study for EGFR-targeted therapeutics. Expert Rev. Proteomics 2016, 13, 817–832. [Google Scholar] [CrossRef]
- Shimizu, N.; Fukuzono, S.; Harada, Y.; Fujimori, K.; Gotoh, K.; Yamazaki, Y. Mass production of human epidermal growth factor using fed-batch cultures of recombinant Escherichia coli. Biotechnol. Bioeng. 1991, 38, 37–42. [Google Scholar] [CrossRef]
- Ferrer Soler, L.; Cedano, J.; Querol, E.; de Llorens, R. Cloning, expression and purification of human epidermal growth factor using different expression systems. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003, 788, 113–123. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, J.; Lin, J.; Wu, S.; Li, S.; Wang, J. High Efficient Expression, Purification, and Functional Characterization of Native Human Epidermal Growth Factor in Escherichia coli. BioMed Res. Int. 2016, 2016, 3758941. [Google Scholar] [CrossRef]
- Feiner, R.C.; Pennè, I.; Müller, B.; Müller, K.M. EGF-mCherry Fusion Protein Expressed in E. coli Shows Product Heterogeneity but a High Biological Activity. Biochemistry 2019, 58, 1043–1047. [Google Scholar] [CrossRef]
- Koushik, S.V.; Chen, H.; Thaler, C.; Puhl, H.L.; Vogel, S.S. Cerulean, venus, and venusY67C FRET reference standards. Biophys. J. 2006, 91, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Thaler, C.; Koushik, S.V.; Blank, P.S.; Vogel, S.S. Quantitative multiphoton spectral imaging and its use for measuring resonance energy transfer. Biophys. J. 2005, 89, 2736–2749. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, M.; Fabritius, A.; Basquin, J.; Griesbeck, O. Targeted In Situ Protein Diversification and Intra-organelle Validation in Mammalian Cells. Cell Chem. Biol. 2020, 27, 610–621.e5. [Google Scholar] [CrossRef] [PubMed]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An Enhanced Monomeric Blue Fluorescent Protein with the High Chemical Stability of the Chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, L.; Schelletter, L.; Hoffrogge, R.; Niehaus, K.; Rudolph, V.; Farr, M. Human Coxsackie- and adenovirus receptor is a putative target of neutrophil elastase-mediated shedding. Mol. Biol. Rep. 2022. [Google Scholar] [CrossRef]
- Brooke, D.; Movahed, N.; Bothner, B. Universal buffers for use in biochemistry and biophysical experiments. AIMS Biophys. 2015, 2, 336–342. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2017, 82, 518–529. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Hung, M.-C. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 2007, 282, 10432–10440. [Google Scholar] [CrossRef]
- Schmitz, J.; Täuber, S.; Westerwalbesloh, C.; von Lieres, E.; Noll, T.; Grünberger, A. Development and application of a cultivation platform for mammalian suspension cell lines with single-cell resolution. Biotechnol. Bioeng. 2021, 118, 992–1005. [Google Scholar] [CrossRef]
- Schmitz, J.; Hertel, O.; Yermakov, B.; Noll, T.; Grünberger, A. Growth and eGFP Production of CHO-K1 Suspension Cells Cultivated From Single Cell to Laboratory Scale. Front. Bioeng. Biotechnol. 2021, 9, 716343. [Google Scholar] [CrossRef]
- Schmitz, J.; Stute, B.; Täuber, S.; Kohlheyer, D.; Lieres, E.V.; Grünberger, A. Reliable cell retention of mammalian suspension cells in microfluidic cultivation chambers Abstract We present a new microfluidic trapping concept to retain randomly moving suspension cells inside a. bioRxiv Prepr. Serv. Biol. 2022. [Google Scholar] [CrossRef]
