Polarized Micro-Raman Spectroscopy and 2D Convolutional Neural Network Applied to Structural Analysis and Discrimination of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Spectral Acquisition
2.3. Spectral Preprocessing
2.4. Data Augmentation and Data Set
2.5. The 2D-CNN Model Building and Training
3. Results
3.1. Raman Spectral Analysis
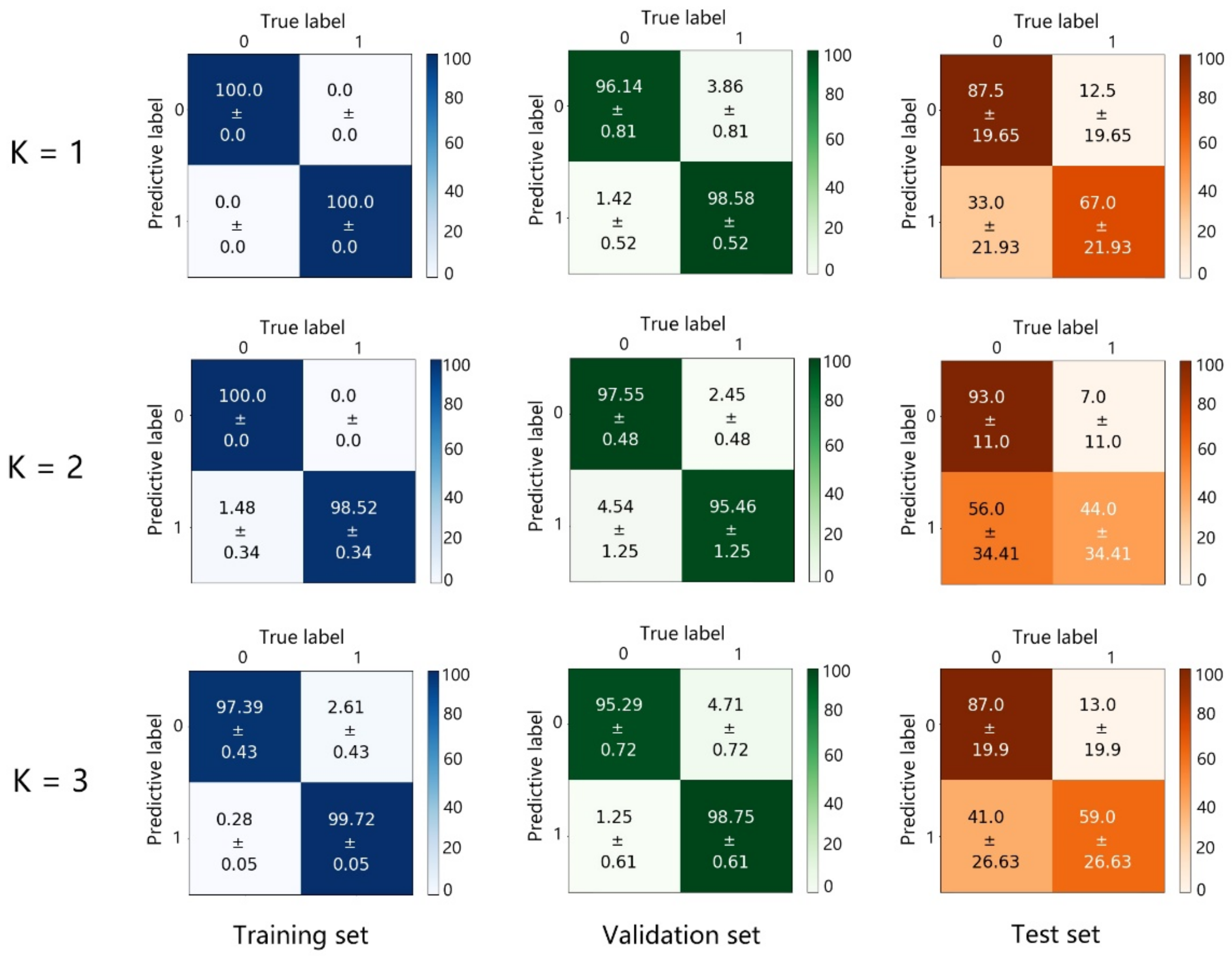
3.2. The 2D-CNN Discrimination Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Bray, F. Cancer statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Daniela, L.P.; Shaaban, A.M.; Rehman, S.; Rehman, I. Raman Spectroscopy of Breast Cancer. Appl. Spectrosc. Rev. 2019, 55, 439–475. [Google Scholar] [CrossRef]
- Takei, J.; Tsunoda-Shimizu, H.; Kikuchi, M.; Kawasaki, T.; Yagata, H.; Tsugawa, K.; Suzuki, K.; Nakamura, S.; Saida, Y. Clinical Implications of Architectural Distortion Visualized by Breast Ultrasonography. Breast Cancer 2009, 16, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Ravert, P.K.; Huffaker, C. Breast Cancer Screening in Women: An Integrative Literature Review. J. Am. Acad. Nurse Pract. 2010, 22, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Rzhevskii, A. The Recent Advances in Raman Microscopy and Imaging Techniques for Biosensors. Biosensors 2019, 9, 25. [Google Scholar] [CrossRef]
- Liu, K.; Zhao, Q.; Li, B.; Zhao, X. Raman Spectroscopy: A Novel Technology for Gastric Cancer Diagnosis. Front. Bioeng. Biotechnol. 2022, 10, 856591. [Google Scholar] [CrossRef]
- Sabtu, S.N.; Sani, S.; Bradley, D.A.; Looi, L.M.; Osman, Z. A Review of the Applications of Raman Spectroscopy for Breast Cancer Tissue Diagnostic and Their Histopathological Classification of Epithelial to Mesenchymal Transition. J. Raman Spectrosc. 2020, 51, 380–389. [Google Scholar] [CrossRef]
- Li, H.; Ning, T.; Yu, F.; Chen, Y.; Zhang, B.; Wang, S. Raman Microspectroscopic Investigation and Classification of Breast Cancer Pathological Characteristics. Molecules 2021, 26, 921. [Google Scholar] [CrossRef]
