Selection of Specific Aptamer against Rivaroxaban and Utilization for Label-Free Electrochemical Aptasensing Using Gold Nanoparticles: First Announcement and Application for Clinical Sample Analysis
Abstract
1. Introduction
2. Experimental Section
2.1. Reagents and Materials
2.2. Instrumentation
2.3. Preparation of RIV Affinity Matrix
2.3.1. Synthesis of Acidic Hydrolysis Product of RIV
2.3.2. Substitution Epoxy-Activated Sepharose 6B with Hexandiamine
2.3.3. Immobilization of RIV Acidic Derivative on the Substituted Sepharose 6B
2.3.4. Characterization of RIV–Sepharose Support
2.4. Generation of Random Library and Primers
2.5. Selection of Specific for Aptamer Sequences RIV Based on ssDNA Library Immobilized SELEX
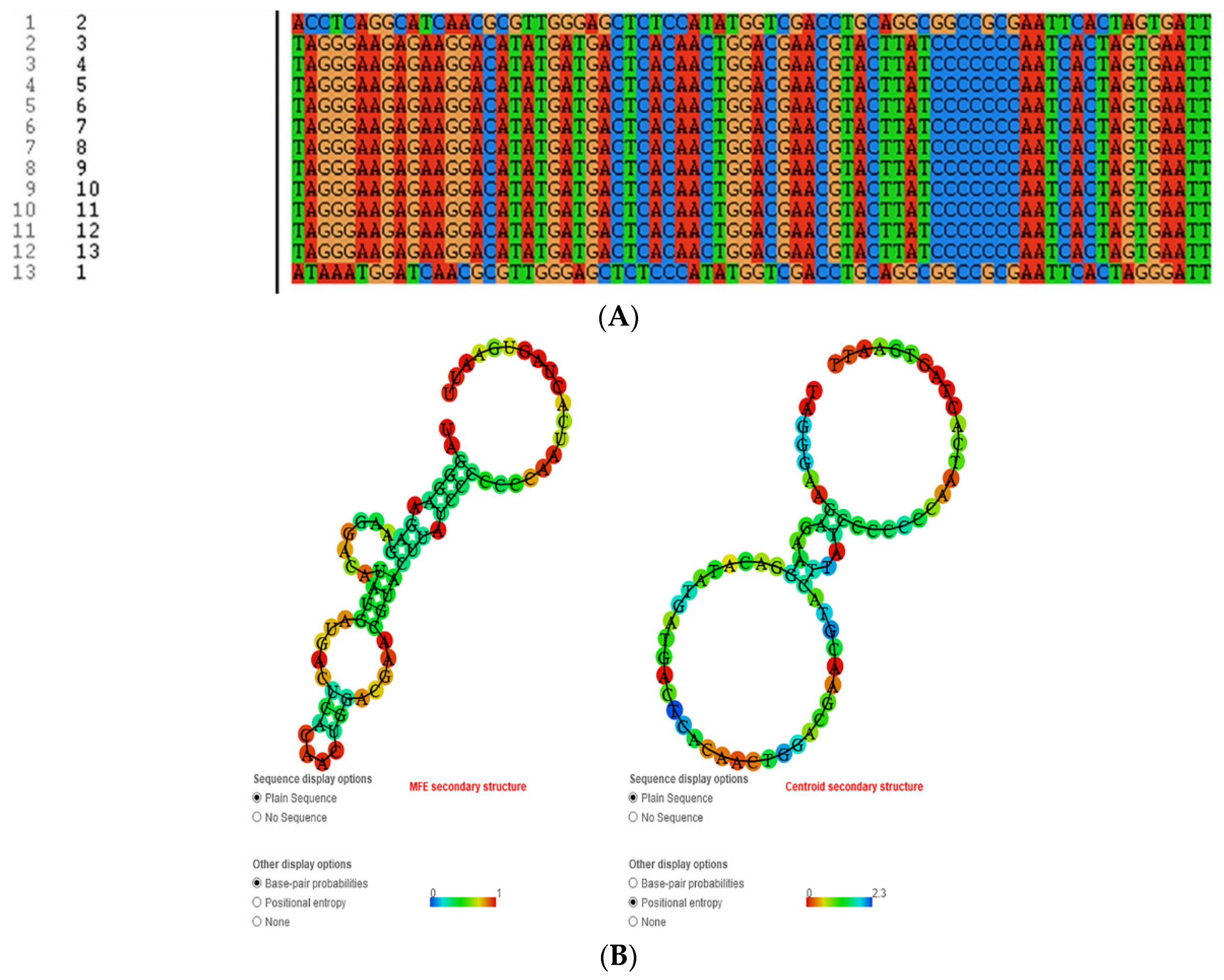
2.6. Sequence Analysis
2.7. Affinity Assay and Kd Measurements Using Surface Plasmon Resonance (SPR)
2.8. Preparation of Plasma Samples
2.9. Preparation of EBC Samples
2.10. Chromatographic Conditions
3. Results and Discussion
3.1. RIV–Sepharose Support Synthesis
3.2. Characterization of RIV–Sepharose
3.3. Selection of Specific Aptamers for Detection of RIV
3.4. Affinity Constants
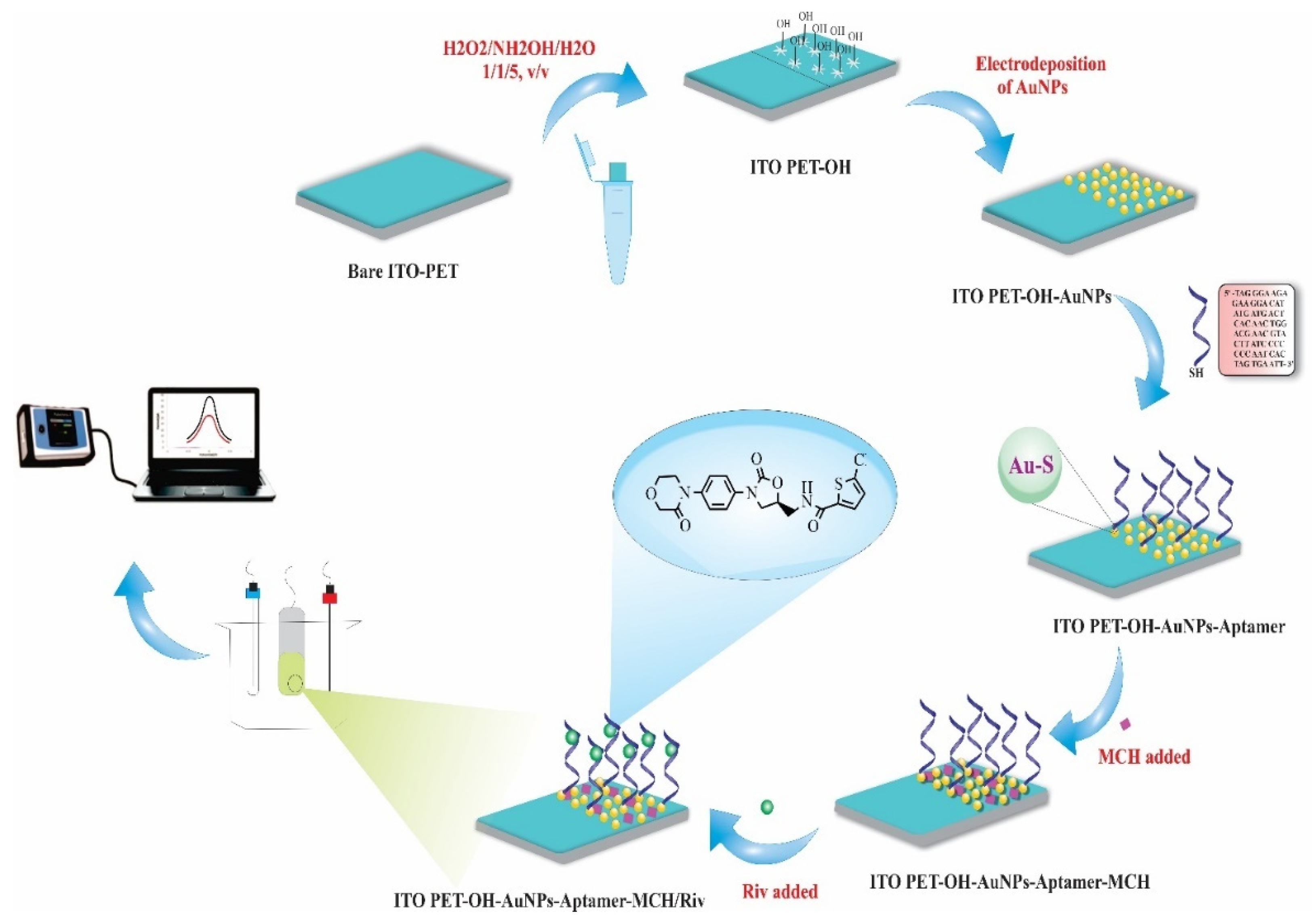
3.5. Fabrication of RIV Aptasensor
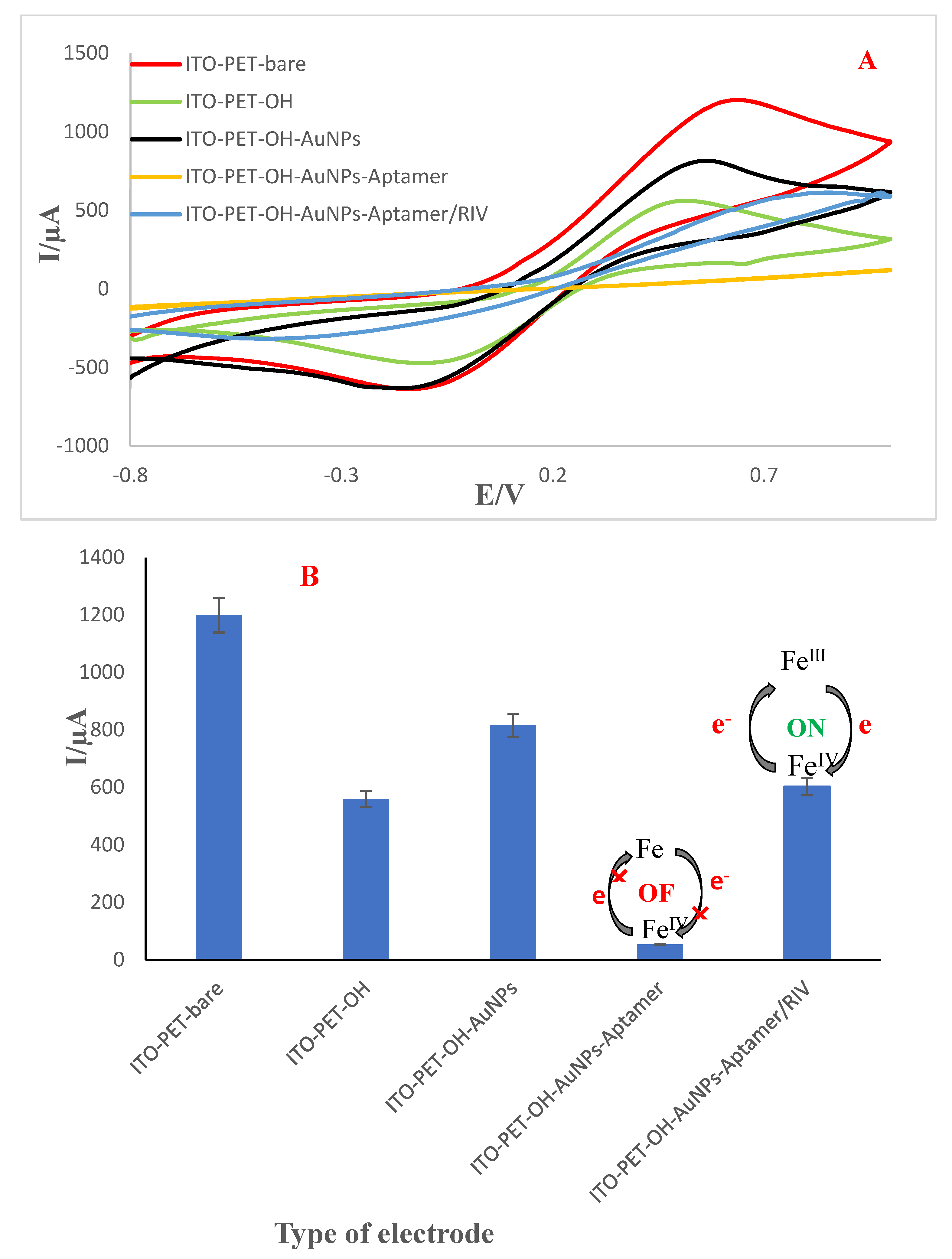
3.6. Electrochemical Behavior of the Engineered Aptasensor
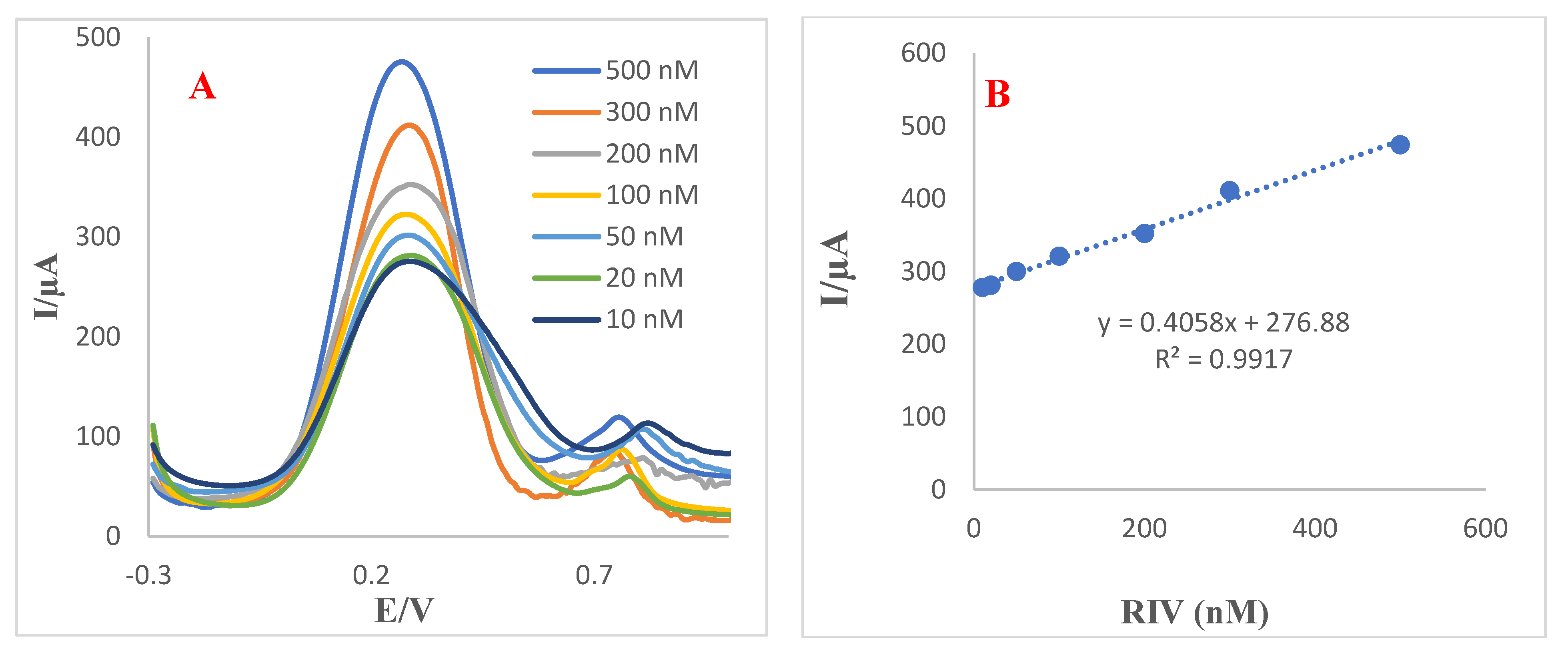
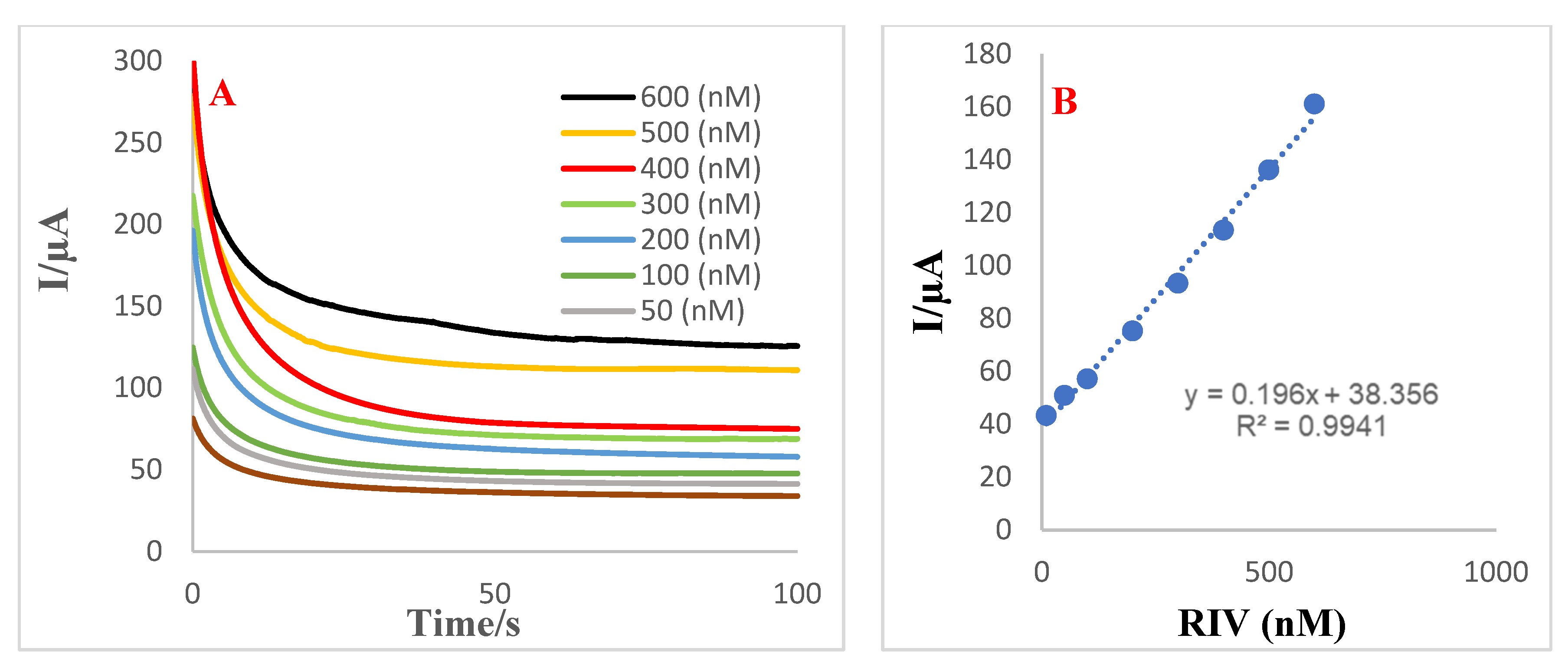
3.7. Analytical Performance of Aptasensor towards Detection of RIV
3.8. Comparison of the Method with Other Procedures
3.9. RIV Measurement in the Plasma and EBC Samples
3.10. Analytical Method Validation
3.10.1. Selectivity of the Proposed Aptasensor
3.10.2. Stability of the Engineered Aptasensor
3.10.3. Intra-Day Repeatability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeda, T.; Ogawa, S.; Kitazono, T.; Nakagawara, J.; Minematsu, K.; Miyamoto, S.; Murakawa, Y.; Iwashiro, S.; Kidani, Y.; Okayama, Y. Outcomes associated with under-dosing of rivaroxaban for management of non-valvular atrial fibrillation in real-world Japanese clinical settings. J. Thromb. Thrombolysis 2019, 48, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.H.; See, L.C.; Tu, H.T.; Yeh, Y.H.; Chang, S.H.; Wu, L.S.; Lee, H.F.; Wang, C.L.; Kuo, C.F.; Kuo, C.T. Efficacy and safety of apixaban, dabigatran, rivaroxaban, and warfarin in Asians with nonvalvular atrial fibrillation. J. Am. Heart Assoc. 2018, 7, e008150. [Google Scholar] [CrossRef] [PubMed]
- Adeboyeje, G.; Sylwestrzak, G.; Barron, J.J.; White, J.; Rosenberg, A.; Abarca, J.; Crawford, G.; Redberg, R. Major bleeding risk during anticoagulation with warfarin, dabigatran, apixaban, or rivaroxaban in patients with nonvalvular atrial fibrillation. J. Manag. Care Spec. Pharm. 2017, 23, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Douketis, J.D. Contra:“Bridging anticoagulation is needed during warfarin interruption when patients require elective surgery”. Thromb. Haemost. 2012, 108, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Phelan, P.; O’Kelly, P.; Holian, J.; Walshe, J.; Delany, C.; Slaby, J.; Winders, S.; O’Toole, D.; Magee, C.; Conlon, P. Warfarin use in hemodialysis patients: What is the risk? Clin. Nephrol. 2011, 75, 204. [Google Scholar] [CrossRef]