- Bolanos-Garcia, V.M.; Davies, O.R. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim. Biophys. Acta-Gen. Subj. 2006, 1760, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Robichon, C.; Luo, J.; Causey, T.B.; Benner, J.S.; Samuelson, J.C. Engineering Escherichia coli BL21(DE3) Derivative Strains To Minimize E. coli Protein Contamination after Purification by Immobilized Metal Affinity Chromatography. Appl. Environ. Microbiol. 2011, 77, 4634–4646. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; von den Driesch, L.; Galiez, C.; Martin, M.J.; Söding, J.; Steinegger, M. ColabFold - Making protein folding accessible to all Milot. bioRxiv Prepr. 2021. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A guide to fluorescent protein FRET pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Patterson, G.H.; Piston, D.W.; Barisas, B.G. Förster Distances between Green Fluorescent Protein Pairs. Anal. Biochem. 2000, 284, 438–440. [Google Scholar] [CrossRef]
- Lam, A.J.; St-Pierre, F.; Gong, Y.; Marshall, J.D.; Cranfill, P.J.; Baird, M.A.; McKeown, M.R.; Wiedenmann, J.; Davidson, M.W.; Schnitzer, M.J.; et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 2012, 9, 1005–1012. [Google Scholar] [CrossRef]
- Davidson, N.E.; Gelmann, E.P.; Lippman, M.E.; Dickson, R.B. Epidermal growth factor receptor gene expression in estrogen receptor-positive and negative human breast cancer cell lines. Mol. Endocrinol. 1987, 1, 216–223. [Google Scholar] [CrossRef]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Bizzarri, R.; Serresi, M.; Luin, S.; Beltram, F. Green fluorescent protein based pH indicators for in vivo use: A review. Anal. Bioanal. Chem. 2009, 393, 1107–1122. [Google Scholar] [CrossRef] [PubMed]
- Grabe, M.; Oster, G. Regulation of Organelle Acidity. J. Gen. Physiol. 2001, 117, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.Q.; Luo, S.; Twomey, J.D.; Zhang, B. Targeting cancer with antibody-drug conjugates: Promises and challenges. MAbs 2021, 13, 1951427. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, C.; Cen, J.; Bai, Y.; He, W.; Guo, Z. Ratiometric detection of pH fluctuation in mitochondria with a new fluorescein/cyanine hybrid sensor. Chem. Sci. 2015, 6, 3187–3194. [Google Scholar] [CrossRef] [PubMed]
- Ke, G.; Zhu, Z.; Wang, W.; Zou, Y.; Guan, Z.; Jia, S.; Zhang, H.; Wu, X.; Yang, C.J. A Cell-Surface-Anchored Ratiometric Fluorescent Probe for Extracellular pH Sensing. ACS Appl. Mater. Interfaces 2014, 6, 15329–15334. [Google Scholar] [CrossRef]
- Xue, Z.; Zhao, H.; Liu, J.; Han, J.; Han, S. Imaging Lysosomal pH Alteration in Stressed Cells with a Sensitive Ratiometric Fluorescence Sensor. ACS Sens. 2017, 2, 436–442. [Google Scholar] [CrossRef]
- Kaur, A.; Haghighatbin, M.A.; Hogan, C.F.; New, E.J. A FRET-based ratiometric redox probe for detecting oxidative stress by confocal microscopy, FLIM and flow cytometry. Chem. Commun. 2015, 51, 10510–10513. [Google Scholar] [CrossRef]
- Valkonen, M.; Mojzita, D.; Penttilä, M.; Benčina, M. Noninvasive High-Throughput Single-Cell Analysis of the Intracellular pH of Saccharomyces cerevisiae by Ratiometric Flow Cytometry. Appl. Environ. Microbiol. 2013, 79, 7179–7187. [Google Scholar] [CrossRef]
- Mahon, M.J. pHluorin2: An enhanced, ratiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol. 2011, 2, 132–137. [Google Scholar] [CrossRef]