- Carter, R.; Martin, A.A.; Netto, M.M.; Soares, F.A. FT-Raman Spectroscopy Study of Human Breast Tissue. Proc. SPIE Int. Soc. Opt. Eng. 2004, 5321, 190–197. [Google Scholar] [CrossRef]
- Vanna, R.; Morasso, C.; Piccotti, F.; Torti, E.; Altamura, D.; Albasini, S.; Agozzino, M.; Villani, L.; Sorrentino, L.; Bunk, O. Raman Spectroscopy Reveals That Biochemical Composition of Breast Microcalcifications Correlates with Histopathologic Features. Cancer Res. 2020, 80, 1762–1772. [Google Scholar] [CrossRef]
- Ma, D.Y.; Shang, L.W.; Tang, J.L.; Bao, Y.L.; Fu, J.J.; Yin, J.H. Classifying Breast Cancer Tissue by Raman Spectroscopy with One-dimensional Convolutional Neural Network. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 256, 119732. [Google Scholar] [CrossRef]
- Shang, L.W.; Ma, D.Y.; Fu, J.J.; Lu, Y.F.; Yin, J.H. Fluorescence Imaging and Raman Spectroscopy Applied for the Accurate Diagnosis of Breast Cancer with Deep Learning Algorithms. Biomed. Opt. Express 2020, 11, 3673–3683. [Google Scholar] [CrossRef]
- Kothari, R.; Fong, Y.; Storrie-Lombardi, M.C. Review of laser Raman spectroscopy for surgical breast cancer detection: Stochastic backpropagation neural networks. Sensors 2020, 20, 6260. [Google Scholar] [CrossRef]
- Ly, E.; Piot, O.; Durlach, A.; Bernard, P.; Manfait, M. Polarized Raman Microspectroscopy Can Reveal Structural Changes of Peritumoral Dermis in Basal Cell Carcinoma. Appl. Spectrosc. 2008, 62, 1088–1094. [Google Scholar] [CrossRef]
- Daniel, A.; Prakasarao, A.; Dornadula, K.; Ganesan, S. Polarized Raman Spectroscopy Unravels the Biomolecular Structural Changes in Cervical Cancer. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 152, 58–63. [Google Scholar] [CrossRef]
- Lin, D.; Huang, H.; Qiu, S.F.; Feng, S.Y.; Chen, G.N.; Chen, R. Diagnostic potential of polarized surface enhanced Raman spectroscopy technology for colorectal cancer detection. Opt. Express 2016, 24, 2222–2234. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluska, B.; Kopec, M. Polarized Raman microscopy imaging: Capabilities and challenges for cancer research. J. Mol. Liq. 2018, 259, 102–111. [Google Scholar] [CrossRef]