- Boulanger, L.; Hauch, O.; Friedman, M.; Foster, T.; Dixon, D.; Wygant, G.; Menzin, J. Warfarin exposure and the risk of thromboembolic and major bleeding events among medicaid patients with atrial fibrillation. Ann. Pharmacother. 2006, 40, 1024–1029. [Google Scholar] [CrossRef]
- Lip, G.Y.; Pan, X.; Kamble, S.; Kawabata, H.; Mardekian, J.; Masseria, C.; Bruno, A.; Phatak, H. Major bleeding risk among non-valvular atrial fibrillation patients initiated on apixaban, dabigatran, rivaroxaban or warfarin: A “real-world” observational study in the United States. Int. J. Clin. Pract. 2016, 70, 752–763. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.S.; Kim, T.-H.; Cha, M.-J.; Lee, J.M.; Park, J.; Park, J.-K.; Kang, K.-W.; Shim, J.; Uhm, J.-S. A prospective survey of the persistence of warfarin or NOAC in nonvalvular atrial fibrillation: A comparison study of Drugs for symptom control and complication prEvention of Atrial Fibrillation (CODE-AF). Korean J. Intern. Med. 2020, 35, 99. [Google Scholar] [CrossRef]
- Saraf, K.; Morris, P.D.; Garg, P.; Sheridan, P.; Storey, R. Non-vitamin K antagonist oral anticoagulants (NOACs): Clinical evidence and therapeutic considerations. Postgrad. Med. J. 2014, 90, 520–528. [Google Scholar] [CrossRef]
- AlShoaibi, N.; Al Harbi, M.; Modaimegh, H.; Al Qubbany, A.; Al Saif, S.; Connolly, D.; Kharabsheh, S.; Fathy, M.; Hegazy, Y.; Tarcha, N. Use of NOACS in high-risk patients with atrial fibrillation in Saudi Arabia: Perspectives on improving patient care. Expert Rev. Cardiovasc. Ther. 2021, 19, 221–236. [Google Scholar] [CrossRef]
- Gu, Z.-C.; Wei, A.-H.; Zhang, C.; Wang, X.-H.; Zhang, L.; Shen, L.; Li, Z.; Pan, M.-M.; Liu, X.-Y.; Pu, J. Risk of major gastrointestinal bleeding with new vs conventional oral anticoagulants: A systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2020, 18, 792–799.e761. [Google Scholar] [CrossRef] [PubMed]
- Costa, O.S.; Beyer-Westendorf, J.; Ashton, V.; Milentijevic, D.; Moore, K.T.; Bunz, T.J.; Coleman, C.I. Effectiveness and safety of rivaroxaban versus warfarin in obese nonvalvular atrial fibrillation patients: Analysis of electronic health record data. Curr. Med. Res. Opin. 2020, 36, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.-C.; Qiao, Z.-Q.; Hao, Z.-Y.; Li, Z.; Jiang, L.