Micro-Raman Analysis of Sperm Cells on Glass Slide: Potential Label-Free Assessment of Sperm DNA toward Clinical Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Semen Sample Collection and Sperm Preparation
2.3. Raman Measurement and Raman Mapping
2.4. Sperm Chromatin Dispersion (SCD) Test
2.5. Raman Spectra Preprocessing and Multivariate Statistical Analysis
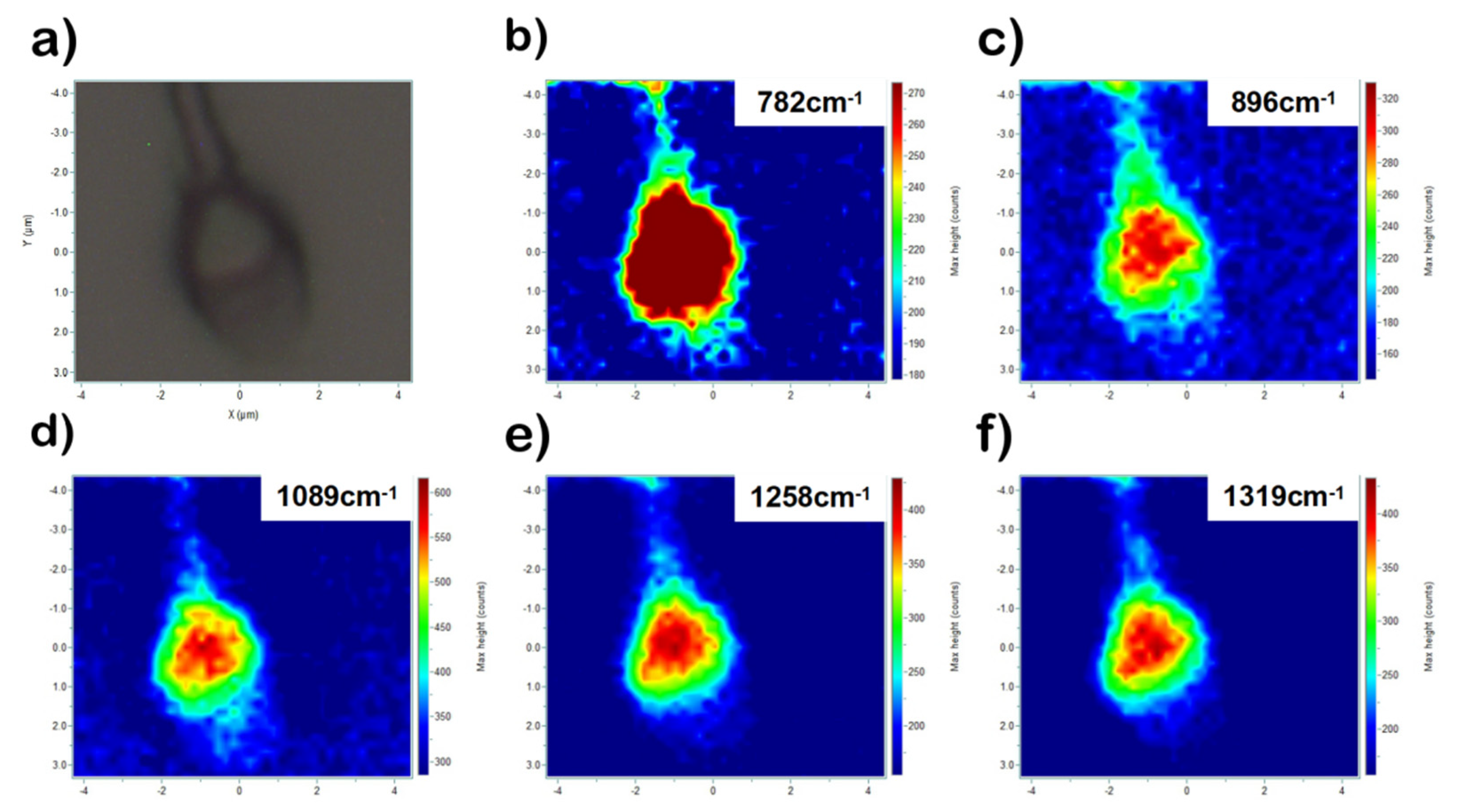
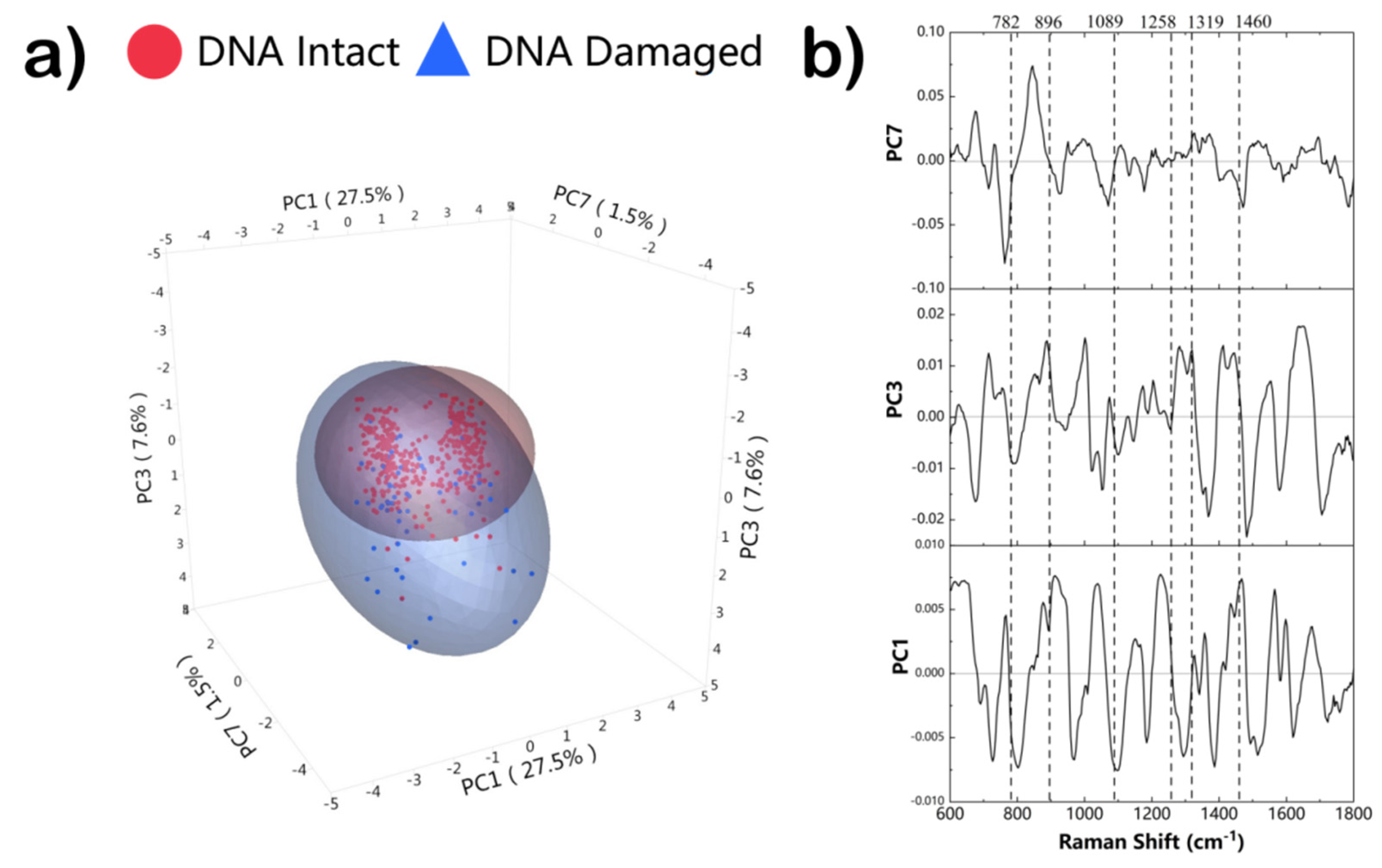
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamilton, B.E.; Martin, J.A.; Osterman, M.J.; Rossen, L.M. Births: Provisional data for 2018. NVDD Vital Stat. Rapid Release 2019, 007. [Google Scholar]
- Kumar, N.; Singh, A.K. Trends of male factor infertility, an important cause of infertility: A review of literature. J. Hum. Reprod. Sci. 2015, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Huang, Y.; Lü, N. Computer-aided sperm analysis: Past, present and future. Andrologia 2014, 46, 329–338. [Google Scholar] [CrossRef]
- Hirano, Y.; Shibahara, H.; Obara, H.; Suzuki, T.; Takamizawa, S.; Yamaguchi, C.; Tsunoda, H.; Sato, I. Andrology: Relationships between sperm motility characteristics assessed by the computer-aided sperm analysis (CASA) and fertilization rates in vitro. J. Assist. Reprod. Genet. 2001, 18, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, D.; Mortimer, S.T. Computer-Aided Sperm Analysis (CASA) of Sperm Motility and Hyperactivation. In Spermatogenesis; Springer: Berlin/Heidelberg, Germany, 2013; pp. 77–87. [Google Scholar]
- Finelli, R.; Leisegang, K.; Tumallapalli, S.; Henkel, R.; Agarwal, A. The validity and reliability of computer-aided semen analyzers in performing semen analysis: A systematic review. Transl. Androl. Urol. 2021, 10, 3069–3079. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Laboratory Manual for the Examination and Processing of Human Semen; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Al Ghedan, M.; Al Matrafi, H.; Al Sufyan, H.; Al Tannir, M. Sperm DNA fragmentation in Saudi infertile men with normal standard semen parameters. Andrology 2015, 4, 2167–0250.10001. [Google Scholar] [CrossRef]
