Abstract
MXene is a two-dimensional (2D) nanomaterial that exhibits several superior properties suitable for fabricating biosensors. Likewise, the nucleic acid (NA) in oligomerization forms possesses highly specific biorecognition ability and other features amenable to biosensing. Hence the combined use of MXene and NA is becoming increasingly common in biosensor design and development. In this review, MXene- and NA-based biosensors are discussed in terms of their sensing mechanisms and fabrication details. MXenes are introduced from their definition and synthesis process to their characterization followed by their use in NA-mediated biosensor fabrication. The emphasis is placed on the detection of various targets relevant to agricultural and food systems, including microbial pathogens, chemical toxicants, heavy metals, organic pollutants, etc. Finally, current challenges and future perspectives are presented with an eye toward the development of advanced biosensors with improved detection performance.
1. Introduction
The detection technologies for quality and safety monitoring in agricultural and food systems help improve our overall quality of life. Conventional methods to detect biological and/or chemical entities include plate-counting tests, enzyme-linked immunosorbent assays (ELISA), liquid and gas chromatography (LC and GC), etc. These methods are slow and expensive, requiring complicated pre-treatment of samples, sophisticated instruments, and highly qualified personnel to perform the tests inside a laboratory. To this end, biosensors are being developed to afford inexpensive, portable, and easy-to-operate devices for on-field detection. Biosensors are usually composed of two functional elements: (1) a bio-recognizer (e.g., enzyme, antibody, or nucleic acid (NA) sequences) that specifically recognizes and/or conjugates with target molecules and (2) a physiochemical transducer that translates the biorecognition event into a corresponding detectable signal [1,2].
NAs in oligomerization forms are usually integrated into biosensors as biorecognition elements due to their high specificity toward different target entities [3]. The most well-known NAs are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). According to the development stage, there are three kinds of NA biosensors, each with a unique function: (1) genosensor, (2) aptasensor, and (3) DNAzyme/aptazyme biosensors [4,5]. These biosensors are now widely explored and used in different areas, e.g., food safety monitoring, environment analysis, etc. [6].
The performance of NA biosensors is significantly increased by incorporating nanomaterials (NMs) that are employed as transducers, which further facilitate the immobilization of NA. NMs can be modified by different ligands that enhance NA–ligand interactions in terms of their superb physicochemical properties. Additionally, NMs with exceptional optical, electrical, and thermal conductivities and mechanical performance help establish various types of sensing devices with high sensing performance. A high surface-area-to-volume ratio of NMs provides a larger surface area for more surface functionalization and consequently increases the NA-loading efficiency and surface immobilization, which is critical to improving the detection sensitivity [7].
Two-dimensional (2D) NMs with sheet-like architectures, which due to their large surface area provide abundant reactive sites for ligand modification to bind with NA, are commonly used in biosensors [8]. Among these are 2D graphene-like MXenes that are hexagonal layered NMs such as carbides, nitrides, and carbonitrides and contain early transition metals [9]. They are usually synthesized from their parent bulk ceramic called the MAX phase, notated as Mn+1AXn, where M is an early transition metal, e.g., Ti, Mo, Nb, etc.; A is an sp element that is primarily in group 13 or 14 from the periodic table; X is a carbon or nitrogen element (Figure 1(Aa,Ac)) [10]. After the sp element is etched, the remaining structures of the MAX phase are collectively called MXene (Figure 1(Ab,Ac)) [11]. Depending on the chemical etching method, various additional elements or groups may be anchored on the surface of MXene via hydrogen bonds or van der Waals bonds. The final MXene is represented as Mn+1XnTx, where T represents different modifiers on the surface (typically fluoride (-F), chloride (-Cl), hydroxyl (-OH), oxygen (=O), etc.) [12,13]. Several unique merits in final MXene products make MXene a better alternative 2D NM in replacing other current 2D NMs. Based on the way they are synthesized, MXenes exhibit high reductivity that can reduce oxidizing reagents and even metal salt precursors without adding extra reducing agents [14]. Due to abundant M and X components, MXenes have a high electrical conductivity (6000–8000 S cm−1), which is highly beneficial to electron (e−) movement throughout the materials [15]. Additionally, the large surface area and the extra chemical terminals of MXene provide a platform for binding bio-recognition elements and modifying the surface that enhances biosensing performance [16]. Notably, with different extra atoms/polar groups terminating, the surface of the MXene products is much more easier to be chemically modified, consequently further facilitating and stabilizing the NA immobilization.
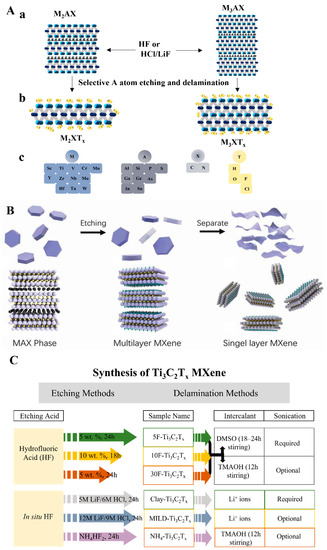
Figure 1.
(A) MAX phase and its etched products. (a) Three typical MAX phase structures with selective etching sites (atoms in red). (b) Selectively etched products (MXene) with surface modification (atoms in yellow). (c) Atoms in MAX phases and MXene structures. Redrawn based on Ref. [11]. (B) Illustration for MXene top-down synthesized process from its precursor. Reproduced from Ref. [18]. (C) General idea for MXene top-down etched from MAX phases with two typical routes. Redrawn with permission from Ref. [21].
Herein we discuss and summarize the research efforts in developing NA biosensors based on 2D MXene for quality and safety monitoring of agri-food systems. The major classes of NA biosensors, such as genosensors, aptasensors, and DNAzyme sensors, are examined emphasizing their sensing mechanisms and fabrication details. We also present the future challenges and opportunities for NA biosensors in the monitoring of agri-food systems.
2. MXene Synthesis and Characterization
There are over 25 top-down and bottom-up approaches for the synthesis of MXene [17]; the top-down approaches are more widely applied (Figure 1B) [18]. As mentioned, MXenes are usually prepared from their MAX phase precursors, where the atomic layer A is removed by chemical or mechanical methods. Owing to covalent, partial ionic, and metallic bonds, the hexagonal layered MAX phase is observed in a stacked form and shows weakly metallic properties [19]. Unlike stacked graphite with π–π bonds, the bulk MAX phase is usually tightly held by partial ionic bonds, making it hard for MXene to separate from its precursors by using mechanical methods such as ultrasonication or mechanical exfoliation alone [20]. Therefore, selective chemical exfoliation is the predominant way to remove the atomic interlayer. The exfoliation is typically done by either hydrofluoric (HF) acid etching or in situ HF etching method (Figure 1C) [21].
Chemical vapor deposition (CVD) is one of the most common ways to scalably and controllably produce 2D NMs in the bottom-up method, where layered high-quality 2D NMs are grown on a substrate at a reasonable cost [22]. Consequently, top-down and bottom-up approaches are extensively explored to develop various MXene composites and structures for NA biosensor applications. This section introduces several popular MXene preparation methods.
2.1. Top-Down Etching of MAX Phase Precursors
2.1.1. HF Etching
HF etching can selectively etch the A interlayer without damaging other adjacent atomic structures [23]. Typically, concentrated aqueous HF solution (50%) is mixed with MAX phase powder at room temperature for several hours (Figure 2(Aa)) [24]. During this, the A layer is selectively removed by the strong acid solution, releasing hydrogen gas (H2) [25]. Concomitantly, other functional groups including -F, -OH, and =O are present on the surface of the remaining structure by van der Waals bonds [26]. The resulting solution is further centrifuged to precipitate the powder products, followed by washing with deionized (DI) water several times to remove excess HF solution. To maximize the abilities of MXene, it is critical to collect the separated MXene nanosheets using different intercalants where the interlayer spacing between the nanosheets is expanded enough to diminish the interaction between each adjacent nanosheet [27]. Dimethyl sulfoxide (DMSO) is usually used as an intercalant solvent that assists in delamination, followed by sonication. Other intercalating compounds, especially tetraalkylammonium salts such as tetrabutylammonium hydroxide (TBAOH), can also be used to separate MXene nanosheets with the aid of sonication and eventually increase d-spacing to achieve thermodynamic stability [27].
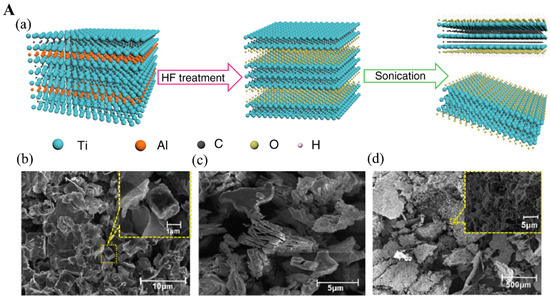
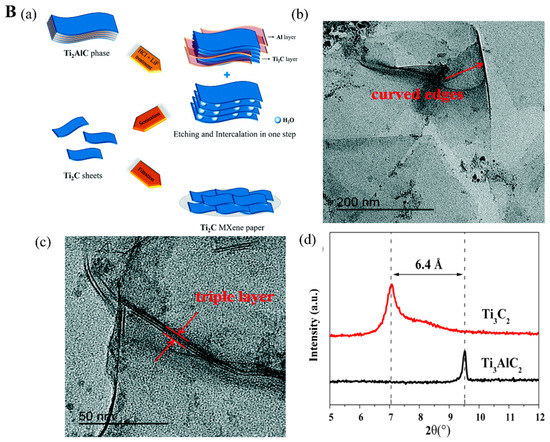
Figure 2.
(A). (a) Schematic of Ti3C2Tx MXene synthesis via HF-etching. The SEM images for (b) Ti3AlC2 MAX phase, (c) multilayer Ti3C2Tx MXene, (d) delaminated Ti3C2Tx MXene nanosheets. Reproduced with permission from Refs. [24,28]. (B). (a) Ti2C MXene synthesis process. (b,c) TEM for exfoliated Ti2C MXene nanosheets. Reproduced with permission from Ref. [30]. (d) X-ray diffraction spectra for Ti3C2 MXene its MAX phase precursor. Reproduced with permission from Ref. [31].
