AuNP Aptasensor for Hodgkin Lymphoma Monitoring
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hypothesis and Plan of Study
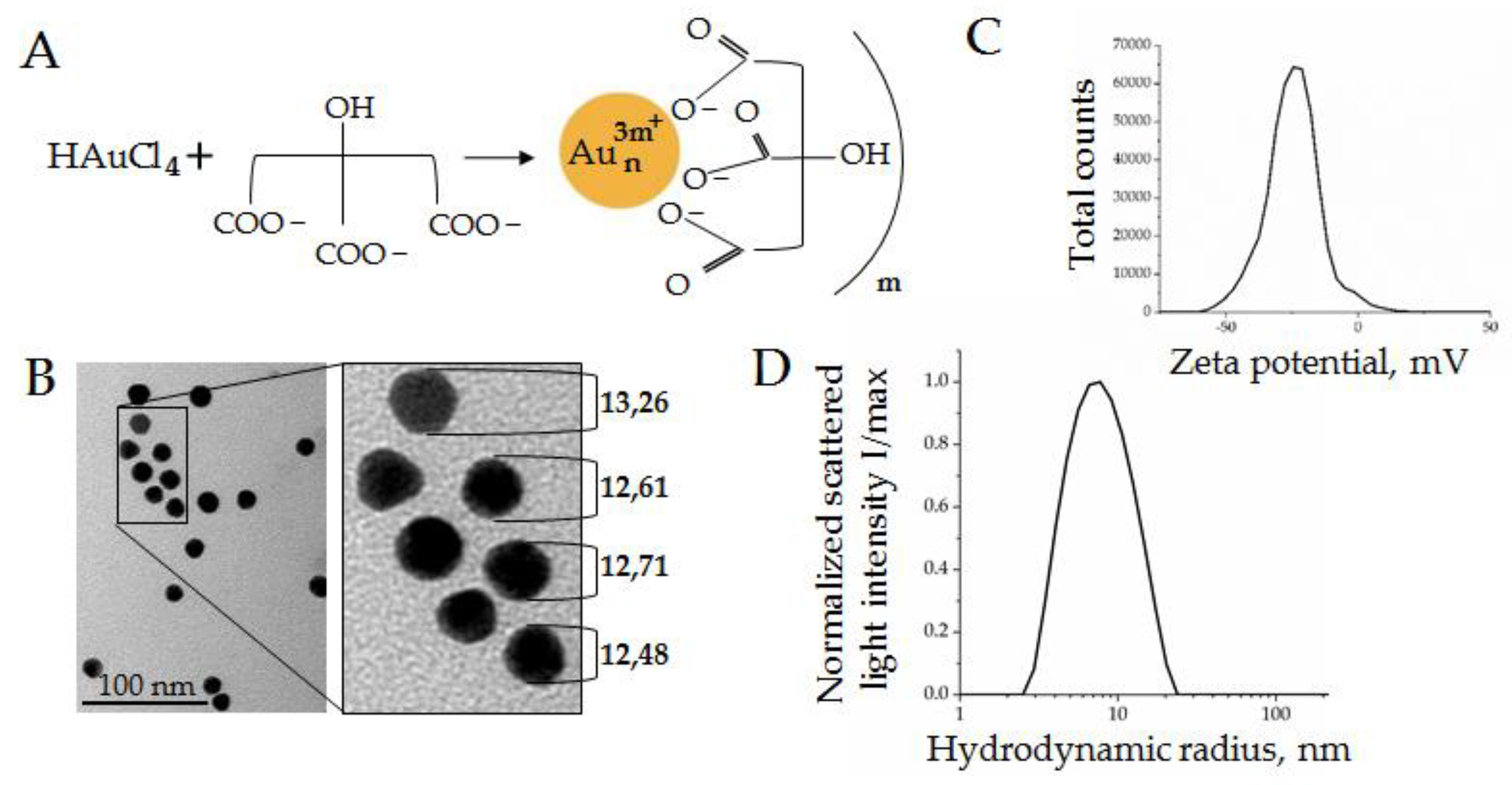
2.2. Aurum Nanoparticle (AuNP) Synthesis
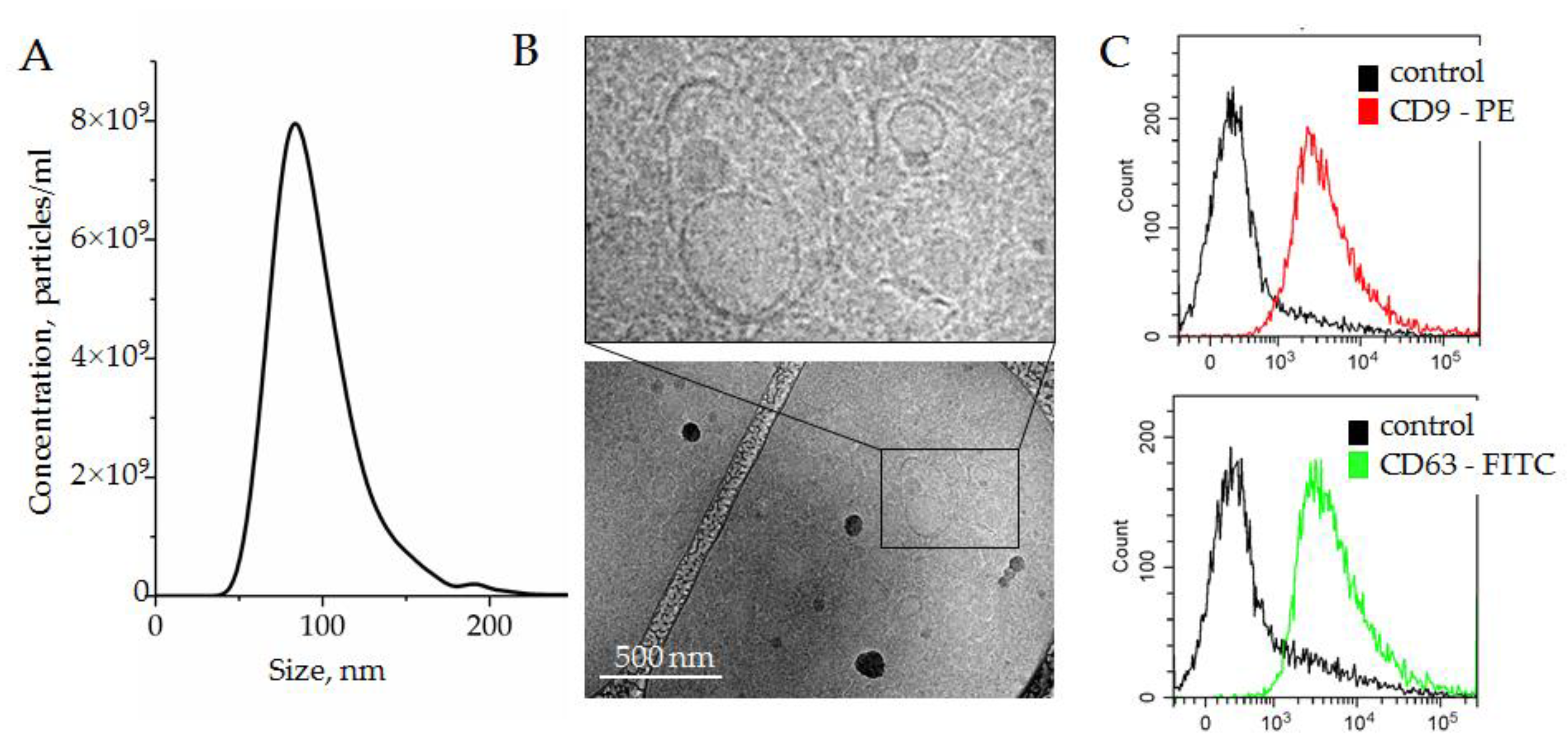
2.3. Isolation of Small Extracellular Nanovesicle from Plasma (SEVs)