- Lin, C.-F.; Lee, G.-B.; Wang, C.-H.; Lee, H.-H.; Liao, W.-Y.; Chou, T.-C. Microfluidic pH-sensing chips integrated with pneumatic fluid-control devices. Biosens. Bioelectron. 2006, 21, 1468–1475. [Google Scholar] [CrossRef]
- Tsai, Y.-T.; Chang, S.-J.; Ji, L.-W.; Hsiao, Y.-J.; Tang, I.-T. Fast Detection and Flexible Microfluidic pH Sensors Based on Al-Doped ZnO Nanosheets with a Novel Morphology. ACS Omega 2019, 4, 19847–19855. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Y.; Han, Y.; Gao, Y.; Wang, C.; Han, L.; Zhang, Y. Graphene-based field-effect transistors integrated with microfluidic chip for real-time pH monitoring of seawater. J. Mater. Sci. Mater. Electron. 2020, 31, 15372–15380. [Google Scholar] [CrossRef]
- Lee, S.; Ibey, B.L.; Coté, G.L.; Pishko, M.V. Measurement of pH and dissolved oxygen within cell culture media using a hydrogel microarray sensor. Sens. Actuators B Chem. 2008, 128, 388–398. [Google Scholar] [CrossRef]
- Magnusson, E.B.; Halldorsson, S.; Fleming, R.M.T.; Leosson, K. Real-time optical pH measurement in a standard microfluidic cell culture system. Biomed. Opt. Express 2013, 4, 1749. [Google Scholar] [CrossRef] [PubMed]
- Abou-Hassan, A.; Dufrêche, J.-F.; Sandre, O.; Mériguet, G.; Bernard, O.; Cabuil, V. Fluorescence Confocal Laser Scanning Microscopy for pH Mapping in a Coaxial Flow Microreactor: Application in the Synthesis of Superparamagnetic Nanoparticles. J. Phys. Chem. C 2009, 113, 18097–18105. [Google Scholar] [CrossRef]
- Shimazu, M.; Mulchandani, A.; Chen, W. Cell Surface Display of Organophosphorus Hydrolase Using Ice Nucleation Protein. Biotechnol. Prog. 2001, 17, 76–80. [Google Scholar] [CrossRef]
- Lee, K.-H.; Lee, K.-Y.; Byun, J.-Y.; Kim, B.-G.; Kim, D.-M. On-bead expression of recombinant proteins in an agarose gel matrix coated on a glass slide. Lab Chip 2012, 12, 1605. [Google Scholar] [CrossRef]
- Suzuki, T.; Arai, S.; Takeuchi, M.; Sakurai, C.; Ebana, H.; Higashi, T.; Hashimoto, H.; Hatsuzawa, K.; Wada, I. Development of Cysteine-Free Fluorescent Proteins for the Oxidative Environment. PLoS ONE 2012, 7, e37551. [Google Scholar] [CrossRef]
- Luria, Y.; Raichlin, D.; Benhar, I. Fluorescent IgG fusion proteins made in E. coli. MAbs 2012, 4, 373–384. [Google Scholar] [CrossRef]
- Haas, A.K.; von Schwerin, C.; Matscheko, D.; Brinkmann, U. Fluorescent Citrine-IgG fusion proteins produced in mammalian cells. MAbs 2010, 2, 648–661. [Google Scholar] [CrossRef][Green Version]
- Kaneyoshi, K.; Yamano-Adachi, N.; Koga, Y.; Uchiyama, K.; Omasa, T. Analysis of the immunoglobulin G (IgG) secretion efficiency in recombinant Chinese hamster ovary (CHO) cells by using Citrine-fusion IgG. Cytotechnology 2019, 71, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Gross, L.A.; Baird, G.S.; Hoffman, R.C.; Baldridge, K.K.; Tsien, R.Y. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 2000, 97, 11990–11995. [Google Scholar] [CrossRef] [PubMed]
- Merzlyak, E.M.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.A.; Lukyanov, S.; Gadella, T.W.J.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, J.; Schenk, A.; Rocker, C.; Girod, A.; Spindler, K.-D.; Nienhaus, G.U. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria). Proc. Natl. Acad. Sci. USA 2002, 99, 11646–11651. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Oh, Y.; Sens, A.; Ataie, N.; Dana, H.; Macklin, J.J.; Laviv, T.; Welf, E.S.; Dean, K.M.; Zhang, F.; et al. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 2016, 34, 760–767. [Google Scholar] [CrossRef]