- Lee, W.; Lenferink, A.; Otto, C.; Offerhaus, H.L. Classifying Raman Spectra of Extracellular Vesicles Based on Convolutional Neural Networks for Prostate Cancer Detection. J. Raman Spectrosc. 2020, 51, 293–300. [Google Scholar] [CrossRef]
- Yan, H.; Yu, M.; Xia, J.; Zhu, L.; Sun, G. Diverse Region-Based CNN for Tongue Squamous Cell Carcinoma Classification with Raman Spectroscopy. IEEE Access 2020, 8, 127313–127328. [Google Scholar] [CrossRef]
- Gao, H.; Wang, X.; Shang, L.W.; Zhao, Y.; Yin, J.H.; Huang, B.K. Design and Application of Small NIR-Raman Spectrometer Based on Dichroic and Transmission Collimating. Spectrosc. Spect. Anal. 2018, 38, 1933–1937. [Google Scholar] [CrossRef]
- Zhao, J.; Lui, H.; Mclean, D.I.; Zeng, H. Automated Autofluorescence Background Subtraction Algorithm for Biomedical Raman Spectroscopy. Appl. Spectrosc. 2007, 61, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, R.; Wang, Y.; Feld, M.S. Histochemical Analysis of Biological Tissues Using Raman Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 1996, 52, 215–249. [Google Scholar] [CrossRef]
- Monaco, M.E. Fatty Acid Metabolism in Breast Cancer Subtypes. Oncotarget 2017, 8, 29487. [Google Scholar] [CrossRef] [PubMed]
- Kinlaw, W.B.; Baures, P.W.; Lupien, L.E.; Davis, W.L.; Kuemmerle, N.B. Fatty Acids and Breast Cancer: Make Them on Site or Have Them Delivered. J. Cell. Physiol. 2016, 231, 2128–2141. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Movasaghi, Z.; Tucker, A.T.; Joel, S.P.; Rehman, I.U. Raman Spectroscopic Analysis of Breast Cancer Tissues: Identifying Differences between Normal, Invasive Ductal Carcinoma and Ductal Carcinoma in Situ of the Breast Tissue. J. Raman Spectrosc. 2010, 38, 1345–1351. [Google Scholar] [CrossRef]
- Kneipp, J.; Schut, T.B.; Kliffen, M.; Menke-Pluijmers, M.; Puppels, G. Characterization of Breast Duct Epithelia: A Raman Spectroscopic Study. Vib. Spectrosc. 2003, 32, 67–74. [Google Scholar] [CrossRef]