The scanning electron microscopy (SEM) pictures of the typical titanium-based MAX phase (Ti3Al1.15C2 powder) with its etched product (Ti3C2 MXene) nanostructures obtained before and after HF etching are shown in Figure 2(Ab,Ac). Figure 2(Ac) shows an accordion-like 2D Ti3C2 nanosheet structure differentiated from Figure 2(Ab). After TBAOH delamination, the exfoliated Ti3C2 nanosheets exhibit several nanobridges at the outer edges, demonstrating that the Ti3C2 nanosheets are successfully exfoliated from their MAX phase precursors (Figure 2(Ad)) [28]. This pristine MXene synthesized by the HF method possesses excellent electrochemical, optical, chemical, and mechanical properties [29].
2.1.2. In Situ HF Etching
Unlike in the HF etching method where pure aqueous HF solution is added, in the in situ HF method, a mixture of molten fluoride or bifluoride-based salts and hydrogen chloride (HCl) is used as etchants. During the etching process, fluoride ion (F−)-containing salts including lithium fluoride (LiF) and ammonium hydrogen difluoride (NH4HF2) are added into the MAX phase to remove the A atomic interlayer resulting from the reactions between F- and A layer [32]. Solvated lithium ions (Li+) intercalated between the interlayer of the MAX phase can also drive the delamination process, ultimately producing exfoliated MXene nanosheets [33]. This method offers several advantages over the HF etching method, such as the release of a low amount of hazardous compounds, mild conditions, and simultaneous delamination as a result of cations (e.g., Li+) [34].
Li et al. [30] fabricated Ti2CTx MXene nanoflake-based paper using the aqueous HCl and LiF salts for etching the Al interlayer of the Ti2AlC precursor (Figure 2(Ba)). Individual exfoliated Ti2CTx MXene nanoflakes were obtained after the mild sonication of the resultant products due to the intercalation of solvated Li+ between the exfoliated layers. Finally, individual Ti2CTx MXene nanoflakes were dispersed with carbon nanotubes to prepare the Ti2CTx MXene paper. The transmission electron microscope (TEM) (Figure 2(Bb,Bc)) images show a majority of LiF/HCl-Ti2CTx MXene with wrinkled and curved edges, which is attributed to the high flexibility of the MXene paper. In another investigation, an aqueous NH4HF2 solution is mixed with MAX phase (Ti3AlC2), which functions as an etchant solution to remove the Al layer and produce the Ti3C2Tx MXene nanosheets [31]. Typically, MAX phases are highly anisotropic and their c-lattice parameter can easily be measured by powder X-ray diffraction (XRD) based on a (002) peak at 2θ around 10° [35,36]. According to Figure 2(Bd), the exfoliated Ti3C2 nanosheets show the (002) peak at 2θ = 7.1° shifted from 2θ = 9.6° (for Ti3AlC2 precursor) with calculated c-lattice parameter reaching 24.9 Å based on Bragg equation due to NH4+ intercalation effect. It indicates that the d-spacing increases while the thickness of Ti3C2 nanosheets reduces [13,31]. The NH4HF2 etching method provides the resulting Ti3C2 nanosheets with large interplanar spacing comparable to other fluoride-based etched methods, which allows the Ti3C2 nanosheets to be obtained in a single-step process. Ti3C2Tx MXene with fewer layers has higher conductivity (4600 ± 1100 S cm−1) with excellent field-effect electron mobility of 2.6 ± 0.7 cm2 V−1 S−1, which facilitates the electron transferring throughout the transducer, finally enhancing the sensitivity of electro-related biosensors [37]. Additionally, ultrathin MXene micropatterned on glass substrates for field-effect transistor fabrication provides a highly sensitive sensing platform for dopamine neurotransmitters analysis [38].
2.2. Bottom-up Synthesis of MXene
CVD is one of the most common bottom-up methods for high-quality and large-area NMs fabrication that is widely used in multiple practical applications, including optoelectronics and solar cell devices [39,40]. Typically, the precursors and substrate are preplaced in a chamber where extremely high temperatures are applied to heat or decompose the precursors, leading to the preheated or decomposed precursors growing on the surface of the substrate [41]. Consequently, more purified NM structures are developed. Several 2D MXenes were achieved via CVD. For example, ultrathin 2D molybdenum carbide (Mo2C) MXene was collected from the graphene-templated growth of Mo2C MXene film by the CVD method (Figure 3A) [42]. In the CVD process, copper (Cu) covered on the Mo foil was placed in a quartz chamber tube where 1100 °C was applied under a continuous flow of gaseous hydrogen (H2) and carbon-source methane (CH4). As a result, the morphology of the final Mo2C crystal MXene is largely dependent on the CH4 flow rate. With a low rate of CH4 flow, homogeneous structures of Mo2C crystal are obtained, while with a high rate of CH4 flow Mo2C/graphene heterostructures are obtained (see Figure 3(Ba,Bc)). When prepared on a Cu surface under a high CH4 flow rate, the final Mo2C crystal MXene had a more uniform structure with significantly lower thickness than when prepared on a graphene surface (Figure 3(Bb,Bd)). This indicates that CVD-synthesized graphene drastically suppresses the growth rate, thus reducing the thickness of as-prepared Mo2C MXene. Though CVD can control the thickness of the MXene nanosheets, a slow synthesis with a lower yield limits its use in MXene preparation.
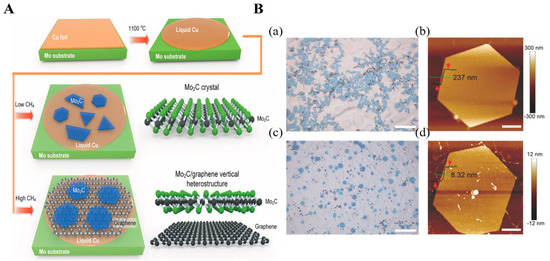
Figure 3.
(A) Illustrative diagram for the Mo2C products growth under the high and low flow rates of CH4 gas. (B) Surface morphology of synthesis crystals: (a,c) Mo2C crystals’ physical distribution on Cu surface and graphene under optical images under low (a) and high (c) CH4 flow rates; (b,d) topological image of hexagonal Mo2C structures on the Cu surface, (b,d) graphene surface. Reproduced with permission from Ref. [42].
Several synthetic methods discussed in this section have been proposed and successfully used to prepare different types of MXene and manipulate their surface, which largely influences the optical/fluorescent and electrochemical properties of the final MXene products. These altered properties further extend the use of MXenes for biosensing applications.
3. MXene in NA Biosensors
The NA sequences are used as biorecognition elements in biosensors to provide target-specific information to the transducer where the biorecognition event is transformed into a measurable signal such as optical, electrochemical, etc. While the specificity of the target depends on the biorecognition element, the sensitivity of the detection signal depends on the properties of the transducer element, for example, its electrical conductivity. Typically, 2D NMs, such as MXene, graphene, etc., that possess a large surface area to volume ratio can provide high signal sensitivity.
Single-strand DNA (ssDNA) or RNA macromolecules naturally hybridize with their complementary strands to form double-stranded molecules with high stability. Thus, NA-based bio-probes are widely used in different biosensors as biorecognition elements [43]. The hybridization performance benefits the genoanalytical device development for NA analysis and monitoring in many areas, particularly in DNA microarray, gene lab-on-a-chip technology, and amplification process in a polymerase chain reaction (PCR) [44,45]. The considerable evolution of NA fragments on high affinity and specificity to non-NAs proceeded when those specific fragments were isolated from large libraries incorporated with the systematic evolution of ligands by the exponential enrichment (SELEX) method [46,47]. Those NA fragments or aptamers have boosted applications of NA-based biosensors which are known as aptasensors [48]. NAs have also been shown to behave as a catalyst since Kruger et al. reported the first ribozymes [49]. Now DNAzymes, RNAzymes, aptazymes, and ribozymes play vital roles in biosensing systems.
NA-based biosensors can be divided into three categories based on their different abilities as bio-recognized probes: (1) genosensor, (2) aptasensor, and (3) DNAzyme/RNAzyme biosensor. An overview of these biosensors, their fabrication details, and the role of MXene in them are presented herein.
3.1. NA-Based Biosensor
3.1.1. Genosensor
A genosensor is developed by taking advantage of the hybridization reaction between selected ssDNA or RNA with their complementary counterpart [50]. In this selected ssDNA or RNA, serving as the biorecognition element is immobilized on the transducer surface, which recognizes and hybridizes with the target DNA or RNA. The hybridization event is transmitted through a transducer and transformed into a measurable signal. While the specificity of the genosensor depends on the selection of the biorecognition element, the sensitivity of the genosensor is largely influenced by the structures and properties of the NMs that constitute the transducer.
Several gene probes in natural or chemical-changed forms have been designed and utilized in genosensors [51,52]. Most commonly, natural nucleonic acid (NNA: DNA/RNA nucleotide) (Figure 4(Aa)) contributes to the major backbone of linear oligonucleotides, either pre- or in situ synthesis [53,54]. Nevertheless, hairpin-like oligonucleotides have been popularly applied recently. A hairpin oligonucleotide has a unique structure with a self-complementary characteristic that is patterned as a closed-stem zone, where base pairs are formed intramolecularly and an opened-loop zone exposed to the external environment with a capture sequence [55,56]. Chemically modified natural DNA/RNA analogs have emerged as promising critical components of the gene probes used in various genosensors. For example, one of the most popular analogs is locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), which possesses superior RNA-binding affinity and excessively stable structure [57]. LNA is a bicyclic-based structure where 2’ oxygen and 4’ carbon are locked via an extra bridge in a flexible ribose motif (Figure 4(Ab)) [58]. With these exceptional properties, LNA is the most used in clinical diagnosis, particularly in microRNA detection. Another well-known NA analog is N-(2-aminoethyl)-glycine-based peptide nucleic acid (PNA) unit, which is a synthetic NA mimic with an achiral and uncharged structure (Figure 4(Ac)). Different bases are linked to this backbone as a side chain by methylene carbonyl groups. As a result, PNA oligonucleotide molecules in uncharged form have high affinity when hybridized with the negatively charged DNA/RNA to form a more stable dsDNA/PNA or RNA/PNA structure in any buffer solution. These hybrid double-stranded NAs have high specificity and accurately matching event, which are beneficial to PNA-based genosensors [59].