- Fabritius, A.; Ng, D.; Kist, A.M.; Erdogan, M.; Portugues, R.; Griesbeck, O. Imaging-Based Screening Platform Assists Protein Engineering. Cell Chem. Biol. 2018, 25, 1554–1561.e8. [Google Scholar] [CrossRef]
- Wannier, T.M.; Gillespie, S.K.; Hutchins, N.; McIsaac, R.S.; Wu, S.-Y.; Shen, Y.; Campbell, R.E.; Brown, K.S.; Mayo, S.L. Monomerization of far-red fluorescent proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E11294–E11301. [Google Scholar] [CrossRef]
- Subach, F.V.; Patterson, G.H.; Renz, M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Bright Monomeric Photoactivatable Red Fluorescent Protein for Two-Color Super-Resolution sptPALM of Live Cells. J. Am. Chem. Soc. 2010, 132, 6481–6491. [Google Scholar] [CrossRef]
- Pennacchietti, F.; Serebrovskaya, E.O.; Faro, A.R.; Shemyakina, I.I.; Bozhanova, N.G.; Kotlobay, A.A.; Gurskaya, N.G.; Bodén, A.; Dreier, J.; Chudakov, D.M.; et al. Fast reversibly photoswitching red fluorescent proteins for live-cell RESOLFT nanoscopy. Nat. Methods 2018, 15, 601–604. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef]
- Wiedenmann, J.; Vallone, B.; Renzi, F.; Nienhaus, K.; Ivanchenko, S.; Röcker, C.; Nienhaus, G.U. Red fluorescent protein eqFP611 and its genetically engineered dimeric variants. J. Biomed. Opt. 2005, 10, 014003. [Google Scholar] [CrossRef] [PubMed]
- Kredel, S.; Oswald, F.; Nienhaus, K.; Deuschle, K.; Röcker, C.; Wolff, M.; Heilker, R.; Nienhaus, G.U.; Wiedenmann, J. mRuby, a Bright Monomeric Red Fluorescent Protein for Labeling of Subcellular Structures. PLoS ONE 2009, 4, e4391. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Kim, J.M.; Cluzel, P. Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 2018, 15, 47–51. [Google Scholar] [CrossRef]
- Subach, O.M.; Vlaskina, A.V.; Agapova, Y.K.; Dorovatovskii, P.V.; Nikolaeva, A.Y.; Ivashkina, O.I.; Popov, V.O.; Piatkevich, K.D.; Khrenova, M.G.; Smirnova, T.A.; et al. LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift. Int. J. Mol. Sci. 2021, 22, 12887. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karsten, L.; Goett-Zink, L.; Schmitz, J.; Hoffrogge, R.; Grünberger, A.; Kottke, T.; Müller, K.M. Genetically Encoded Ratiometric pH Sensors for the Measurement of Intra- and Extracellular pH and Internalization Rates. Biosensors 2022, 12, 271. https://doi.org/10.3390/bios12050271
Karsten L, Goett-Zink L, Schmitz J, Hoffrogge R, Grünberger A, Kottke T, Müller KM. Genetically Encoded Ratiometric pH Sensors for the Measurement of Intra- and Extracellular pH and Internalization Rates. Biosensors. 2022; 12(5):271. https://doi.org/10.3390/bios12050271
Chicago/Turabian StyleKarsten, Lennard, Lukas Goett-Zink, Julian Schmitz, Raimund Hoffrogge, Alexander Grünberger, Tilman Kottke, and Kristian M. Müller. 2022. "Genetically Encoded Ratiometric pH Sensors for the Measurement of Intra- and Extracellular pH and Internalization Rates" Biosensors 12, no. 5: 271. https://doi.org/10.3390/bios12050271
APA StyleKarsten, L., Goett-Zink, L., Schmitz, J., Hoffrogge, R., Grünberger, A., Kottke, T., & Müller, K. M. (2022). Genetically Encoded Ratiometric pH Sensors for the Measurement of Intra- and Extracellular pH and Internalization Rates. Biosensors, 12(5), 271. https://doi.org/10.3390/bios12050271