-S.; Ge, H.; He, B.; Pu, J. Initial anticoagulation experience with standard-dose rivaroxaban after Watchman left atrial appendage occlusion. Ann. Transl. Med. 2020, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Baral, N.; Katel, A.; Adhikari, G.; Khan, M.R.; Khan, H.M.; Rauniyar, R.; Akanbi, M.; Malik, B.; Ahmad, M.; Khan, A. Rivaroxaban is Comparable to Warfarin in Prevention of Thromboembolism in Patients with Non-Valvular Atrial Fibrillation with Valvular Heart Disease: A Systematic Review and Meta-analysis. medRxiv 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Brouwer, M.A.; van Vugt, S.P.; Focks, J.J.; Damen, S.A.; Verheugt, F.W. Rivaroxaban Plasma Levels and Levetiracetam. Ann. Intern. Med. 2020, 173, 770–771. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, Y.; Ma, H.; Zeng, H.; Lv, J. Effects of rivaroxaban and warfarin on the risk of gastrointestinal bleeding and intracranial hemorrhage in patients with atrial fibrillation: Systematic review and meta-analysis. Clin. Cardiol. 2021, 44, 1208–1215. [Google Scholar] [CrossRef]
- Hua, Y.; Sun, J.-Y.; Su, Y.; Qu, Q.; Wang, H.-Y.; Sun, W.; Kong, X.-Q. The safety and efficacy of rivaroxaban compared with warfarin in patients with atrial fibrillation and diabetes: A systematic review and meta-analysis. Am. J. Cardiovasc. Drugs 2021, 21, 51–61. [Google Scholar] [CrossRef]
- Fattah, T.A.; Saeed, A. A review on the synthetic approaches of rivaroxaban: An anticoagulant drug. Tetrahedron Asymmetry 2017, 28, 485–504. [Google Scholar] [CrossRef]
- Yang, Y.-P.; Wang, H.-B.; Ma, Q.-W. Improved synthesis of rivaroxaban. Chin. J. Med. Chem. 2013, 1, 169–177. [Google Scholar]
- Zhai, L.; Zhang, Z.; Guo, L.; Zhu, Z.; Hu, C.; Zhang, G. Synthesis, Characterization, and Properties of Rivaroxaban New Crystalline Forms. Cryst. Res. Technol. 2021, 56, 2000243. [Google Scholar] [CrossRef]
- Singh, A.K.; Noronha, V.; Gupta, A.; Singh, D.; Singh, P.; Singh, A.; Singh, A. Rivaroxaban: Drug review. Cancer Res. Stat. Treat. 2020, 3, 264. [Google Scholar] [CrossRef]
- Weitz, J.I.; Raskob, G.E.; Spyropoulos, A.C.; Spiro, T.E.; De Sanctis, Y.; Xu, J.; Lu, W.; Suh, E.; Argenti, D.; Yang, H. Thromboprophylaxis with rivaroxaban in acutely ill medical patients with renal impairment: Insights from the MAGELLAN and MARINER trials. Thromb. Haemost. 2020, 120, 515–524. [Google Scholar] [CrossRef]
- Cheong, E.J.Y.; Goh, J.J.N.; Hong, Y.; Kojodjojo, P.; Chan, E.C.Y. Rivaroxaban with and without amiodarone in renal impairment. J. Am. Coll. Cardiol. 2018, 71, 1395–1397. [Google Scholar] [CrossRef]
- Bonnemeier, H.; Huelsebeck, M.; Kloss, S. Comparative effectiveness of rivaroxaban versus a vitamin K antagonist in patients with renal impairment treated for non-valvular atrial fibrillation in Germany—A retrospective cohort study. IJC Heart Vasc. 2019, 23, 100367. [Google Scholar] [CrossRef]
- Fox, K.A.; Eikelboom, J.W.; Shestakovska, O.; Connolly, S.J.; Metsarinne, K.P.; Yusuf, S. Rivaroxaban plus aspirin in patients with vascular disease and renal dysfunction: From the COMPASS trial. J. Am. Coll. Cardiol. 2019, 73, 2243–2250. [Google Scholar] [CrossRef]