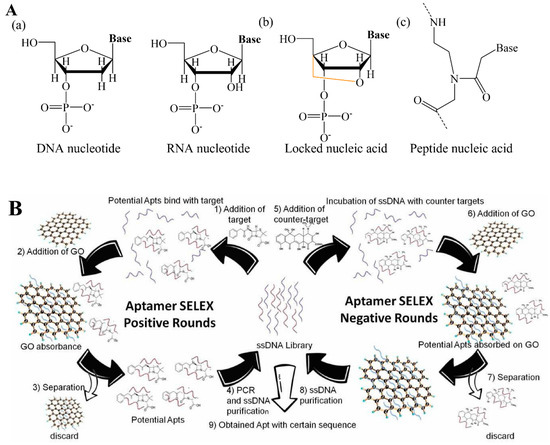
Figure 4.
(A) Chemical structures of nucleic acids: (a) DNA and RNA nucleotide, (b) locked nucleic acid, (c) peptide nucleic acid. (B) Schematic illustration of aptamer selection by positive and negative SELEX methods. Reproduced with permission from Ref. [60]. Copyright 2022, Elsevier.
3.1.2. Aptasensor
An aptasensor uses the aptamer, an ssDNA sequence with specific recognition ability to one type of chemical target. Similar to the construction of the genosensor, the aptasensor also consists of the specific structure of the ssDNA/RNA ligand that is immobilized on the transducer surface to provide measurable signals for targets that have substantial health indications to humans or animals [61].
Most aptamers are selected using the SELEX method. In principle, the SELEX requires a large library containing more than 1015 oligonucleotide sequence structures with a maximum of 60 random synthetic oligonucleotide regions flanked by two constant short regions as primers for PCR amplification [47]. After the target is incubated with oligonucleotide pools under suitable conditions for a while, the unbound aptamers are removed, and the target/aptamers are separated. The separated aptamers are either directly amplified by PCR or amplified by PCR after in vitro transcription and reversed transcription, based on the type of NA (DNA/RNA). The amplified DNA/RNA PCR sequences then enter the subsequent selection round. The final selected aptamer is characterized by its binding kinetics via multiple methods [46]. Depending on the integration methods of SELEX, several SELEX methods have emerged for preferred aptamer selection, including cell-SELEX, microfluidic-SELEX, capillary electrophoresis-SELEX methods, etc. [62,63]. In our group, we obtained the aptamer specific against antibiotic penicillin (PenG) via graphene oxide (MAX)-SELEX method described in Figure 4B [60]. The total, selection rounds are generally divided into (1) positive rounds and (2) negative rounds. During the positive rounds, PenG was incubated with an ssDNA pool under proper conditions where PenG-ssDNA conjugation forms. Subsequently, MAX was introduced to remove unbounded ssDNA, taking advantage of the π–π staking between MAX and free-ssDNA, which are then discarded. The resulting bounded ssDNA were recovered via alcohol to obtain a ssDNA pool for PCR amplification and a subsequent couple of selection processes. The final obtained ssDNA mixture was further repeatedly selected via negative rounds where other interfering antibiotics were introduced to attach and separate less specific ssDNA. Three PenG aptamers were finally obtained (PenG-1, PenG-2, PenG-3), among which PenG-1 showed the lowest dissociation constant (Kd) as 105.15 ± 1.94 nM, indicating that this aptamer has the highest affinity to PenG due to the inverse relationship between the Kd value and binding affinity. Based on the SELEX method, more than a thousand aptamers have been selected to bind different targets, including pathogens, antibiotics, mycotoxins, pollutants, etc., which further extends the applications of aptasensors [64].
3.1.3. NA Enzyme (NAzyme) Biosensor
The notion that all enzymes are proteins has permanently changed since the discovery of RNAs with enzyme function (ribozymes) [65,66]. However, natural ribozymes can only catalyze a limited number of biological reactions, mostly in phosphodiester bond cleavage, splicing, and peptide bond formation [67]. In contrast, artificial ribozymes synthesized and selected by in vitro selection or in vitro evolution have broad catalytic abilities in additional chemical reactions such as phosphodiester bond formation, aminoacylation in coenzyme A for multiple metabolic processes, etc. [68,69]. Later on, artificial DNA molecules were also identified with enzymatic activities (DNAzyme) that allow them to mediate RNA/DNA cleavage and ligation, porphyrin metalation, redox reactions as peroxidases do, etc. [70]. Currently, a new generation of NAs known as aptazymes is being used for biosensing, which integrates aptamer and DNAzyme technologies to specifically identify a target and catalyze biochemical reactions [71].
Once the NAzyme biosensors are assembled for the detection of different targets, the target molecules usually act as cofactors either in accelerating or inhibiting biochemical reactions triggered by the NAzyme probe [72]. Therefore, an appropriate NAzyme biosensor design needs to be considered, as well as the way the catalytic reaction only occurs in the presence of the target. Theoretically, all DNAzyme and aptazyme systems can be employed in biosensors. The most common DNAzyme biosensors are based on the RNA-cleavage DNAzyme probe, taking advantage of both RNA-cleavage DNAzyme and catalytic products released from the RNA-cleavage reaction [73]. The metal ions or amino acids are two critical elements in the RNA-cleavage activity due to the mechanism of the catalytic reaction that requires a specific cofactor during the DNAzyme catalysis. As a result, the preference for using the RNA-cleavage DNAzyme biosensor is for the metal ion analysis or amino acids and peptides detection [74]. An RNA-cleavage fluorogenic DNAzymes (RFDs) for sensing Legionella pneumphila was selected and isolated from a DNA pool of 1014 oligonucleotide structures containing 40 random nucleotides by utilizing the counter selection method. The structure of the library used for this is shown in Figure 5A [75]. The random domain of RFDs was examined, as shown in Figure 5B. These DNAzymes were tailed with thymidine nucleotide-based fluorophore substrates at the 5′-end and quenchers at the 3′-end (Figure 5C). This unique feature elicits a fluorescence signal in the biosensor. The DNAzymes are highly sensitive to Legionella pneumophila strains due to the activation of those bacterial pathogens by specific RNase I protein sequences that were listed as the identity of these strains [76].
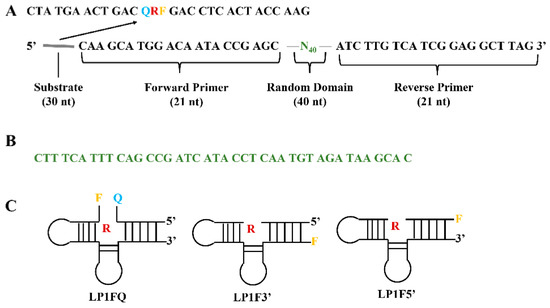
Figure 5.
(A). The DNA general construct in the library for selection of DNAzyme specific to Legionella pneumphila. (B) The random domain of selective Legionella pneumphila DNAzyme. (C) Representative construction of selected DNAzyme. R: Adenosine ribonucleotide, F: Fluorescein tail molecules, Q: quencher tail molecules (DABCYL). Redrawn based on Ref. [75].
3.2. Role of MXene in NA-Based Biosensors
The MXene nanoflakes are used in biosensors not only for their large surface area to facilitate NA immobilization for target capture but also their ability to transduce the physiochemical interaction into measurable signals. With the surface chemically terminated by several external single atoms and polar groups, MXene nanosheets are capable of interacting with NA by van der Waals forces, hydrogen bonds, electrostatic attraction, and perhaps coordination bonds, which allows them to be a better material for biosensing system than other 2D NMs [77]. In addition, good biocompatibility and enzyme-responsive biodegradability of MXene nanosheets extend their applications for NA immobilization in the biological area [78,79]. The long-term durability of MXene nanosheets also benefits the reproducibility of biosensor measurements. The merit of MXene nanocomposites is anticipated to be significantly expanded and enhanced as they incorporate other nano(bio)materials. Kashefi-Kheyrabadi et al. [80] designed an electrochemical aptasensor for thyroxine analysis by immobilizing thyroxine aptamer on the MoS2/Ti3C2Tx MXene/Au nanosheets, with a limit of detection (LOD) of 0.39 pg/mL and a dynamic range from 7.8 × 10−1 to 7.8 × 106 pg/mL.
4. Application of MXene-Based NA Biosensors in the Agricultural Food System
As an emerging class of 2D NMs, MXene nanosheets have proven to be suitable transducer materials for biosensor applications. Hence, various NA biosensors with MXene as transducer NM have been developed for quality and safety monitoring of food products as listed in Table 1.

Table 1.
Summary of MXene-based NA biosensors for agri-food systems.
4.1. Pathogen Detection
The presence of pathogenic microorganisms in agri-food systems is a growing global concern for human and animal health. If not promptly detected, the spread of these pathogens through the food supply chain can become a major problem causing illnesses and loss of lives. Hence, simple and rapid biosensors are being developed for the detection of pathogens.
Wang et al. [82] developed an electrochemical and colorimetric signal-producing biosensor for the on-site detection of Vibrio parahaemolyticus (VP). VP is a gram-negative bacteria found in the ocean and estuaries that can cause inflammatory gastroenteritis in humans [112]. The VP detection system was developed using phenylboronic acid and ferrocene-modified platinum (Pt)-doped Ti3C2 MXene nanocomposites (PBA-Fc@Pt@MXene). The use of PBA-Fc@Pt@MXene provided high electrical conductivity and significantly enhanced the electrochemical signal intensity and produced a color change when the existence of hydrogen peroxide (H2O2) and 3,3′,5,5′-tetramethylbenzidine (TMB) was detected. Incorporated with a VP aptamer, this biosensor can both voltammetrically determine and colorimetrically visualize the presence of VP as low as 5 CFU/mL and 30 CFU/mL, respectively. The linear range for electrochemical measurement and colorimetric determination is 10–108 CFU/mL and 102–108 CFU/mL, respectively. This fabricated biosensing device provides a dual-mode detection method for highly effective VP detection and measurement.