- Haka, A.S.; Volynskaya, Z.I.; Gardecki, J.A.; Nazemi, J.; Shenk, R.; Wang, N.; Rao Dasari, R.; Fitzmaurice, M.; Feld, M.S. Diagnosing Breast Cancer Using Raman Spectroscopy: Prospective Analysis. J. Biomed. Opt. 2009, 14, 054023. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman Spectroscopy for Identification of Epithelial Cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Evans, C.A.; Holen, I.; Coleman, R.E.; Ur Rehman, I. Raman Spectroscopic Analysis Differentiates between Breast Cancer Cell Lines. J. Raman Spectrosc. 2015, 46, 421–427. [Google Scholar] [CrossRef]
- You, S.; Tu, H.; Zhao, Y.; Liu, Y.; Chaney, E.J.; Marjanovic, M.; Boppart, S.A. Raman Spectroscopic Analysis Reveals Abnormal Fatty Acid Composition in Tumor Micro- and Macroenvironments in Human Breast and Rat Mammary Cancer. Sci. Rep. 2016, 6, 32922. [Google Scholar] [CrossRef]
- Han, B.; Du, Y.; Fu, T.; Fan, Z.; Xu, S.; Hu, C.; Bi, L.; Gao, T.; Zhang, H.; Xu, W. Differences and Relationships between Normal and Atypical Ductal Hyperplasia, Ductal Carcinoma in Situ, and Invasive Ductal Carcinoma Tissues in the Breast Based on Raman Spectroscopy. Appl. Spectrosc. 2017, 71, 300–307. [Google Scholar] [CrossRef]
- Parker, F.S. Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry; Plenum Press: New York, NY, USA, 1983. [Google Scholar]
- Dehring, K.A.; Crane, N.J.; Smukler, A.R.; Mchugh, J.B.; Morris, M.D. Identifying Chemical Changes in Subchondral Bone Taken from Murine Knee Joints Using Raman Spectroscopy. Appl. Spectrosc. 2006, 60, 1134–1141. [Google Scholar] [CrossRef]
- Dehring, K.A.; Smukler, A.R.; Roessler, B.J.; Morris, M.D. Correlating Changes in Collagen Secondary Structure with Aging and Defective Type II Collagen by Raman Spectroscopy. Appl. Spectrosc. 2006, 60, 366–372. [Google Scholar] [CrossRef]
- Wisniewski, M.; Sionkowska, A.; Kaczmarek, H.; Lazare, S.; Tokarev, V.; Belin, C. Spectroscopic Study of a KrF Excimer Laser Treated Surface of the Thin Collagen Films. J. Photochem. Photobiol. A Chem. 2007, 188, 192–199. [Google Scholar] [CrossRef]