- Agarwal, A.; Said, T.M. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum. Reprod. Update 2003, 9, 331–345. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.; Wang, L.; Zhou, Z.; Sha, J.; Mao, Y.; Cai, L.; Feng, T.; Yan, Z.; Ma, L. The clinical significance of sperm DNA damage detection combined with routine semen testing in assisted reproduction. Mol. Med. Rep. 2008, 1, 617–624. [Google Scholar] [CrossRef]
- Tejada, R.I.; Mitchell, J.C.; Norman, A.; Marik, J.J.; Friedman, S. A test for the practical evaluation of male fertility by acridine orange (AO) fluorescence. Fertil. Steril. 1984, 42, 87–91. [Google Scholar] [CrossRef]
- Talebi, A.; Moein, M.; Tabibnejad, N.; Ghasemzadeh, J. Effect of varicocele on chromatin condensation and DNA integrity of ejaculated spermatozoa using cytochemical tests. Andrologia 2008, 40, 245–251. [Google Scholar] [CrossRef]
- Evenson, D.P. The Sperm Chromatin Structure Assay (SCSA®) and other sperm DNA fragmentation tests for evaluation of sperm nuclear DNA integrity as related to fertility. Anim. Reprod. Sci. 2016, 169, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.L.; Johnston, S.; Gosálvez, J. Sperm Chromatin Dispersion (SCD) Assay. In A Clinician’s Guide to Sperm DNA and Chromatin Damage; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2018; pp. 137–152. [Google Scholar]
- Hayashi, M.; Sofuni, T.; Ishidate, M., Jr. An application of acridine orange fluorescent staining to the micronucleus test. Mutat. Res. Lett. 1983, 120, 241–247. [Google Scholar] [CrossRef]
- Fernández, J.L.; Muriel, L.; Goyanes, V.; Segrelles, E.; Gosálvez, J.; Enciso, M.; LaFromboise, M.; De Jonge, C. Simple determination of human sperm DNA fragmentation with an improved sperm chromatin dispersion test. Fertil. Steril. 2005, 84, 833–842. [Google Scholar] [CrossRef]