Salmonella typhimurium (ST) is another dangerous foodborne pathogen that causes high morbidity and mortality [113]. A novel dual-target fluorescent aptasensor was fabricated for live ST and VP analysis [81]. As shown in Figure 6A, this fluorescent device was assembled by two magnetic bars (1 and 2), where bar 1 was used as a biorecognizer for target recognition and capture, and bar 2 served as a signal amplifier that released the enhanced fluorescent signal. Bar 1 was coated with aptamers capable of recognizing and capturing two bacteria strains. Bar 2, modified with Ti3C2 MXene nanosheets, served as a platform to support and immobilize complementary DNA (cDNA) tailed with polyhedral oligomeric silsesquioxane-perovskite quantum dots probes (cDNA-POSS-PQDs). When the two target bacteria were present, they were captured by their specific aptamers on bar 1 and subsequently released with their aptamers into the aqueous supernatant. Due to the agitation, bacteria–aptamer conjugate attached to the cDNA on bar 2 and then tore away the cDNA-POSS-PQDs from bar 2, which finally provided a fluorescent response in the solution. The live bacteria can be released from the aptamer and become free again due to the reaction between the aptamer and cDNA-POSS-PQD with low Gibbs free energy (∆G < 0), leading them to trigger the next detection cycle. Ultimately, the fluorescent response was amplified and reached a maximum that significantly increased the biosensor sensitivity. LODs of 30 CFU/mL and 10 CFU/mL, respectively, for ST and VP, and a linear range of 102 to 106 CFU/mL were obtained (Figure 6(Ba,Bb)). Therefore, this biosensor can sensitively detect the target bacteria and differentiate the live from the dead ones.
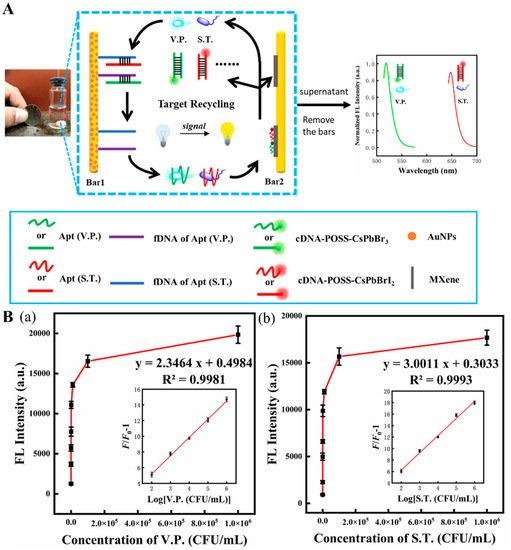
Figure 6.
(A) Scheme of fluorescent aptasensor for dual-targets detection with its signal amplification strategy. (B) (a,b) the fluorescence intensity of the aptasensor in the presence of VP (a) and ST (b). Inset: linear range for this aptasensor for two corresponding foodborne pathogens. Reproduced with permission from Ref. [81].
Simply using MXene and MXene-based nanocomposites for the NA immobilization yields only weak, unstable signals due to the activity of the live pathogens which may (1) escape or detach from the aptamer capture and stay free in the solution, and (2) disturb the signal when they are struggling with the aptamer capture. Hence, a MXene-based signal amplifier is used in NA biosensors to facilitate the on-site determination of live pathogens.
4.2. Mycotoxins Detection
Mycotoxins are toxic secondary fungal metabolites found in plant and animal foods. The consumption of some mycotoxin-containing foods beyond certain recommended amounts can cause serious health issues, including death. A sensor for the detection of mycotoxins in food and feed products will help enable a strategy to mitigate adverse health effects in humans and animals.
Zhang et al. [87] developed a switchable signal electrochemical aptasensor for the detection of mycotoxin Ochratoxin A (OTA) via a substrate-aptamer-signal amplifier sandwich structure. The OTA aptamer cDNA with a thiol group (-SH) was immobilized on the surface of MXene/Au nanocomposites-coated paper device via Au-S covalent bond (Figure 7(Aa)), employed as the transducer substrate coated on the working electrode. OTA aptamers tailed with peroxidase mimic nanostructure based on Pt nanoparticles-doped NiCo hollow layer double hydroxides (Pt@NiCo-LDH) were adopted to hybridize with their cDNA to form a rigid dsDNA helix, which kept the “signal-on” state (Figure 7(Aa)). As the OTA-contaminated sample is tested, OTA aptamers conjugating with OTA on their active sites result in the detachment of OTA aptamers from their cDNA. Without the nanostructure amplifier, this NA biosensor significantly decreased its electrochemical performance and switched to a “signal-off” state, thus lowering the electrical signals. This biosensor can detect OTA concentration as low as 8.9 fg/mL in aqueous samples (Figure 7(Ab,Ac)). A surface-enhanced Raman scattering aptasensor has also been assembled for OTA measurement based on the Ti3C2O2 MXene. The Raman signal was enhanced by plasmonic OTA aptamers modified with Au-Ag Janus nanocomposites [88]. In the absence of OTA, the OTA aptamer/Au–Ag conjugates bind to the MXene surface by hydrogen bond and chelation interaction between the phosphate group of ssDNA and Ti ions, providing an amplified Raman signal. However, in the presence of OTA, the OTA aptamer/Au–Ag conjugates dissociate from the MXene surface since the active sites of ssDNA are blocked by OTA. The Raman signal thus was attenuated to MXene internal signal without Au–Ag Janus nanocomposites. Consequently, an ultralow LOD of 1.28 pM was achieved and was used to test red wine samples for OTA. The presence of OTA was also detected by a DNAzyme-based biosensor [89]. The Ti3C2 MXene-TiO2 nanocomposites were doped with Au@PtAg nanoparticles that were a part of the platform for facilitating the photogenerated electron transfer, supporting the ferrocene-labeled duplex DNA probe. With DNAzyme cascade amplification, this photoelectrochemical DNAzyme-based biosensor can rapidly measure the concentration of OTA from 5 fg/mL to 10 ng/mL with a LOD of 1.73 fg/mL. Therefore, this device might be more effective for on-site detection of OTA.
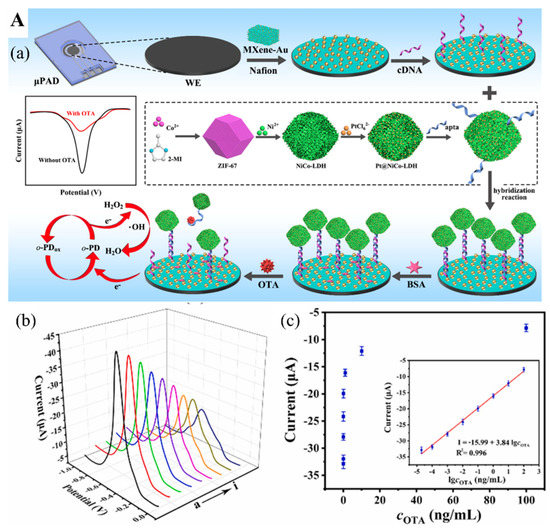
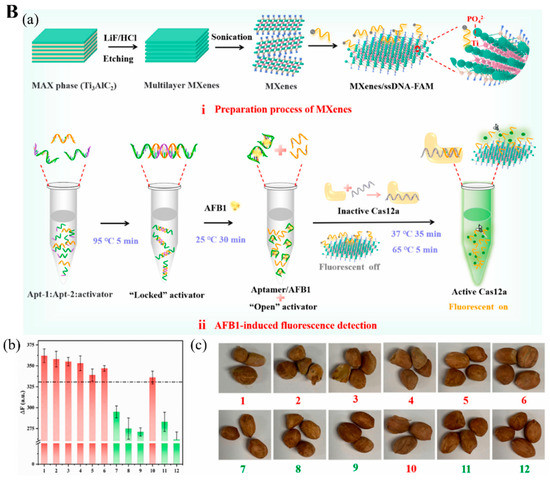
Figure 7.
(A) (a) Diagram of the assembly process of electrochemical aptasensor for OTA analysis. (b) Voltammetric response of aptasensor in the presence of different OTA concentrations. (c) Relationship between the electrochemical response and OTA concentration. Inset. The linear relationship between the response and OTA concentration. Reproduced with permission from Ref. [87]. Copyright 2022, Elsevier. (B) (a) The construction of CRISPR/Cas12a-based fluorescent biosensor for AFB1 determination. (b) Measurement of AFB1 level in 12 peanut samples by the constructed fluorescent biosensor. (c) Pictures of 12 peanut samples tested positive (red) and negative (green). Reproduced from Ref. [92].
The clustered regularly interspaced short palindromic repeats and specific proteins (CRISPR-Cas) technique was also used for detecting aflatoxin B1 (AFB1) and deoxynivalenol (DON) [92,94]. AFB1 is a secondary toxic metabolite released from Aspergillus flavus and A. parasiticus, and A. nominus, frequently found in various foods, including peanuts, grains, and animal feed [114]. Wu et al. [92] explored a MXene-based fluorescence biosensor incorporated with the CRISPER-Cas12 technique for AFB1 measurement. When AFB1 is present in the sample, it conjugates with one ssDNA, a locked activator in the form of helical double-strand aptamer opens and releases an aptamer to activate the inactive Cas12a protein linked with guide RNA (crRNA) and thus cleaves the quenched fluorophore-modified ssDNA immobilized on the MXene as small fragments (Figure 7(Ba)). These fragments then leave the MXene and are dispersed into the solution, recovering the fluorescence signal to the aqueous environment. Their tests in 12 peanut samples for AFB1 were highly accurate (Figure 7(Bb,Bc)).
Lin et al. [94] designed a luminescent aptasensor for the detection of DON using an aptamer to activate Cas12 protein trans-cleavage activity that facilitated the luminophore released from the MXene-Au platform but was suppressed in the presence of DON, which finally produced an on/off signal.
Other mycotoxins, such as gliotoxin containing di- or polysulfide bridges, can induce cytotoxicity in humans and animals [115]. A tetrahedral DNA nanostructure (TDN) was grown on the surface of Ti3C2 MXene with one extended capture aptamer in a stand-up posture from a vertex. Combined with the merit of signal enhancers, this MXene-based TDN biosensor can measure gliotoxin in human serum samples as low as 5 pM [96].
4.3. Antibiotics Detection
The presence of antibiotics and antibiotic-based medicines is also among the public concerns regarding food safety. The use of antibiotics and related drugs in agri-food systems is often necessary to curb bacterial invasion. Nevertheless, excessive or improper usage of antibiotics leads to food and feed sources laced with antibiotic residues. Consumption of such food and feed products can not only cause several adverse side effects in humans and animals, but also enhance antibacterial resistance [116,117].