- Bonifacio, A.; Sergo, V. Effects of Sample Orientation in Raman Microspectroscopy of Collagen Fibers and Their Impact on the Interpretation of the Amide III Band. Vib. Spectrosc. 2010, 53, 314–317. [Google Scholar] [CrossRef]
- Han, W.; Chen, S.; Wei, Y.; Fan, Q.; Liu, L. Oriented Collagen Fibers Direct Tumor Cell Intravasation. Proc. Natl. Acad. Sci. USA 2016, 113, 11208. [Google Scholar] [CrossRef]
- Holmes, D.F.; Lu, Y.; Starborg, T.; Kadler, K.E. Collagen Fibril Assembly and Function. Curr. Top. Dev. Biol. 2018, 130, 107–142. [Google Scholar] [CrossRef]
- Xu, Z.B.; Wu, J.J.; Ding, L.; Wang, Z.H.; Zhou, S.W.; Shang, H.; Wang, H.J.; Yin, J.H. Intelligent Diagnosis of Breast Cancer Based on Polarization and Bright-field Multimodal Microscopic Imaging. Chin. J. Lasers 2022, 49, 2407102. [Google Scholar] [CrossRef]




| Raman Shift (cm−1) of Normal (Cancerous) Tissues | Mode of Vibration | Assignment | Spectral Difference and Cancer vs. Normal Breast |
|---|---|---|---|
| 875 | ν(C—C) | Hydroxyproline in collagen | Decrease, more obvious in polarized spectra |
| 921 | ν(C—C) | Proline in collagen | Decrease, more obvious in polarized spectra |
| 1003 | ν(C—C) | Phenylalanine | Decrease |
| 1032 | δ(CH2CH3) | Phenylalanine in collagen | Decrease |
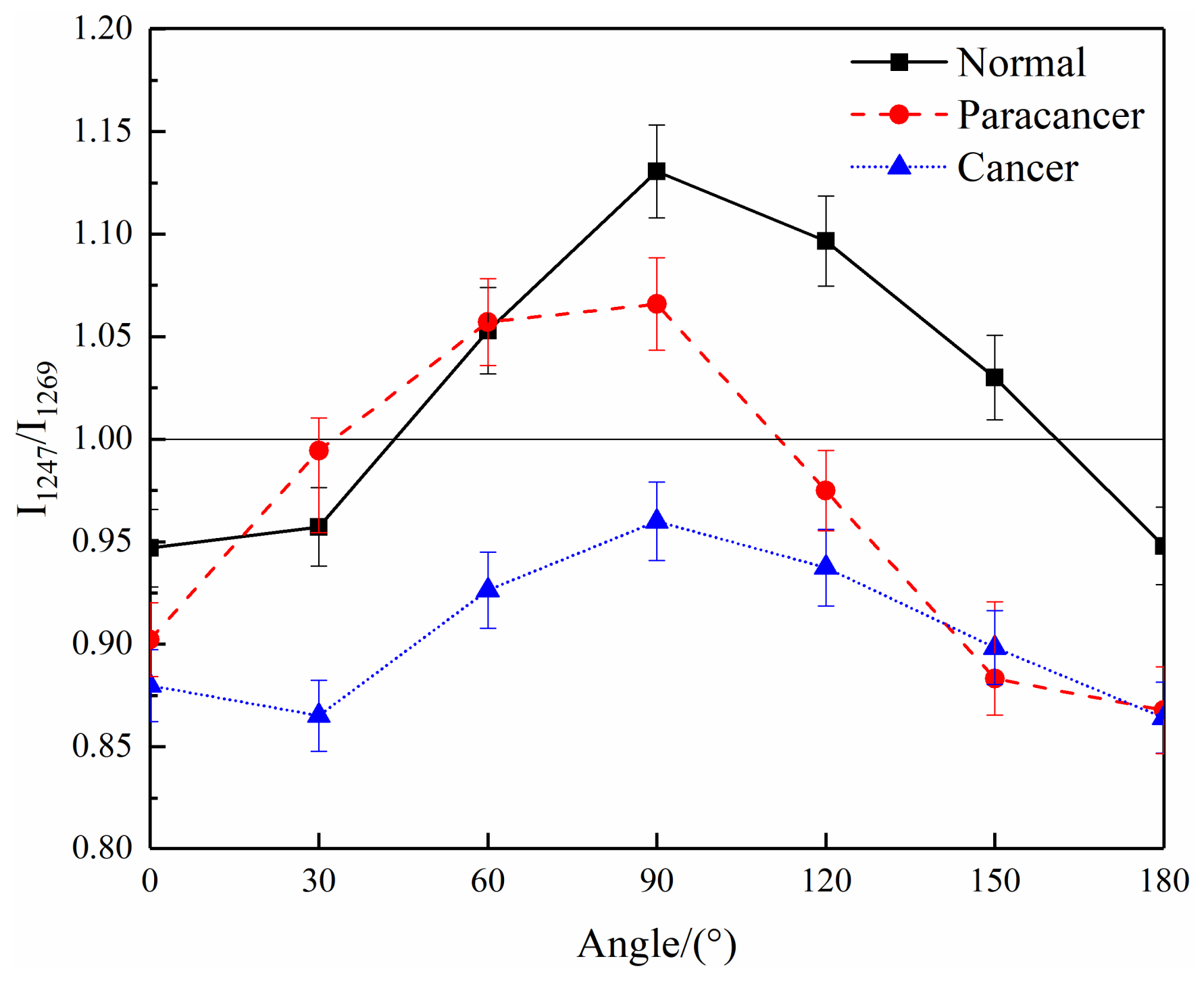
| 1247 | δ(N—H) | Amide III | Increase |
| 1269 | ν(C—N) | Amide III | \ |
| 1302 | γt(CH2) | Collagen | Increase, more obvious in polarized spectra |
| 1318 | γt(CH2) | Collagen | Decrease, more obvious in polarized spectra |
| 1450 | δ(CH2, CH3) | Proteins | Decrease |
| 1660 (1656) | ν(C=O) | Amide I, α-helix | Red shift, increase, more obvious in conventional spectra |
| Method | Raman Band (cm−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 875 | 921 | 1003 | 1032 | 1247 | 1269 | 1302 | 1318 | 1450 | 1660 | |
| Polarized | 0.019 | 0.008 | 0.026 | 0.034 | 0.562 | 0.397 | 0.035 | 0.067 | 0.319 | 0.072 |
| Conventional | 0.023 | 0.113 | 0.025 | 0.96 | 0.681 | 0.685 | 0.063 | 0.167 | 0.271 | 0.02 |
| Raman Band (cm−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 875 | 921 | 1003 | 1032 | 1247 | 1269 | 1302 | 1318 | 1450 | 1660 | ||