- Kim, P.Y.; Yeh, C.H.; Dale, B.J.; Leslie, B.A.; Stafford, A.R.; Fredenburgh, J.C.; Hirsh, J.; Weitz, J.I. Mechanistic basis for the differential effects of rivaroxaban and apixaban on global tests of coagulation. TH Open 2018, 2, e190–e201. [Google Scholar] [CrossRef]
- Rocha, B.M.L.; da Cunha, G.J.L.; Aguiar, C.M.T. A narrative review of low-dose rivaroxaban in patients with atherothrombotic cardiovascular disease: Vascular protection beyond anticoagulation. Cardiovasc. Diagn. Ther. 2021, 11, 130. [Google Scholar] [CrossRef]
- Fernández, M.S.; Marín, F.; Rafols, C.; Arribas, F.; Barrios, V.; Cosín-Sales, J.; Sánchez, M.A. Thromboembolic and bleeding events with rivaroxaban in clinical practice in Spain: Impact of inappropriate doses (the EMIR study). J. Comp. Eff. Res. 2021, 10, 583–593. [Google Scholar] [CrossRef]
- Lobato, S.M.; Tarrazo, C.T.; Fernández, E.G.; Alcalá, M.M. Clinical profile, adequacy of dosage and thromboembolic and bleeding outcomes in patients with nonvalvular atrial fibrillation treated with rivaroxaban in a regional hospital of Asturias, Spain. Future Cardiol. 2018, 14, 17–24. [Google Scholar] [CrossRef]
- Slavik, L.; Jacova, J.; Friedecky, D.; Ulehlova, J.; Tauber, Z.; Prochazkova, J.; Hlusi, A.; Palova, M. Evaluation of the DOAC-Stop procedure by LC-MS/MS assays for determining the residual activity of dabigatran, rivaroxaban, and apixaban. Clin. Appl. Thromb./Hemost. 2019, 25, 1076029619872556. [Google Scholar] [CrossRef]
- Studt, J.D.; Alberio, L.; Angelillo-Scherrer, A.; Asmis, L.; Fontana, P.; Korte, W.; Mendez, A.; Schmid, P.; Stricker, H.; Tsakiris, D. Accuracy and consistency of anti-Xa activity measurement for determination of rivaroxaban plasma levels. J. Thromb. Haemost. 2017, 15, 1576–1583. [Google Scholar] [CrossRef]
- Wiesen, M.H.; Blaich, C.; Streichert, T.; Michels, G.; Müller, C. Paramagnetic micro-particles as a tool for rapid quantification of apixaban, dabigatran, edoxaban and rivaroxaban in human plasma by UHPLC-MS/MS. Clin. Chem. Lab. Med. (CCLM) 2017, 55, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Gouin-Thibault, I.; Freyburger, G.; de Maistre, E.; Susen, S.; Delavenne, X.; Golmard, J.-L.; Gruel, Y.; Sié, P.; Abecassis, L.; Aillaud, M.-F. Evaluation of dabigatran, rivaroxaban and apixaban target-specific assays in a multicenter French study. Thromb. Res. 2017, 158, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Kim, D.Y.; Choi, M.-J.; Jin, S.K.; Choi, Y.S. A Simple and Efficient Method to Determine Rivaroxaban in Rat Plasma Using Liquid-Liquid Extraction and LC-MRM. Mass Spectrom. Lett. 2019, 10, 66–70. [Google Scholar]
- Bhavyasri, K.; Dhanalakshmi, C.; Sumakanth, M. Development and Validation of Ultra Violet-Visible Spectrophotometric Method for Estimation of Rivaroxaban in Spiked Human Plasma. J. Pharm. Sci. Res. 2020, 12, 1215–1219. [Google Scholar]