Chloramphenicol (CAP) is one of the common antibiotics banned from food production due to its potential for bone marrow aplasia, gray-baby syndrome, etc., but it still probably exists in foods of animal origin [118]. Yang et al. [100] developed an electrochemical aptasensor using CAP aptamer immobilized on the MXene surface for the detection of CAP in honey as low as 0.03 pM. In another effort, Jiang et al. [101] employed ZnO quantum dots/nitrogen (N)-doped MXene as a sensing framework that supported aptamers to interact with CAP specifically. Their electrochemiluminescence aptasensor could detect CAP in water and milk with a LOD of 0.019 ng/mL and a linear range from 0.1 to 100 ng/mL.
Another important antibiotic family is aminoglycosides, which are natural or semisynthetic actinomycetes derivatives, including streptomycin (STR), kanamycin, neomycin, tobramycin, etc. [119]. You et al. [102] devised a photoelectrochemical “on-off-on” aptasensor for STR measurement with STR aptamer-modified Bi4VO8Br/Ti3C2 nanostructure. The ability of the Bi4VO8Br/Ti3C2 nanostructure to generate photogenerated signals is efficiently inhibited by the STR aptamer, resulting in an “on-off” signal framework. When STR is present, the STR aptamer–STR conjugate lowers the inhibition ability of STR. Hence, the photoelectrochemical signal from Bi4VO8Br/Ti3C2 is again generated. This “on-off-on” aptasensing scheme was able to measure STR concentration in honey in the range of 1 to 1000 nM. In another electrochemical aptasensor, polyethyleneimine (PEI) was used in a metal–organic framework (PEI-UIO-66-NH3) to coat the polyaniline-Ti3C2 MXene surface (PANI-MXene), which was used to modify a glassy electrode (Figure 8(Aa)) [103]. After incubating with cDNA, the electrode was ready to immobilize the STR aptamer-methylene blue (MB) structure, which is released from the electrode in the presence of STR. This aptasensor successfully detected 0.01 to 200 nM STR in milk (Figure 8(Ab–Ad)).
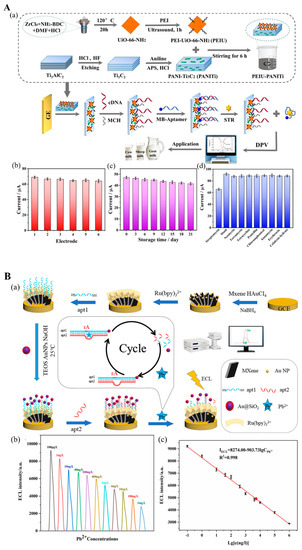
Figure 8.
(A) (a) Illustration of STR electrochemical aptasensor construction. (b) Electrochemical response for six replications of STR detection. (c) Aptasensor stability after storing for 21 days. (d) Response of electrochemical signal for STR and interference evaluation. Reproduced with permission from Ref. [103]. Copyright 2022, Elsevier. (B) (a) Illustration of electrochemiluminescent (ECL) DNAzyme-based biosensor assembly for Pb2+ detection. (b) Responses of ECL of assembled DNAzyme-based biosensor in the presence of various Pb2+ concentrations. (c) The linear range of ECL biosensors for Pb2+ measurement. Reproduced with permission from Ref. [107].
MXene-based NA biosensors have also been developed for the detection of enrofloxacin and ciprofloxacin [104,105]. Enrofloxacin and ciprofloxacin are synthetic fluoroquinolone antibiotics and are used in veterinary drugs. Jiang et al. [104] fabricated an electrochemiluminescence aptasensor based on O-terminated Ti3C2 MXene doped with AgBr nanocrystals. The LOD of this sensor for enrofloxacin in pond water was 5.97 10−13 mol/L. The aptamer/Ti3C2-Bi4VO8Br-TiO2 nano-construction was used for photoelectrochemical detection of ciprofloxacin, which provides the effectiveness of health implications to milk samples [105].
4.4. Other Targets
MXene-based NA biosensors have also been developed for the detection of other target analytes in the agri-food systems, such as the presence of heavy metals. Metal ions are essential to life. However, overconsumption of heavy metal-containing food and feed products causes health issues, e.g., cancer, renal damage, nervous system disruption, etc. Mercury (Hg2+) and lead (Pb2+) poisoning are responsible for several diseases and even death due to the ingestion of contaminated food or drinks [1]. In an electrochemical biosensor designed by Liu et al. [106], Ti3C2Tx MXene/Nafion was modified by GR5 DNAzyme that can cleave the ribo-adenine (rA) site of the DNA substrate in the presence of Pb2+. Here, Nafion with high viscosity can function as an adhesive not only for Ti3C2Tx MXene-GR5 DNAzyme binding but also for Ti3C2Tx MXene immobilization over the sensing substrate. After DNA substrate cleavage, the absorptivity of MXene is largely improved, facilitating the adoption of GR5 DNAzyme and ion intercalation, thus increasing its electrochemical performance. This DNAzyme-based biosensor can detect the Pb2+ effectively and selectively for food safety and environment monitoring. Zhai et al.[107] employed an electrochemiluminescent biosensor for Pb2+ in four water samples. They used MXene@Au loaded with tris(2,2-bipyridyl) ruthenium (II) (Ru(bpy)32+) as a transducer platform to support the aptazyme and its rA-containing DNA substrate labeled with Au@SiO2 luminophore (Figure 8(Ba)). Pb2+ ions activated the endonuclease behavior of the aptazyme that cut the rA sites of the DNA substrate, leading to the luminophore-tailed sequence of DNA substrate releasing into the solution, thus reducing the electrochemiluminescence signal generated from the biosensor. Based on this, they achieved an LOD of 0.059 ng/L over the detection rage of 0.1 to 106 ng/L for Pb2+ (Figure 8(Bb,Bc)).
A DNAzyme-based fluorometric biosensor was proposed by Lu et al. [110] for the detection of Hg2+. Principally, Ti3C2 MXene nanosheets adsorbed the fluorophore-labeled ssDNA by metal chelation interaction and hydrogen bond but quenching the fluorescence effect of a fluorophore. The introduction of H2+ ions initiated the hybridization reaction between the exposed sequence of hairpin DNA and free primers (hairpin-Hg2+-primer) that were further digested by exonuclease III. Then, the hairpin-Hg2+-primer structure was broken down into several small ssDNA sequences and nickers that hybridized with fluorophore-tailed ssDNA, thus facilitating the fluorophore-tailed ssDNA separated from the MXene surface to generate a fluorescent signal. This sensor was capable of detecting Hg2+ in water as low as 42.5 pM.
Other deleterious contaminants in the agri-food systems include pesticide residues. As an organophosphorus insecticide and acaricide, isocarbophos (ICP) can induce several acute toxicities and are carcinogenic to humans. Zhi et al. [111] developed a Ti3C2 MXene@Au sol support ICP aptamer probe using the catalytic activity of the sol to reduce mandelic acid-chloroauric acid (MA-HAuCl4) for a dual-mode measurement of the ICP concentration in farmland wastewater with high stability, reproducibility, and specificity.
5. Summary and Future Perspectives
MXene-based NA biosensors offer high sensitivity that can help quantitively measure the presence of various analytes down to picomolar concentrations. This has prompted the development of various biosensors for different applications, enabling the implementation of preventive strategies at the source of the agri-food systems.
MXenes are usually employed as transducer elements due to their unique physical and electrical properties. However, the effectiveness of signal transmission may vary depending on the quality of MXene. In addition, the short life of NA impedes the development of robust MXene-based NA biosensors. Furthermore, the oxidation of MXene may impair the long-term functionality and stability of MXene biosensors, especially those that are employed in hot and humid environments, which is often the case for on-site testing of agri-food products. The cost of the MAX phase, the MXene precursor, is rather high compared to other 2D NM precursors such as graphite. In addition to the above challenges, the biggest challenge currently faced is the real-world application of MXene-NA biosensors, e.g., in food or environmental sample matrices such as corn or wheat flour, fruit juice, milk, or soil where a lot of interfering miro/nanoparticles may generate noise signals when they nonspecifically attach to the biosensor.
To improve the performance of NA biosensors based on MXene and address the challenges mentioned above, we offer the following. First, it is essential to develop a complete understanding of how MXene and NA interact. Though several investigations have suggested that certain ssDNA can chemically conjugate with MXene, there is still a lack of definitive confirmation of this. Second, there is a need to develop highly stable ssDNA mimics that have specific recognition abilities. Third, since in agri-food systems, on-site testing is a high priority, portable MXene-based NA biosensors such as lateral flow assays and lab-on-a-chip devices are needed. Some MXenes other than Ti3C2Tx such as Nb4C3 may be a suitable substitute that can be widely adopted in the NA biosensors for chemical target detection.
In summary, despite these recent developments, the technology of MXene-based NA biosensors is still in its relative infancy. We expect many exciting developments in the next five to ten years, responding to the need for on-site testing of complex real matrices in agri-food systems.