| Cancerous Polarized | Area | 1.03 | 1.17 | 4.16 | 2.66 | 5.88 | 1.64 | 0.37 | 0.18 | 40.82 | 49.99 |
| Std | 0.64 | 0.54 | 0.85 | 0.87 | 1.64 | 0.86 | 0.57 | 0.28 | 3.94 | 9.23 | |
| Normal Polarized | Area | 1.22 | 1.46 | 4.33 | 2.72 | 5.76 | 1.72 | 0.13 | 0.27 | 43.94 | 48.21 |
| Std | 0.62 | 0.5 | 0.79 | 0.78 | 1.56 | 0.86 | 0.55 | 0.31 | 4.38 | 8.26 | |
| Cancerous Conventional | Area | 1.09 | 1.34 | 4.78 | 2.73 | 6.42 | 1.42 | 0.32 | 0.24 | 39.07 | 56.52 |
| Std | 0.63 | 0.51 | 1.28 | 0.77 | 1.53 | 0.7 | 0.68 | 0.26 | 5.38 | 7.43 | |
| Normal Conventional | Area | 1.15 | 1.49 | 4.9 | 2.82 | 6.24 | 1.52 | 0.25 | 0.29 | 42.2 | 48.68 |
| Std | 0.49 | 0.53 | 1.31 | 0.94 | 2.18 | 0.97 | 0.95 | 0.34 | 5.61 | 8.94 | |
| Raman Shift (cm−1) of Normal Tissues | Mode of Vibration | Assignment | Spectral Difference and Cancer vs. Normal Breast |
|---|---|---|---|
| 871 | ν(N+(CH3)3) | Phospholipids | Decrease, more obvious in polarized spectra |
| 971 | ν(C—C) | Phospholipids | Decrease, more obvious in polarized spectra |
| 1032 | δ(CH2CH3) | Phospholipids | Decrease |
| 1084 | ν(C—O—C) | Phospholipids | Decrease, more obvious in polarized spectra |
| 1269 | ν(PO2), δ(=C—H) | Lipids | \ |
| 1302 | δ(=C—H) | Lipids | \ |
| 1442 | δ(CH2) | Lipids | \ |
| 1652 | ν(C=C) | Unsaturated bonds of lipids | Increase |
| 1745 | ν(C=O) | Lipids | Increase |
| Method | Raman Band (cm−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 871 | 971 | 1032 | 1084 | 1269 | 1302 | 1442 | 1652 | 1745 | |
| Polarized | 0.005 | 0.011 | 0.052 | 0.043 | 0.68 | 0.375 | 0.214 | 0.033 | 0.008 |
| Conventional | 0.767 | 0.539 | 0.852 | 0.781 | 0.726 | 0.454 | 0.139 | 0.077 | 0.045 |
| Raman Band (cm−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 871 | 971 | 1032 | 1084 | 1269 | 1302 | 1442 | 1652 | 1745 | ||
| Cancerous Polarized | Area | 0.24 | 1.06 | 1.18 | 1.36 | 6.94 | 8.47 | 37.55 | 17.54 | 2.98 |
| Std | 0.28 | 1.07 | 0.76 | 0.83 | 3.41 | 2.30 | 5.74 | 4.38 | 1.09 | |