- Sarkis, N.; Bitar, Y.; Sarraj, M.M. Development and Validation of RP-HPLC Method for Simultaneous Estimation of Aspirin and Rivaroxaban in Synthetic Mixture. Res. J. Pharm. Technol. 2020, 13, 5459–5465. [Google Scholar]
- Yadav, S.; Dubey, N. Development and validation of bioanalytical method for estimation of rivaroxaban using HPLC-PDA in human blood plasma. J. Drug Deliv. Ther. 2017, 7, 123–125. [Google Scholar]
- Varga, A.; Șerban, R.C.; Muntean, D.L.; Tătar, C.M.; Farczadi, L.; Tilea, I. Rapid liquid chromatography tandem mass spectrometry determination of rivaroxaban levels in human plasma for therapeutic drug monitoring. Rev. Română Med. Lab. 2017, 25, 145–155. [Google Scholar] [CrossRef]
- Derogis, P.B.M.; Sanches, L.R.; de Aranda, V.F.; Colombini, M.P.; Mangueira, C.L.P.; Katz, M.; Faulhaber, A.C.L.; Mendes, C.E.A.; Ferreira, C.E.d.S.; França, C.N. Determination of rivaroxaban in patient’s plasma samples by anti-Xa chromogenic test associated to High Performance Liquid Chromatography tandem Mass Spectrometry (HPLC-MS/MS). PLoS ONE 2017, 12, e0171272. [Google Scholar] [CrossRef]
- Lagoutte-Renosi, J.; Le Poupon, J.; Girard, A.; Montange, D.; Davani, S. A simple and fast HPLC-MS/MS method for simultaneous determination of direct oral anticoagulants apixaban, dabigatran, rivaroxaban in human plasma. J. Chromatogr. B 2018, 1100, 43–49. [Google Scholar] [CrossRef]
- Wang, L.; Gai, S.; Zhang, X.; Xu, X.; Gou, N.; Wang, X.; Zhou, N.; Feng, T. Simultaneous determination of Rivaroxaban and TAK-438 in rat plasma by LC–MS/MS: Application to pharmacokinetic interaction study. Bioanalysis 2020, 12, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, S.; Dyrkorn, R.; Spigset, O.; Hegstad, S. Quantification of apixaban, dabigatran, edoxaban, and rivaroxaban in human serum by UHPLC-MS/MS—method development, validation, and application. Ther. Drug Monit. 2018, 40, 369. [Google Scholar] [CrossRef] [PubMed]
- Von Horn, H.; Rasmusson, A.; Söderblom, L.; Malmström, R.E.; Antovic, J. Using a low–molecular weight heparin–calibrated anti–factor Xa assay to assess the concentration of apixaban and rivaroxaban. Int. J. Lab. Hematol. 2021, 44, 163–167. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Kochan, J.; Lin, M.; Vandell, A.; Brown, K.; Depasse, F. Determination of edoxaban equivalent concentrations in human plasma by an automated anti-factor Xa chromogenic assay. Thromb. Res. 2017, 155, 121–127. [Google Scholar] [CrossRef]
- Kim, B.; Jang, S.; Lee, Y.-J.; Park, N.; Cho, Y.-U.; Park, C.-J. Determination of the cut-off prothrombin time to estimate plasma rivaroxaban overdose status. J. Thromb. Thrombolysis 2020, 49, 245–250. [Google Scholar] [CrossRef]
- Kim, D.D.; Wilkinson, C.L.; Pope, E.F.; Chambers, J.D.; Cohen, J.T.; Neumann, P.J. The influence of time horizon on results of cost-effectiveness analyses. Expert Rev. Pharm. Outcomes Res. 2017, 17, 615–623. [Google Scholar] [CrossRef]