Author Contributions
Conceptualization, W.W. and S.G.; data curation, W.W.; writing—original draft preparation, W.W.; writing—review and editing, S.G.; supervision and project administration, S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, W.; You, Y.; Gunasekaran, S. LSPR-based colorimetric biosensing for food quality and safety. Compr. Rev. Food Sci. Food Saf. 2021, 20, 5829–5855. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.; Wichers, J.; Xu, M.; Van Hoof, R.; Van Dooremalen, C.; Van Amerongen, A.; Peters, J. Biosensing Chlorpyrifos in Environmental Water Samples by a Newly Developed Carbon Nanoparticle-Based Indirect Lateral Flow Assay. Biosensors 2022, 12, 735. [Google Scholar] [CrossRef] [PubMed]
- Lubin, A.A.; Plaxco, K.W. Folding-based electrochemical biosensors: The case for responsive nucleic acid architectures. Acc. Chem. Res. 2010, 43, 496–505. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Cozma, I.; Morrison, D.; Li, Y. Biosensors made of synthetic functional nucleic acids toward better human health. Anal. Chem. 2019, 92, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Manzanares-Palenzuela, C.L.; Martín-Fernández, B.; López, M.S.-P.; López-Ruiz, B. Electrochemical genosensors as innovative tools for detection of genetically modified organisms. TrAC Trends Anal. Chem. 2015, 66, 19–31. [Google Scholar] [CrossRef]
- Tian, C.; Zhao, L.; Zhu, J.; Zhang, S. Ultrasensitive detection of trace Hg2+ by SERS aptasensor based on dual recycling amplification in water environment. J. Hazard. Mater. 2021, 416, 126251. [Google Scholar] [CrossRef]
- Ullah, S.; Shahzad, F.; Qiu, B.; Fang, X.; Ammar, A.; Luo, Z.; Zaidi, S.A. MXene-Based Aptasensors: Advances, Challenges, and Prospects. Prog. Mater. Sci. 2022, 129, 100967. [Google Scholar]
- Zhu, C.; Du, D.; Lin, Y. Graphene-like 2D nanomaterial-based biointerfaces for biosensing applications. Biosens. Bioelectron. 2017, 89, 43–55. [Google Scholar] [CrossRef]
- Kurra, N.; Ahmed, B.; Gogotsi, Y.; Alshareef, H.N. MXene-on-paper coplanar microsupercapacitors. Adv. Energy Mater. 2016, 6, 1601372. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Q.; Huang, H.; Mao, L.; Liu, M.; Zhang, X.; Wei, Y. Recent progress and advances in the environmental applications of MXene related materials. Nanoscale 2020, 12, 3574–3592. [Google Scholar] [CrossRef]
- Hong, W.; Wyatt, B.C.; Nemani, S.K.; Anasori, B. Double transition-metal MXenes: Atomistic design of two-dimensional carbides and nitrides. MRS Bull. 2020, 45, 850–861. [Google Scholar] [CrossRef]
- Kalambate, P.K.; Gadhari, N.S.; Li, X.; Rao, Z.; Navale, S.T.; Shen, Y.; Patil, V.R.; Huang, Y. Recent advances in MXene–based electrochemical sensors and biosensors. TrAC Trends Anal. Chem. 2019, 120, 115643. [Google Scholar] [CrossRef]
- Wang, W.; Yin, Y.; Gunasekaran, S. Oxygen-terminated few-layered Ti3C2Tx MXene nanosheets as peroxidase-mimic nanozyme for colorimetric detection of kanamycin. Biosens. Bioelectron. 2022, 218, 114774. [Google Scholar] [CrossRef] [PubMed]
- Morales-García, A.N.; Calle-Vallejo, F.; Illas, F. MXenes: New horizons in catalysis. ACS Catal. 2020, 10, 13487–13503. [Google Scholar] [CrossRef]
- Fan, Z.; Wang, D.; Yuan, Y.; Wang, Y.; Cheng, Z.; Liu, Y.; Xie, Z. A lightweight and conductive MXene/graphene hybrid foam for superior electromagnetic interference shielding. Chem. Eng. J. 2020, 381, 122696. [Google Scholar] [CrossRef]
- Yoon, J.; Shin, M.; Lim, J.; Lee, J.-Y.; Choi, J.-W. Recent advances in MXene nanocomposite-based biosensors. Biosensors 2020, 10, 185. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, P.; Soomro, R.A.; Zhu, Q.; Xu, B. Advances in the synthesis of 2D MXenes. Adv. Mater. 2021, 33, 2103148. [Google Scholar] [CrossRef]
- Chen, N.; Yang, W.; Zhang, C. Perspectives on preparation of two-dimensional MXenes. Sci. Technol. Adv. Mater. 2021, 22, 917–930. [Google Scholar] [CrossRef]
- Ronchi, R.M.; Arantes, J.T.; Santos, S.F. Synthesis, structure, properties and applications of MXenes: Current status and perspectives. Ceram. Int. 2019, 45, 18167–18188. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, D.; Yang, J.; Zhou, S.; Wang, H.; Yuan, X.; Liang, J.; Li, X.; Chen, Y.; Li, H. 2D Single-and Few-Layered MXene: Synthesis, applications and perspectives. J. Mater. Chem. A 2022, 10, 13651–13672. [Google Scholar] [CrossRef]
- Alhabeb, M.; Maleski, K.; Anasori, B.; Lelyukh, P.; Clark, L.; Sin, S.; Gogotsi, Y. Guidelines for synthesis and processing of two-dimensional titanium carbide (Ti3C2T x MXene). Chem. Mater. 2017, 29, 7633–7644. [Google Scholar] [CrossRef]
- Cai, Z.; Liu, B.; Zou, X.; Cheng, H.-M. Chemical vapor deposition growth and applications of two-dimensional materials and their heterostructures. Chem. Rev. 2018, 118, 6091–6133. [Google Scholar] [CrossRef] [PubMed]
- Naguib, M.; Mashtalir, O.; Carle, J.; Presser, V.; Lu, J.; Hultman, L.; Gogotsi, Y.; Barsoum, M.W. Two-dimensional transition metal carbides. ACS Nano 2012, 6, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, L.; Milligan, C.; Ma, T.; Zhou, L.; Cui, Y.; Qi, Z.; Libretto, N.; Xu, B.; Luo, J. Two-dimensional transition metal carbides as supports for tuning the chemistry of catalytic nanoparticles. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Z.; Wang, J.; Yang, Y.; Wu, S.; You, C.; Tian, N.; Li, Y. Structural Evolution of MXenes and Their Composites for Electromagnetic Interference Shielding Applications. Nanoscale 2022, 14, 9218–9247. [Google Scholar] [CrossRef]
- Zhou, J.; Zha, X.; Zhou, X.; Chen, F.; Gao, G.; Wang, S.; Shen, C.; Chen, T.; Zhi, C.; Eklund, P. Synthesis and electrochemical properties of two-dimensional hafnium carbide. ACS Nano 2017, 11, 3841–3850. [Google Scholar] [CrossRef]
- Boota, M.; Pasini, M.; Galeotti, F.; Porzio, W.; Zhao, M.-Q.; Halim, J.; Gogotsi, Y. Interaction of polar and nonpolar polyfluorenes with layers of two-dimensional titanium carbide (MXene): Intercalation and pseudocapacitance. Chem. Mater. 2017, 29, 2731–2738. [Google Scholar] [CrossRef]
- Vaghasiya, J.V.; Mayorga-Martinez, C.C.; Sofer, Z.; Pumera, M. MXene-based flexible supercapacitors: Influence of an organic ionic conductor electrolyte on the performance. ACS Appl. Mater. Interfaces 2020, 12, 53039–53048. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Feng, M.; Xia, J.; Zhang, F.; Wang, Z. An electrochemical biosensor based on AuNPs/Ti3C2 MXene three-dimensional nanocomposite for microRNA-155 detection by exonuclease III-aided cascade target recycling. J. Electroanal. Chem. 2020, 878, 114669. [Google Scholar] [CrossRef]
- Li, L.; Wang, F.; Zhu, J.; Wu, W. The facile synthesis of layered Ti 2 C MXene/carbon nanotube composite paper with enhanced electrochemical properties. Dalton Trans. 2017, 46, 14880–14887. [Google Scholar] [CrossRef]
- Feng, A.; Yu, Y.; Jiang, F.; Wang, Y.; Mi, L.; Yu, Y.; Song, L. Fabrication and thermal stability of NH4HF2-etched Ti3C2 MXene. Ceram. Int. 2017, 43, 6322–6328. [Google Scholar] [CrossRef]
- Ihsanullah, I. MXenes as next-generation materials for the photocatalytic degradation of pharmaceuticals in water. J. Environ. Chem. Eng. 2022, 10, 107381. [Google Scholar] [CrossRef]
- Zhu, X.; Cao, Z.; Li, X.-L.; Pei, L.; Jones, J.; Zhou, Y.-N.; Dong, P.; Wang, L.; Ye, M.; Shen, J. Ion-intercalation regulation of MXene-derived hydrated vanadates for high-rate and long-life Zn-Ion batteries. Energy Storage Mater. 2022, 45, 568–577. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Kim, S.J.; Seo, D.; Chae, Y.; Anayee, M.; Lee, Y.; Gogotsi, Y.; Ahn, C.W.; Jung, H.-T. Etching mechanism of monoatomic aluminum layers during MXene synthesis. Chem. Mater. 2021, 33, 6346–6355. [Google Scholar] [CrossRef]
- Shekhirev, M.; Shuck, C.E.; Sarycheva, A.; Gogotsi, Y. Characterization of MXenes at every step, from their precursors to single flakes and assembled films. Prog. Mater. Sci. 2021, 120, 100757. [Google Scholar] [CrossRef]
- Pinto, D.; Anasori, B.; Avireddy, H.; Shuck, C.E.; Hantanasirisakul, K.; Deysher, G.; Morante, J.R.; Porzio, W.; Alshareef, H.N.; Gogotsi, Y. Synthesis and electrochemical properties of 2D molybdenum vanadium carbides–solid solution MXenes. J. Mater. Chem. A 2020, 8, 8957–8968. [Google Scholar] [CrossRef]
- Lipatov, A.; Alhabeb, M.; Lukatskaya, M.R.; Boson, A.; Gogotsi, Y.; Sinitskii, A. Effect of synthesis on quality, electronic properties and environmental stability of individual monolayer Ti3C2 MXene flakes. Adv. Electron. Mater. 2016, 2, 1600255. [Google Scholar] [CrossRef]
- Xu, B.; Zhu, M.; Zhang, W.; Zhen, X.; Pei, Z.; Xue, Q.; Zhi, C.; Shi, P. Ultrathin MXene-micropattern-based field-effect transistor for probing neural activity. Adv. Mater. 2016, 28, 3333–3339. [Google Scholar] [CrossRef]
- Zhai, X.; Xu, X.; Peng, J.; Jing, F.; Zhang, Q.; Liu, H.; Hu, Z. Enhanced optoelectronic performance of CVD-grown metal–semiconductor NiTe2/MoS2 heterostructures. ACS Appl. Mater. Interfaces 2020, 12, 24093–24101. [Google Scholar] [CrossRef]