| Normal Polarized | Area | 0.43 | 1.49 | 1.31 | 1.72 | 6.81 | 8.68 | 36.89 | 14.21 | 1.89 |
| Std | 0.63 | 1.13 | 1.2 | 2.45 | 4.71 | 4.49 | 8.64 | 4.59 | 1.76 | |
| Cancerous Conventional | Area | 0.63 | 1.23 | 0.76 | 2.12 | 7.89 | 8.72 | 38.91 | 19.93 | 3.98 |
| Std | 0.30 | 0.58 | 0.96 | 0.63 | 3.82 | 1.70 | 1.80 | 1.83 | 0.98 | |
| Normal Conventional | Area | 0.64 | 1.31 | 0.83 | 2.25 | 8.08 | 8.52 | 37.33 | 18.43 | 3.15 |
| Std | 0.46 | 0.63 | 0.50 | 0.35 | 1.43 | 1.22 | 2.77 | 0.93 | 0.46 | |
| Algorithm | Training Set | Validation Set | Test Set |
|---|---|---|---|
| 2D-CNN | 97.71% | 97.75% | 96.01% |
| 1D-CNN | 92.0% | 92.8% | 92.0% |
| Ratios (I875/I921) | ||
|---|---|---|
| Conventional | Polarized | |
| Cancerous | 0.79 ± 0.15 | 0.81 ± 0.22 |
| Normal | 0.74 ± 0.09 | 0.76 ± 0.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, L.; Tang, J.; Wu, J.; Shang, H.; Huang, X.; Bao, Y.; Xu, Z.; Wang, H.; Yin, J. Polarized Micro-Raman Spectroscopy and 2D Convolutional Neural Network Applied to Structural Analysis and Discrimination of Breast Cancer. Biosensors 2023, 13, 65. https://doi.org/10.3390/bios13010065
Shang L, Tang J, Wu J, Shang H, Huang X, Bao Y, Xu Z, Wang H, Yin J. Polarized Micro-Raman Spectroscopy and 2D Convolutional Neural Network Applied to Structural Analysis and Discrimination of Breast Cancer. Biosensors. 2023; 13(1):65. https://doi.org/10.3390/bios13010065
Chicago/Turabian StyleShang, Linwei, Jinlan Tang, Jinjin Wu, Hui Shang, Xing Huang, Yilin Bao, Zhibing Xu, Huijie Wang, and Jianhua Yin. 2023. "Polarized Micro-Raman Spectroscopy and 2D Convolutional Neural Network Applied to Structural Analysis and Discrimination of Breast Cancer" Biosensors 13, no. 1: 65. https://doi.org/10.3390/bios13010065
APA StyleShang, L., Tang, J., Wu, J., Shang, H., Huang, X., Bao, Y., Xu, Z., Wang, H., & Yin, J. (2023). Polarized Micro-Raman Spectroscopy and 2D Convolutional Neural Network Applied to Structural Analysis and Discrimination of Breast Cancer. Biosensors, 13(1), 65. https://doi.org/10.3390/bios13010065