- Alnajrani, M.N.; Aljohani, M.M.; Chinnappan, R.; Zourob, M.; Alsager, O.A. Highly sensitive and selective lateral flow aptasensor for anti-coagulant dabigatran etexilate determination in blood. Talanta 2021, 236, 122887. [Google Scholar] [CrossRef]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, antibody scFv, and antibody Fab’fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef]
- Hosseinzadeh, L.; Mazloum-Ardakani, M. Advances in aptasensor technology. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 99, pp. 237–279. [Google Scholar]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef]
- Kaur, H. Recent developments in cell-SELEX technology for aptamer selection. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2018, 1862, 2323–2329. [Google Scholar] [CrossRef]
- Lyu, C.; Khan, I.M.; Wang, Z. Capture-SELEX for aptamer selection: A short review. Talanta 2021, 229, 122274. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.K.; Sharma, V.; Mishra, R.K. Electrochemical aptasensors for food and environmental safeguarding: A review. Biosensors 2018, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.-Y.; Zhu, Q.-Q.; Zhang, H.-W.; Yuan, R.; He, H. A porous organic framework composite embedded with Au nanoparticles: An ultrasensitive electrochemical aptasensor toward detection of oxytetracycline. J. Mater. Chem. C 2020, 8, 14075–14082. [Google Scholar] [CrossRef]
- Ghorbani, F.; Abbaszadeh, H.; Dolatabadi, J.E.N.; Aghebati-Maleki, L.; Yousefi, M. Application of various optical and electrochemical aptasensors for detection of human prostate specific antigen: A review. Biosens. Bioelectron. 2019, 142, 111484. [Google Scholar] [CrossRef] [PubMed]
- Esfahani, S.L.; Rouhani, S.; Ranjbar, Z. Electrochemical solid-state nanosensor based on a dual amplification strategy for sensitive detection of (FeIII-dopamine). Electrochim. Acta 2019, 299, 1011–1023. [Google Scholar] [CrossRef]
- Mazzara, F.; Patella, B.; Ganci, F.; O’Riordan, A.; Aiello, G.; Torino, C.; Vilasi, A.; Sunseri, C.; Inguanta, R. Flexible electrode based on gold nanoparticles and reduced graphene oxide for uric acid detection using linear sweep voltammetry. Chem. Eng. Trans. 2021, 87, 421–426. [Google Scholar]
- Burcu Aydın, E. A label-free and sensitive impedimetric immunosensor for TNF α biomarker detection based on epoxysilane-modified disposable ITO-PET electrode. Int. J. Environ. Anal. Chem. 2020, 100, 363–377. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Daoud, W.A. High power-output mechanical energy harvester based on flexible and transparent Au nanoparticle-embedded polymer matrix. Nano Energy 2019, 55, 433–440. [Google Scholar] [CrossRef]
- US7932278B2; 2-Aminoethoxyacetic Acid Dervatives and Their Use. Bayer Intellectual Property GmbH: Monheim am Rhein, Germany, 2011.