- Wang, H.; Malvadkar, N.; Koytek, S.; Bylander, J.; Reeves, W.B.; Demirel, M.C. Quantitative analysis of creatinine in urine by metalized nanostructured parylene. J. Biomed. Opt. 2010, 15, 027004. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Paidi, S.K.; Valdez, T.A.; Zhang, C.; Spegazzini, N.; Dasari, R.R.; Barman, I. Noninvasive Monitoring of Blood Glucose with Raman Spectroscopy. Acc. Chem. Res. 2017, 50, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Gao, Y.; Wu, Y.P.; Guo, X.Y.; Ying, Y.; Wen, Y.; Yang, H.F. Raman tracking the activity of urease in saliva for healthcare. Biosens. Bioelectron. 2019, 129, 24–28. [Google Scholar] [CrossRef]
- Calado, G.; Behl, I.; Daniel, A.; Byrne, H.J.; Lyng, F.M. Raman spectroscopic analysis of saliva for the diagnosis of oral cancer: A systematic review. Transl. Biophotonics 2019, 1, e201900001. [Google Scholar] [CrossRef]
- Atkins, C.G.; Buckley, K.; Blades, M.W.; Turner, R.F.B. Raman Spectroscopy of Blood and Blood Components. Appl. Spectrosc. 2017, 71, 767–793. [Google Scholar] [CrossRef]
- Mallidis, C.; Wistuba, J.; Bleisteiner, B.; Damm, O.; Gross, P.; Wübbeling, F.; Fallnich, C.; Burger, M.; Schlatt, S. In situ visualization of damaged DNA in human sperm by Raman microspectroscopy. Hum. Reprod. 2011, 26, 1641–1649. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, X.; Chen, Y.; Feng, S.; Chen, R.; Chen, J.; Dou, M.; Zeng, H. Raman spectroscopic characterization and differentiation of seminal plasma. J. Biomed. Opt. 2011, 16, 110501. [Google Scholar] [CrossRef]
- Fullwood, L.M.; Griffiths, D.; Ashton, K.; Dawson, T.; Lea, R.W.; Davis, C.; Bonnier, F.; Byrne, H.J.; Baker, M.J. Effect of substrate choice and tissue type on tissue preparation for spectral histopathology by Raman microspectroscopy. Analyst 2014, 139, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Huser, T.; Chan, J. Raman spectroscopy for physiological investigations of tissues and cells. Adv. Drug Deliv. Rev. 2015, 89, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Kerr, L.T.; Byrne, H.J.; Hennelly, B.M. Optimal choice of sample substrate and laser wavelength for Raman spectroscopic analysis of biological specimen. Anal. Methods 2015, 7, 5041–5052. [Google Scholar] [CrossRef]