- Jiang, Y.; Leyden, M.R.; Qiu, L.; Wang, S.; Ono, L.K.; Wu, Z.; Juarez-Perez, E.J.; Qi, Y. Combination of Hybrid CVD and Cation Exchange for Upscaling Cs-Substituted Mixed Cation Perovskite Solar Cells with High Efficiency and Stability. Adv. Funct. Mater. 2018, 28, 1703835. [Google Scholar] [CrossRef]
- Sonawane, S.L.; Labhane, P.K.; Sonawane, G.H. Carbon-based nanocomposite membranes for water purification. In Handbook of Nanomaterials for Wastewater Treatment; Elsevier: Amsterdam, The Netherlands, 2021; pp. 555–574. [Google Scholar]
- Geng, D.; Zhao, X.; Chen, Z.; Sun, W.; Fu, W.; Chen, J.; Liu, W.; Zhou, W.; Loh, K.P. Direct synthesis of large-area 2D Mo2C on in situ grown graphene. Adv. Mater. 2017, 29, 1700072. [Google Scholar] [CrossRef] [PubMed]
- Umapathi, R.; Park, B.; Sonwal, S.; Rani, G.M.; Cho, Y.; Huh, Y.S. Advances in optical-sensing strategies for the on-site detection of pesticides in agricultural foods. Trends Food Sci. Technol. 2022, 119, 69–89. [Google Scholar] [CrossRef]
- Yen, T.M.; Zhang, T.; Chen, P.-W.; Ku, T.-H.; Chiu, Y.-J.; Lian, I.; Lo, Y.-H. Self-Assembled Pico-Liter Droplet Microarray for Ultrasensitive Nucleic Acid Quantification. ACS Nano 2015, 9, 10655–10663. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, A.C.; Svedlund, J.; Darai, E.; Lee, Y.; Lee, D.; Lee, H.-B.; Kim, S.-M.; Kim, O.; Bae, H.J.; Choi, A. OPENchip: An on-chip in situ molecular profiling platform for gene expression analysis and oncogenic mutation detection in single circulating tumour cells. Lab Chip 2020, 20, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent advances in SELEX technology and aptamer applications in biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef]
- Lan, Y.; He, B.; Tan, C.S.; Ming, D. Applications of Smartphone-Based Aptasensor for Diverse Targets Detection. Biosensors 2022, 12, 477. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Bahrani, S.; Yousefi, K.; Behbudi, G.; Babapoor, A.; Omidifar, N.; Lai, C.W.; Gholami, A.; Chiang, W.-H. Recent advancements in polythiophene-based materials and their biomedical, geno sensor and DNA detection. Int. J. Mol. Sci. 2021, 22, 6850. [Google Scholar] [CrossRef]
- Singhal, C.; Pundir, C.; Narang, J. A genosensor for detection of consensus DNA sequence of Dengue virus using ZnO/Pt-Pd nanocomposites. Biosens. Bioelectron. 2017, 97, 75–82. [Google Scholar] [CrossRef]
- Ye, Y.; Mao, S.; He, S.; Xu, X.; Cao, X.; Wei, Z.; Gunasekaran, S. Ultrasensitive electrochemical genosensor for detection of CaMV35S gene with Fe3O4-Au@ Ag nanoprobe. Talanta 2020, 206, 120205. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xu, J.; Yao, B.; Yang, L.; Yao, L.; Liu, G.; Chen, W. Facile design of multifunction-integrated linear oligonucleotide probe with multiplex amplification effect for label-free and highly sensitive GMO biosensing. Talanta 2022, 236, 122821. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Li, Y.; Qiao, Z.; Song, W.; Bi, S. Rolling circle replication for biosensing, bioimaging, and biomedicine. Trends Biotechnol. 2021, 39, 1160–1172. [Google Scholar] [CrossRef] [PubMed]
- Abolhasan, R.; Mehdizadeh, A.; Rashidi, M.R.; Aghebati-Maleki, L.; Yousefi, M. Application of hairpin DNA-based biosensors with various signal amplification strategies in clinical diagnosis. Biosens. Bioelectron. 2019, 129, 164–174. [Google Scholar] [CrossRef]
- Chen, C.; Feng, Y.; Xia, X.; La, M.; Zhou, B. Recent Advances in Nanomaterials-Based Electrochemical Biosensors for MicroRNAs Detection. Int. J. Electrochem. Sci. 2019, 14, 5174–5187. [Google Scholar] [CrossRef]
- Wang, F.; Li, P.; Chu, H.C.; Lo, P.K. Nucleic acids and their analogues for biomedical applications. Biosensors 2022, 12, 93. [Google Scholar] [CrossRef]
- Hagedorn, P.H.; Persson, R.; Funder, E.D.; Albæk, N.; Diemer, S.L.; Hansen, D.J.; Møller, M.R.; Papargyri, N.; Christiansen, H.; Hansen, B.R. Locked nucleic acid: Modality, diversity, and drug discovery. Drug Discov. Today 2018, 23, 101–114. [Google Scholar] [CrossRef]
- Siddiquee, S.; Rovina, K.; Azriah, A. A review of peptide nucleic acid. Adv. Tech. Biol. Med. 2015, 3, 1000131. [Google Scholar] [CrossRef]
- Guan, J.; He, K.; Gunasekaran, S. Selection of ssDNA aptamer using GO-SELEX and development of DNA nanostructure-based electrochemical aptasensor for penicillin. Biosens. Bioelectron. X 2022, 12, 100220. [Google Scholar] [CrossRef]
- Umapathi, R.; Ghoreishian, S.M.; Sonwal, S.; Rani, G.M.; Huh, Y.S. Portable electrochemical sensing methodologies for on-site detection of pesticide residues in fruits and vegetables. Coord. Chem. Rev. 2022, 453, 214305. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.; Yang, G.; Qu, F. Investigating the Influences of Random-Region Length on Aptamer Selection Efficiency Based on Capillary Electrophoresis–SELEX and High-Throughput Sequencing. Anal. Chem. 2021, 93, 17030–17035. [Google Scholar] [CrossRef] [PubMed]
- Rosch, J.C.; Neal, E.H.; Balikov, D.A.; Rahim, M.; Lippmann, E.S. CRISPR-mediated isogenic cell-SELEX approach for generating highly specific aptamers against native membrane proteins. Cell. Mol. Bioeng. 2020, 13, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; He, K.; Gunasekaran, S. Self-assembled tetrahedral DNA nanostructures-based ultrasensitive label-free detection of ampicillin. Talanta 2022, 243, 123292. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. The ribosome is a ribozyme. Science 2000, 289, 878–879. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Maurel, M.-C.; Leclerc, F.; Hervé, G. Ribozyme Chemistry: To be or not to be under high pressure. Chem. Rev. 2019, 120, 4898–4918. [Google Scholar] [CrossRef]
- Ishida, S.; Terasaka, N.; Katoh, T.; Suga, H. An aminoacylation ribozyme evolved from a natural tRNA-sensing T-box riboswitch. Nat. Chem. Biol. 2020, 16, 702–709. [Google Scholar] [CrossRef]
- Walton, T.; DasGupta, S.; Duzdevich, D.; Oh, S.S.; Szostak, J.W. In vitro selection of ribozyme ligases that use prebiotically plausible 2-aminoimidazole–activated substrates. Proc. Natl. Acad. Sci. USA 2020, 117, 5741–5748. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, M.; Yang, T.; Chang, Y.; Peng, S.; Xu, Q.; Wang, D.; Zhou, X.; Shao, Y. A catalytic triplex DNAzyme for porphyrin metalation. Chem. Commun. 2021, 57, 6499–6502. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. Aptazyme–gold nanoparticle sensor for amplified molecular probing in living cells. Anal. Chem. 2016, 88, 5981–5987. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Cozma, I.; Mou, Q.; Brennan, J.D.; Lu, Y.; Li, Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Li, G.; Xu, L.; Yu, Q.; Yue, X.; Wu, Y.; Ye, B. Peptide-conjugated hemin/G-quadruplex as a versatile probe for “signal-on” electrochemical peptide biosensor. Talanta 2020, 209, 120611. [Google Scholar] [CrossRef] [PubMed]
- Rothenbroker, M.; McConnell, E.M.; Gu, J.; Urbanus, M.L.; Samani, S.E.; Ensminger, A.W.; Filipe, C.D.; Li, Y. Selection and Characterization of an RNA-Cleaving DNAzyme Activated by Legionella pneumophila. Angew. Chem. Int. Ed. 2021, 60, 4782–4788. [Google Scholar] [CrossRef]
- Chang, D.; Chang, T.; Salena, B.; Li, Y. An Unintentional Discovery of a Fluorogenic DNA Probe for Ribonuclease I. ChemBioChem. 2020, 21, 464–468. [Google Scholar] [CrossRef]
- Mathew, M.; Rout, C.S. Electrochemical biosensors based on Ti3C2Tx MXene: Future perspectives for on-site analysis. Curr. Opin. Electrochem. 2021, 30, 100782. [Google Scholar] [CrossRef]
- Feng, W.; Wang, R.; Zhou, Y.; Ding, L.; Gao, X.; Zhou, B.; Hu, P.; Chen, Y. Ultrathin molybdenum carbide MXene with fast biodegradability for highly efficient theory-oriented photonic tumor hyperthermia. Adv. Funct. Mater. 2019, 29, 1901942. [Google Scholar] [CrossRef]
- Yang, G.; Zhao, J.; Yi, S.; Wan, X.; Tang, J. Biodegradable and photostable Nb2C MXene quantum dots as promising nanofluorophores for metal ions sensing and fluorescence imaging. Sens. Actuators B Chem. 2020, 309, 127735. [Google Scholar] [CrossRef]
- Kashefi-Kheyrabadi, L.; Koyappayil, A.; Kim, T.; Cheon, Y.-P.; Lee, M.-H. A MoS2@Ti3C2Tx MXene hybrid-based electrochemical aptasensor (MEA) for sensitive and rapid detection of Thyroxine. Bioelectrochemistry 2021, 137, 107674. [Google Scholar] [CrossRef]
- Liu, L.; Hong, J.; Wang, W.; Xiao, S.; Xie, H.; Wang, Q.; Gan, N. Fluorescent aptasensor for detection of live foodborne pathogens based on multicolor perovskite-quantum-dot-encoded DNA probes and dual-stirring-bar-assisted signal amplification. J. Pharm. Anal. 2022, in press. [Google Scholar] [CrossRef]
- Wang, W.; Xiao, S.; Jia, Z.; Xie, H.; Li, T.; Wang, Q.; Gan, N. A dual-mode aptasensor for foodborne pathogens detection using Pt, phenylboric acid and ferrocene modified Ti3C2 MXenes nanoprobe. Sens. Actuators B Chem. 2022, 351, 130839. [Google Scholar] [CrossRef]