- Teng, S.F.; Sproule, K.; Husain, A.; Lowe, C.R. Affinity chromatography on immobilized “biomimetic” ligands: Synthesis, immobilization and chromatographic assessment of an immunoglobulin G-binding ligand. J. Chromatogr. B Biomed. Sci. Appl. 2000, 740, 1–15. [Google Scholar] [CrossRef]
- Szumilak, M.; Galdyszynska, M.; Dominska, K.; Bak-Sypien, I.I.; Merecz-Sadowska, A.; Stanczak, A.; Karwowski, B.T.; Piastowska-Ciesielska, A.W. Synthesis, biological activity and preliminary in Silico ADMET screening of polyamine conjugates with bicyclic systems. Molecules 2017, 22, 794. [Google Scholar] [CrossRef]
- Kim, C.-H.; Lee, L.-P.; Min, J.-R.; Lim, M.-W.; Jeong, S.-H. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk. Biosens. Bioelectron. 2014, 51, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Ramisetti, N.R.; Kuntamukkala, R. Development and validation of a stability indicating LC-PDA-MS/MS method for separation, identification and characterization of process related and stress degradation products of rivaroxaban. RSC Adv. 2014, 4, 23155–23167. [Google Scholar] [CrossRef]
- Arous, B.; Al-Mardini, M.A.; Ghazal, H.; Al-Lahham, F. Stability-Indicating Method for the Determination of Rivaroxaban and its Degradation Products using LC-MS and TLC. Res. J. Pharm. Technol. 2018, 11, 212–220. [Google Scholar] [CrossRef]
- Roop-ngam, P.; Thongboonkerd, V. Development of an oxalate-affinity chromatographic column to isolate oxalate-binding proteins. Anal. Methods 2010, 2, 1051–1055. [Google Scholar] [CrossRef]
- Furniss, B.S. Vogel’s Textbook of Practical Organic Chemistry; Pearson Education India: Bengaluru, India, 1989. [Google Scholar]
- Caramelo-Nunes, C.; Gabriel, M.; Almeida, P.; Marcos, J.; Tomaz, C. Negative pseudo-affinity chromatography for plasmid DNA purification using berenil as ligand. J. Chromatogr. B 2014, 944, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Caramelo-Nunes, C.; Bicho, D.; Almeida, P.; Marcos, J.; Tomaz, C. Dynamic binding capacity and specificity of 3, 8-diamino-6-phenylphenanthridine-Sepharose support for purification of supercoiled plasmid deoxyribonucleic acid. J. Chromatogr. A 2013, 1307, 91–98. [Google Scholar] [CrossRef]
- Yagati, A.K.; Ngoc Le, H.T.; Cho, S. Bioelectrocatalysis of Hemoglobin on Electrodeposited Ag Nanoflowers toward H2O2 Detection. Nanomaterials 2020, 10, 1628. [Google Scholar] [CrossRef]
- Barman, S.C.; Zahed, M.A.; Sharifuzzaman, M.; Ko, S.G.; Yoon, H.; Nah, J.S.; Xuan, X.; Park, J.Y. A polyallylamine anchored amine-rich laser-ablated graphene platform for facile and highly selective electrochemical IgG biomarker detection. Adv. Funct. Mater. 2020, 30, 1907297. [Google Scholar] [CrossRef]
- Daniel, M.-C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef]
- Vais, R.D.; Sattarahmady, N.; Heli, H. Green electrodeposition of gold nanostructures by diverse size, shape, and electrochemical activity. Gold Bull. 2016, 49, 95–102. [Google Scholar] [CrossRef]
- Komsiyska, L.; Staikov, G. Electrocrystallization of Au nanoparticles on glassy carbon from HClO4 solution containing [AuCl4]−. Electrochim. Acta 2008, 54, 168–172. [Google Scholar] [CrossRef]
- Sattarahmady, N.; Movahedpour, A.; Heli, H.; Hatam, G. Gold nanoparticles-based biosensing of Leishmania major kDNA genome: Visual and spectrophotometric detections. Sens. Actuators B Chem. 2016, 235, 723–731. [Google Scholar] [CrossRef]
- Mobed, A.; Hasanzadeh, M.; Babaie, P.; Aghazadeh, M.; Mokhtarzadeh, A.; Rezaee, M.A. Cetyltrimethyl ammonium bromide modified gold nanostructure supported by chitosan as a novel scaffold for immobilization of DNA and ultra-sensitive bioassay of legionella pneumophila. Microchem. J. 2019, 149, 103961. [Google Scholar] [CrossRef]
- Hui, X.; Xuan, X.; Kim, J.; Park, J.Y. A highly flexible and selective dopamine sensor based on Pt-Au nanoparticle-modified laser-induced graphene. Electrochim. Acta 2019, 328, 135066. [Google Scholar] [CrossRef]
- Xuan, X.; Kim, J.Y.; Hui, X.; Das, P.S.; Yoon, H.S.; Park, J.-Y. A highly stretchable and conductive 3D porous graphene metal nanocomposite based electrochemical-physiological hybrid biosensor. Biosens. Bioelectron. 2018, 120, 160–167. [Google Scholar] [CrossRef]
- Kim, G.-I.; Kim, K.-W.; Oh, M.-K.; Sung, Y.-M. Electrochemical detection of vascular endothelial growth factors (VEGFs) using VEGF antibody fragments modified Au NPs/ITO electrode. Biosens. Bioelectron. 2010, 25, 1717–1722. [Google Scholar] [CrossRef]
- Cini, M.; Legnani, C.; Padrini, R.; Cosmi, B.; Dellanoce, C.; De Rosa, G.; Marcucci, R.; Pengo, V.; Poli, D.; Testa, S. DOAC plasma levels measured by chromogenic anti-Xa assays and HPLC-UV in apixaban-and rivaroxaban-treated patients from the START-Register. Int. J. Lab. Hematol. 2020, 42, 214–222. [Google Scholar] [CrossRef]
- Gouveia, F.; Bicker, J.; Santos, J.; Rocha, M.; Alves, G.; Falcão, A.; Fortuna, A. Development, validation and application of a new HPLC-DAD method for simultaneous quantification of apixaban, dabigatran, edoxaban and rivaroxaban in human plasma. J. Pharm. Biomed. Anal. 2020, 181, 113109. [Google Scholar] [CrossRef]
- Foerster, K.I.; Huppertz, A.; Mueller, O.J.; Rizos, T.; Tilemann, L.; Haefeli, W.E.; Burhenne, J. Simultaneous quantification of direct oral anticoagulants currently used in anticoagulation therapy. J. Pharm. Biomed. Anal. 2018, 148, 238–244. [Google Scholar] [CrossRef]
- Li, W.; Jian, W.; Fu, Y. Basic Sample Preparation Techniques in LC-MS Bioanalysis: Protein Precipitation, Liquid–Liquid Extraction, and Solid-Phase Extraction. Sample Prep. LC-MS Bioanal. 2019, 1, 1–30. [Google Scholar]