- Zhao, J.; Lui, H.; McLean, D.I.; Zeng, H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. Spectrosc. 2007, 61, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Kerr, L.; Hennelly, B. A multivariate statistical investigation of background subtraction algorithms for Raman spectra of cytology samples recorded on glass slides. Chemom. Intell. Lab. 2016, 158, 61–68. [Google Scholar] [CrossRef]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Izenman, A.J. Linear discriminant analysis. In Modern Multivariate Statistical Techniques; Springer: Berlin/Heidelberg, Germany, 2013; pp. 237–280. [Google Scholar]
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: Taking the magic away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Huser, T.; Orme, C.A.; Hollars, C.W.; Corzett, M.H.; Balhorn, R. Raman spectroscopy of DNA packaging in individual human sperm cells distinguishes normal from abnormal cells. J. Biophotonics 2009, 2, 322–332. [Google Scholar] [CrossRef]
- Huang, Z.F.; Du, S.R.; Liang, X.Z.; Sun, Y.; Chen, X.W.; Lin, J.Q.; Li, Y.Z.; Chen, G.N.; Feng, S.Y. Head-attached live sperm cell for label-free micro-Raman evaluation of sperm DNA integrity: A preliminary study. J. Raman. Spectrosc. 2020, 51, 591–595. [Google Scholar] [CrossRef]
- Martens, H.; Stark, E. Extended multiplicative signal correction and spectral interference subtraction: New prepro-cessing methods for near infrared spectroscopy. J. Pharm. Biomed. 1991, 9, 625–635. [Google Scholar] [CrossRef]
- Sánchez, V.; Redmann, K.; Wistuba, J.; Wübbeling, F.; Burger, M.; Oldenhof, H.; Wolkers, W.F.; Kliesch, S.; Schlatt, S.; Mallidis, C. Oxidative DNA damage in human sperm can be detected by Raman microspectroscopy. Fertil. Steril. 2012, 98, e1123. [Google Scholar] [CrossRef] [PubMed]


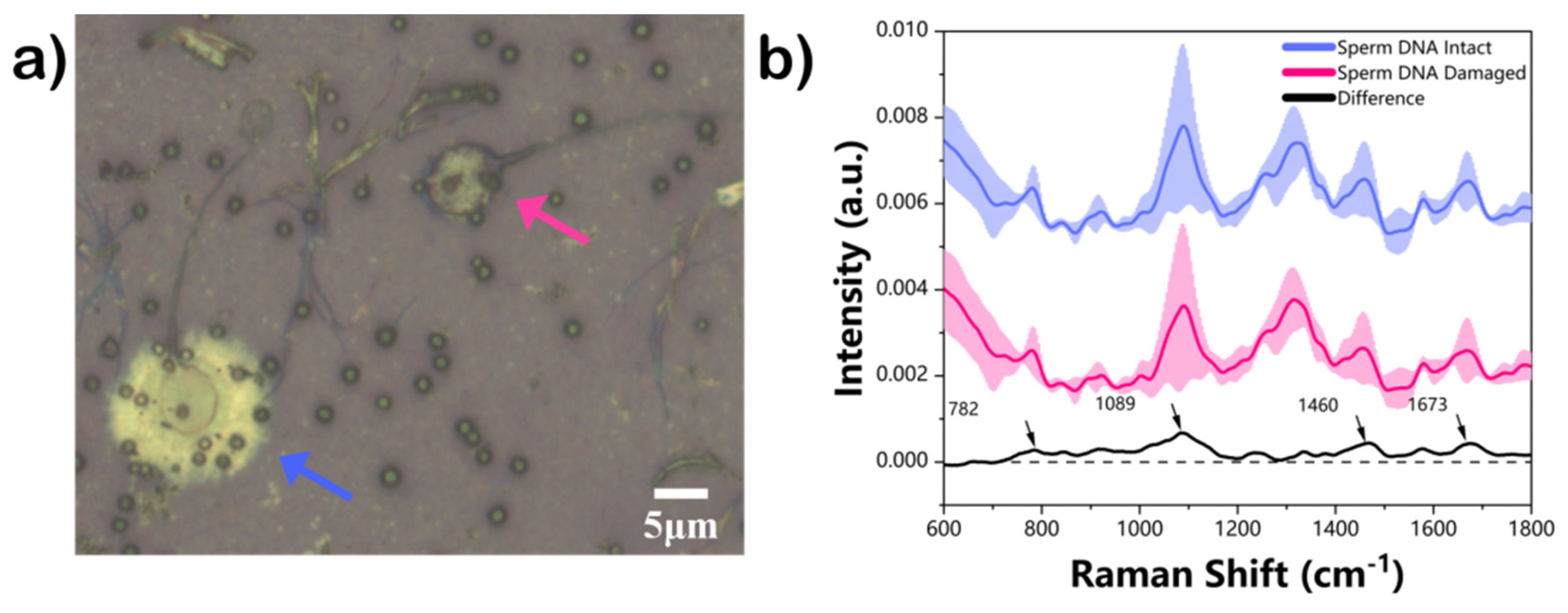

| Wavenumber (cm−1) | Vibrational Modes | Molecular Origin |