- Hong, J.; Wang, W.; Wang, J.; Wang, X.; Xie, H.; Li, T.; Gan, N. A turn-on–type fluorescence resonance energy transfer aptasensor for vibrio detection using aptamer-modified polyhedral oligomeric silsesquioxane-perovskite quantum dots/Ti3C2 MXenes composite probes. Microchim. Acta 2021, 188, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Duan, S.; He, F. Highly electrically conductive two-dimensional Ti3C2 Mxenes-based 16S rDNA electrochemical sensor for detecting Mycobacterium tuberculosis. Anal. Chim. Acta 2020, 1123, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Rizi, K.S.; Hatamluyi, B.; Darroudi, M.; Meshkat, Z.; Aryan, E.; Soleimanpour, S.; Rezayi, M. PCR-free electrochemical genosensor for Mycobacterium tuberculosis complex detection based on two-dimensional Ti3C2 Mxene-polypyrrole signal amplification. Microchem. J. 2022, 179, 107467. [Google Scholar] [CrossRef]
- Sheng, A.; Wang, P.; Yang, J.; Tang, L.; Chen, F.; Zhang, J. MXene Coupled with CRISPR-Cas12a for analysis of endotoxin and bacteria. Anal. Chem. 2021, 93, 4676–4681. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, F.; Zhi, H.; Zhao, J.; Wan, P.; Feng, L. Electrochemical “signal on/off” paper-based aptasensor for ochratoxin A detection based on MXene-Au and Pt@ NiCo-LDH-catalyzed signal amplification. Sens. Actuators B Chem. 2022, 368, 132161. [Google Scholar] [CrossRef]
- Zheng, F.; Ke, W.; Shi, L.; Liu, H.; Zhao, Y. Plasmonic Au–Ag janus nanoparticle engineered ratiometric surface-enhanced raman scattering aptasensor for Ochratoxin A detection. Anal. Chem. 2019, 91, 11812–11820. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, X.; Wang, Z.; Han, J.; Chen, S. Core–shell Au@ PtAg modified TiO2–Ti3C2 heterostructure and target-triggered DNAzyme cascade amplification for photoelectrochemical detection of ochratoxin A. Anal. Chim. Acta 2022, 1216, 339943. [Google Scholar] [CrossRef]
- Li, Y.L.; Xie, F.-T.; Yao, C.; Zhang, G.; Guan, Y.; Yang, Y.; Yang, J.-M.; Hu, R. DNA tetrahedral-based dual-signal ratiometric electrochemical aptasensor for the detection of ochratoxin A in corn kernels samples. Analyst 2022, 147, 4578–4586. [Google Scholar] [CrossRef]
- Al-Dhahebi, A.M.; Jose, R.; Mustapha, M.; Saheed, M.S.M. Ultrasensitive aptasensor using electrospun MXene/polyvinylidene fluoride nanofiber composite for Ochratoxin A detection. Food Chem. 2022, 390, 133105. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, D.-W.; Pu, H.; Wei, Q. A novel fluorescence biosensor based on CRISPR/Cas12a integrated MXenes for detecting Aflatoxin B1. Talanta 2022, 252, 123773. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, D.-W.; Pu, H.; Wei, Q.; Lin, X. Ti3C2Tx MXenes loaded with Au nanoparticle dimers as a surface-enhanced Raman scattering aptasensor for AFB1 detection. Food Chem. 2022, 372, 131293. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, C.; Meng, X.; Yu, W.; Duan, N.; Wang, Z.; Wu, S. CRISPR-Cas12a-mediated luminescence resonance energy transfer aptasensing platform for deoxynivalenol using gold nanoparticle-decorated Ti3C2Tx MXene as the enhanced quencher. J. Hazard. Mater. 2022, 433, 128750. [Google Scholar] [CrossRef] [PubMed]
- Sangu, S.S.; Illias, N.M.; Ong, C.C.; Gopinath, S.C.B.; Saheed, M.S.M. MXene-based aptasensor: Characterization and high-performance voltammetry detection of deoxynivalenol. BioNanoScience 2021, 11, 314–323. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Huang, Y.; Xiong, M.; Wang, F.; Li, C. A label-free electrochemical biosensor for highly sensitive detection of gliotoxin based on DNA nanostructure/MXene nanocomplexes. Biosens. Bioelectron. 2019, 142, 111531. [Google Scholar] [CrossRef]
- Sangu, S.S.; Gopinath, S.C.B.; Shukur, M.F.A.; Saheed, M.S.M. An Electrochemical Approach for Ultrasensitive Detection of Zearalenone in Commodity Using Disposable Screen-Printed Electrode Coated with MXene/Chitosan Film. BioNanoScience 2022, 12, 814–823. [Google Scholar] [CrossRef]
- Fan, L.; Huang, J.J.; Liao, J. Competitive smartphone-based portable electrochemical aptasensor system based on an MXene/cDNA-MB probe for the determination of Microcystin-LR. Sens. Actuators B Chem. 2022, 369, 132164. [Google Scholar] [CrossRef]
- Ullah, N.; Chen, W.; Noureen, B.; Tian, Y.; Du, L.; Wu, C.; Ma, J. An electrochemical Ti3C2Tx aptasensor for sensitive and label-free detection of marine biological toxins. Sensors 2021, 21, 4938. [Google Scholar] [CrossRef]
- Yang, J.; Zhong, W.; Yu, Q.; Zou, J.; Gao, Y.; Liu, S.; Zhang, S.; Wang, X.; Lu, L. MXene–AuNP-Based Electrochemical Aptasensor for Ultra-Sensitive Detection of Chloramphenicol in Honey. Molecules 2022, 27, 1871. [Google Scholar] [CrossRef]
- Jiang, D.; Wei, M.; Du, X.; Qin, M.; Shan, X.; Chen, Z. One-pot synthesis of ZnO quantum dots/N-doped Ti3C2 MXene: Tunable nitrogen-doping properties and efficient electrochemiluminescence sensing. Chem. Eng. J. 2022, 430, 132771. [Google Scholar] [CrossRef]
- You, F.; Wei, J.; Cheng, Y.; Wen, Z.; Ding, C.; Hao, N.; Wang, K. Selective and sensitive photoelectrochemical aptasensor for streptomycin detection based on Bi4VO8Br/Ti3C2 nanohybrids. J. Hazard. Mater. 2021, 414, 125539. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Hui, Y.; Zhang, L.; Zhang, F.; Liu, Y.; Zheng, J.; Jia, R.; Song, Y.; Wang, B. A novel electrochemical aptasensor based on Ti3C2-MOFs nanocomposites for rapid streptomycin detection in milk samples. Sens. Actuators B Chem. 2022, 368, 132119. [Google Scholar] [CrossRef]
- Jiang, D.; Wei, M.; Du, X.; Qin, M.; Shan, X.; Wang, W.; Chen, Z. Ultrasensitive near-infrared aptasensor for enrofloxacin detection based on wavelength tunable AgBr nanocrystals electrochemiluminescence emission triggered by O-terminated Ti3C2 MXene. Biosens. Bioelectron. 2022, 200, 113917. [Google Scholar] [PubMed]
- You, F.; Wen, Z.; Yuan, R.; Ding, L.; Wei, J.; Qian, J.; Long, L.; Wang, K. Selective and ultrasensitive detection of ciprofloxacin in milk using a photoelectrochemical aptasensor based on Ti3C2/Bi4VO8Br/TiO2 nanocomposite. J. Electroanal. Chem. 2022, 914, 116285. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, R.; Zhang, Z.; Chen, D.; Gao, Y.; Liu, Z.; Li, H.; Wang, C. Label-free electrochemical biosensor based on GR5 DNAzyme/Ti3C2Tx Mxenes for Pb2+ detection. J. Electroanal. Chem. 2022, 905, 115979. [Google Scholar] [CrossRef]
- Zhai, H.; Wang, Y.; Yin, J.; Zhang, Y.; Guo, Q.; Sun, X.; Guo, Y.; Yang, Q.; Li, F.; Zhang, Y. Electrochemiluminescence biosensor for determination of lead (II) ions using signal amplification by Au@ SiO2 and tripropylamine-endonuclease assisted cycling process. Microchim. Acta 2022, 189, 1–11. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Wang, B.; Liu, Z.; Yang, C.; Wang, J.; Ma, X.; Li, H.; Sun, C. Engineering DNAzyme strategies for fluorescent detection of lead ions based on RNA cleavage-propelled signal amplification. J. Hazard. Mater. 2022, 440, 129712. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Pandey, R.P.; Jabbar, K.A.; Mahmoud, K.A. Nb4C3Tx (MXene)/Au/DNA aptasensor for the ultraselective electrochemical detection of lead in water samples. Electroanalysis 2022, 34, 1540–1546. [Google Scholar] [CrossRef]
- Lu, L.; Han, X.; Lin, J.; Zhang, Y.; Qiu, M.; Chen, Y.; Li, M.; Tang, D. Ultrasensitive fluorometric biosensor based on Ti3C2 MXenes with Hg2+-triggered exonuclease III-assisted recycling amplification. Analyst 2021, 146, 2664–2669. [Google Scholar] [CrossRef]
- Zhi, S.; Shi, J.; Liang, A.; Jiang, Z. MXene nanosheet loaded gold nanocluster catalytic amplification–aptamer SERS quantitative assay platform for isocarbophos. Talanta 2022, 251, 123771. [Google Scholar] [CrossRef]
- Matsuda, S.; Hiyoshi, H.; Tandhavanant, S.; Kodama, T. Advances on Vibrio parahaemolyticus research in the postgenomic era. Microbiol. Immunol. 2020, 64, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E. Salmonella Typhimurium and inflammation: A pathogen-centric affair. Nat. Rev. Microbiol. 2021, 19, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gunasekaran, S. Nanozymes-based biosensors for food quality and safety. TrAC Trends Anal. Chem. 2020, 126, 115841. [Google Scholar] [CrossRef]
- Ye, W.; Liu, T.; Zhang, W.; Zhang, W. The Toxic Mechanism of Gliotoxins and Biosynthetic Strategies for Toxicity Prevention. Int. J. Mol. Sci. 2021, 22, 13510. [Google Scholar] [CrossRef]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef]
- Paige, J.C.; Tollefson, L.; Miller, M.A. Health implications of residues of veterinary drugs and chemicals in animal tissues. Vet. Clin. N. Am. Food Anim. Pract. 1999, 15, 31–43. [Google Scholar] [CrossRef]
- Dinos, G.P.; Athanassopoulos, C.M.; Missiri, D.A.; Giannopoulou, P.C.; Vlachogiannis, I.A.; Papadopoulos, G.E.; Papaioannou, D.; Kalpaxis, D.L. Chloramphenicol derivatives as antibacterial and anticancer agents: Historic problems and current solutions. Antibiotics 2016, 5, 20. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).