- Medvedovici, A.; Bacalum, E.; David, V. Sample preparation for large-scale bioanalytical studies based on liquid chromatographic techniques. Biomed. Chromatogr. 2018, 32, e4137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Couchman, L.; Kipper, K.; Arya, R.; Patel, J.P. A UHPLC-MS/MS method to simultaneously quantify apixaban, edoxaban and rivaroxaban in human plasma and breast milk: For emerging lactation studies. J. Chromatogr. B 2020, 1144, 122095. [Google Scholar] [CrossRef] [PubMed]
- Schellings, M.; Boonen, K.; Schmitz, E.; Jonkers, F.; Van Den Heuvel, D.; Besselaar, A.; Hendriks, J.; van de Kerkhof, D. Determination of dabigatran and rivaroxaban by ultra-performance liquid chromatography-tandem mass spectrometry and coagulation assays after major orthopaedic surgery. Thromb. Res. 2016, 139, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Çelebier, M.; Reçber, T.; Koçak, E.; Altınöz, S.; Kır, S. Determination of rivaroxaban in human plasma by solid-phase extraction–high performance liquid chromatography. J. Chromatogr. Sci. 2016, 54, 216–220. [Google Scholar] [CrossRef]
- Gous, T.; Couchman, L.; Patel, J.P.; Paradzai, C.; Arya, R.; Flanagan, R.J. Measurement of the direct oral anticoagulants apixaban, dabigatran, edoxaban, and rivaroxaban in human plasma using turbulent flow liquid chromatography with high-resolution mass spectrometry. Ther. Drug Monit. 2014, 36, 597–605. [Google Scholar] [CrossRef]
- Kuhn, J.; Gripp, T.; Flieder, T.; Dittrich, M.; Hendig, D.; Busse, J.; Knabbe, C.; Birschmann, I. UPLC-MRM mass spectrometry method for measurement of the coagulation inhibitors dabigatran and rivaroxaban in human plasma and its comparison with functional assays. PLoS ONE 2015, 10, e0145478. [Google Scholar] [CrossRef]
- United Nations Office on Drugs, Crime. Laboratory, Scientific Section. Guidance for the Validation of Analytical Methodology and Calibration of Equipment Used for Testing of Illicit Drugs in Seized Materials and Biological Specimens: A Commitment to Quality and Continuous Improvement; United Nations Publications: New York, NY, USA, 2009. [Google Scholar]
- Sennesael, A.-L.; Larock, A.-S.; Douxfils, J.; Elens, L.; Stillemans, G.; Wiesen, M.; Taubert, M.; Dogné, J.-M.; Spinewine, A.; Mullier, F. Rivaroxaban plasma levels in patients admitted for bleeding events: Insights from a prospective study. Thromb. J. 2018, 16, 28. [Google Scholar] [CrossRef]

| Method | Biological Matrix | LOD/LOQ/LLOQ | Linear RANG | Ref |
|---|---|---|---|---|
| HPLC-UV | Human plasma | 11.6 nM (LOD) | 11.6 to 34.8 nM | [80] |
| HPLC-DAD | Human plasma | 0.0165 μg/mL (LLOQ) | 0.017 to 5.28 μ/mL | [81] |
| (UPLC/MS/MS) | Human plasma | 2.3 nM (LLOQ) | 2.3 to 2325 nM | [82] |
| UPLC/MS/MS | Human plasma | 2.3 (LLOQ) | 2.3 to 2325 nM | [83] |
| SALDI-MS | Human urine and serum | Urine: 6 nM (LOD) Serum: 60 nM (LOD) | 5 to 500 nM in plasma | [84] |
| UHPLC-MS/MS | Human plasma and breast milk | 10.4 nM in plasma (LLOQ) 11.16 nM in breast milk (LLOQ) | 11.6 to 1163nM in plasma 11.6 to 581.4 nM in breast milk | [85] |
| UPLC-MS/MS | Human plasma | 55.8 nM (LOD) | 39.3 to 465.8 nM | [86] |
| UPLC/MS/MS | Human plasma | 5.8 nM (LLOQ) | 5.8 to 1744.1 nM | [82] |
| UPLC/MS/MS | Human plasma | 1.14 nM (LOD) 3.42 nM (LOQ) | 4.65 to 1162.7 nM | [32] |
| LC-MS/MS | Human plasma | 55.8 nM (LLOQ) | 55.81 to 2232.5 nM | [38] |
| SPE–HPLC–UV | Human plasma | 11.6 nM (LOD) | 23.25 to 9302 nM | [87] |
| LC-MS (High resolution) | Human plasma | 2.3 nM (LLOQ) | 2.3 to 1162.7 nM | [88] |
| UPLC-MS/MS | Human plasma | 1.25 nM (LLOQ) | 1.86 to 1860.4 nM | [89] |
| Electrochemical aptamer biosensor | Human plasma and EBC | 6.03 nM in EBC (LOD) 14.08 nM in plasma (LOD) | 10 to 600 nM in plasma and EBC | This Study |
| LCMS/MS | Human plasma and EBC | 0.5 nM in EBC (LOD) 1.1 nM in plasma (LOD) | 10 to 500 nM in plasma and EBC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebrahimi, R.; Barzegari, A.; Teimuri-Mofrad, R.; Kordasht, H.K.; Hasanzadeh, M.; Khoubnasabjafari, M.; Jouyban-Gharamaleki, V.; Rad, A.A.; Shadjou, N.; Rashidi, M.-R.; et al. Selection of Specific Aptamer against Rivaroxaban and Utilization for Label-Free Electrochemical Aptasensing Using Gold Nanoparticles: First Announcement and Application for Clinical Sample Analysis. Biosensors 2022, 12, 773. https://doi.org/10.3390/bios12100773
Ebrahimi R, Barzegari A, Teimuri-Mofrad R, Kordasht HK, Hasanzadeh M, Khoubnasabjafari M, Jouyban-Gharamaleki V, Rad AA, Shadjou N, Rashidi M-R, et al. Selection of Specific Aptamer against Rivaroxaban and Utilization for Label-Free Electrochemical Aptasensing Using Gold Nanoparticles: First Announcement and Application for Clinical Sample Analysis. Biosensors. 2022; 12(10):773. https://doi.org/10.3390/bios12100773
Chicago/Turabian StyleEbrahimi, Rokhsareh, Abolfazl Barzegari, Reza Teimuri-Mofrad, Houman Kholafazad Kordasht, Mohammad Hasanzadeh, Maryam Khoubnasabjafari, Vahid Jouyban-Gharamaleki, Abbas Afrasiabi Rad, Nasrin Shadjou, Mohammad-Reza Rashidi, and et al. 2022. "Selection of Specific Aptamer against Rivaroxaban and Utilization for Label-Free Electrochemical Aptasensing Using Gold Nanoparticles: First Announcement and Application for Clinical Sample Analysis" Biosensors 12, no. 10: 773. https://doi.org/10.3390/bios12100773
APA StyleEbrahimi, R., Barzegari, A., Teimuri-Mofrad, R., Kordasht, H. K., Hasanzadeh, M., Khoubnasabjafari, M., Jouyban-Gharamaleki, V., Rad, A. A., Shadjou, N., Rashidi, M.-R., Afshar Mogaddam, M. R., & Jouyban, A. (2022). Selection of Specific Aptamer against Rivaroxaban and Utilization for Label-Free Electrochemical Aptasensing Using Gold Nanoparticles: First Announcement and Application for Clinical Sample Analysis. Biosensors, 12(10), 773. https://doi.org/10.3390/bios12100773