|---|---|---|
| 782 | Ring breathing modes in the DNA/RNA bases | DNA, thymine, cytosine, uracil |
| 896 | C-O-C | Phosphodiester, Deoxyribose |
| 920 | C-C | Ribose-phosphate |
| 1001 | C-C | Phenylalanine |
| 1089 | C-C, PO2− stretch | DNA |
| 1208 | υ (C-C6H5) | Tryptophan, phenylalanine |
| 1258 | C-C | Proteins, DNA |
| 1319 | CH3CH2 twisting | Guanine, collagen |
| 1367 | υs(CH3) | Phosphplipids |
| 1460 | CH2/CH3 | Thymine |
| 1580 | δ(C=C) | Phenylalanine |
| 1673 | C=C | Amide I |
| 1748 | C=O | Lipids |
| Model | Sensitivity | Specificity | Total Accuracy | Validation |
|---|---|---|---|---|
| PCA-LDA | 74.8 | 72.3 | 74.4 | LOOCV |
| PLS-DA | 77.0 | 81.5 | 77.7 | |
| PCA-LDA | 73.1 | 75.4 | 73.5 | k-fold (k = 5) |
| PLS-DA | 78.4 | 87.7 | 79.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, S.; Zhang, Q.; Guan, H.; Chen, G.; Wang, S.; Sun, Y.; Li, Y.; Chen, R.; He, Y.; Huang, Z. Micro-Raman Analysis of Sperm Cells on Glass Slide: Potential Label-Free Assessment of Sperm DNA toward Clinical Applications. Biosensors 2022, 12, 1051. https://doi.org/10.3390/bios12111051
Du S, Zhang Q, Guan H, Chen G, Wang S, Sun Y, Li Y, Chen R, He Y, Huang Z. Micro-Raman Analysis of Sperm Cells on Glass Slide: Potential Label-Free Assessment of Sperm DNA toward Clinical Applications. Biosensors. 2022; 12(11):1051. https://doi.org/10.3390/bios12111051
Chicago/Turabian StyleDu, Shengrong, Qun Zhang, Haohao Guan, Guannan Chen, Sisi Wang, Yan Sun, Yuling Li, Rong Chen, Youwu He, and Zufang Huang. 2022. "Micro-Raman Analysis of Sperm Cells on Glass Slide: Potential Label-Free Assessment of Sperm DNA toward Clinical Applications" Biosensors 12, no. 11: 1051. https://doi.org/10.3390/bios12111051
APA StyleDu, S., Zhang, Q., Guan, H., Chen, G., Wang, S., Sun, Y., Li, Y., Chen, R., He, Y., & Huang, Z. (2022). Micro-Raman Analysis of Sperm Cells on Glass Slide: Potential Label-Free Assessment of Sperm DNA toward Clinical Applications. Biosensors, 12(11), 1051. https://doi.org/10.3390/bios12111051