2.4. Scanning Electron Microscopy (SEM) and Transmission Cryo-Electron Microscopy (Cryo-TEM)
2.5. Nanoparticle Tracking Analysis (NTA)
2.6. Bead-Assisted Flow Cytometry
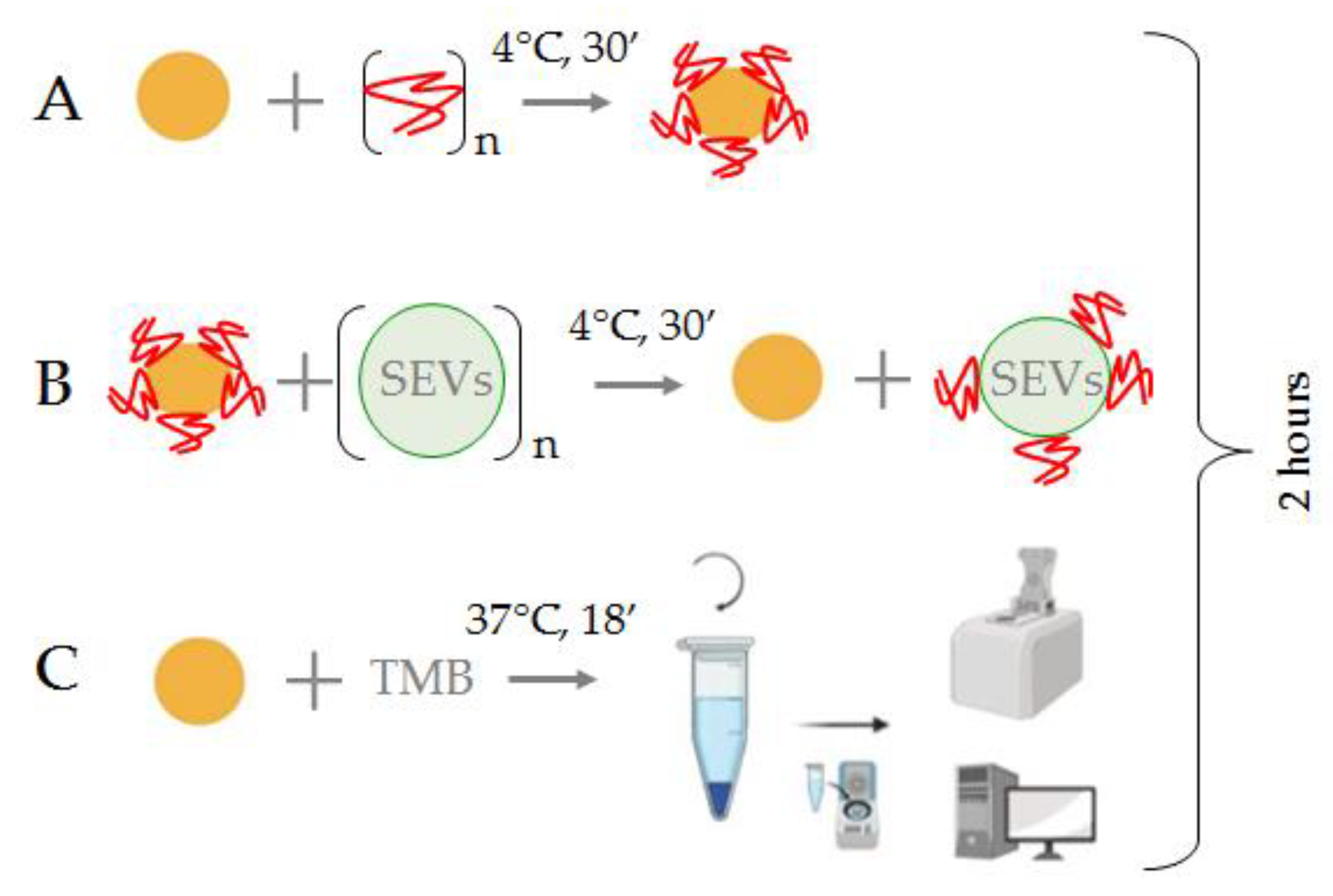
2.7. Interaction of AuNPs, ssDNA Aptamers, and Extracellular Nanovesicles (SEVs)
2.8. Spectrophotometry
2.9. Dynamic Light Scattering
2.10. Patients
2.11. The Protocol of CD30 + SEV Quantification in Plasma Samples
3. Results
3.1. Components of the Analytic System
3.1.1. Gold Nanoparticle (AuNP) Synthesis and Characteristics
3.1.2. Plasma SEV Isolation and Characterization
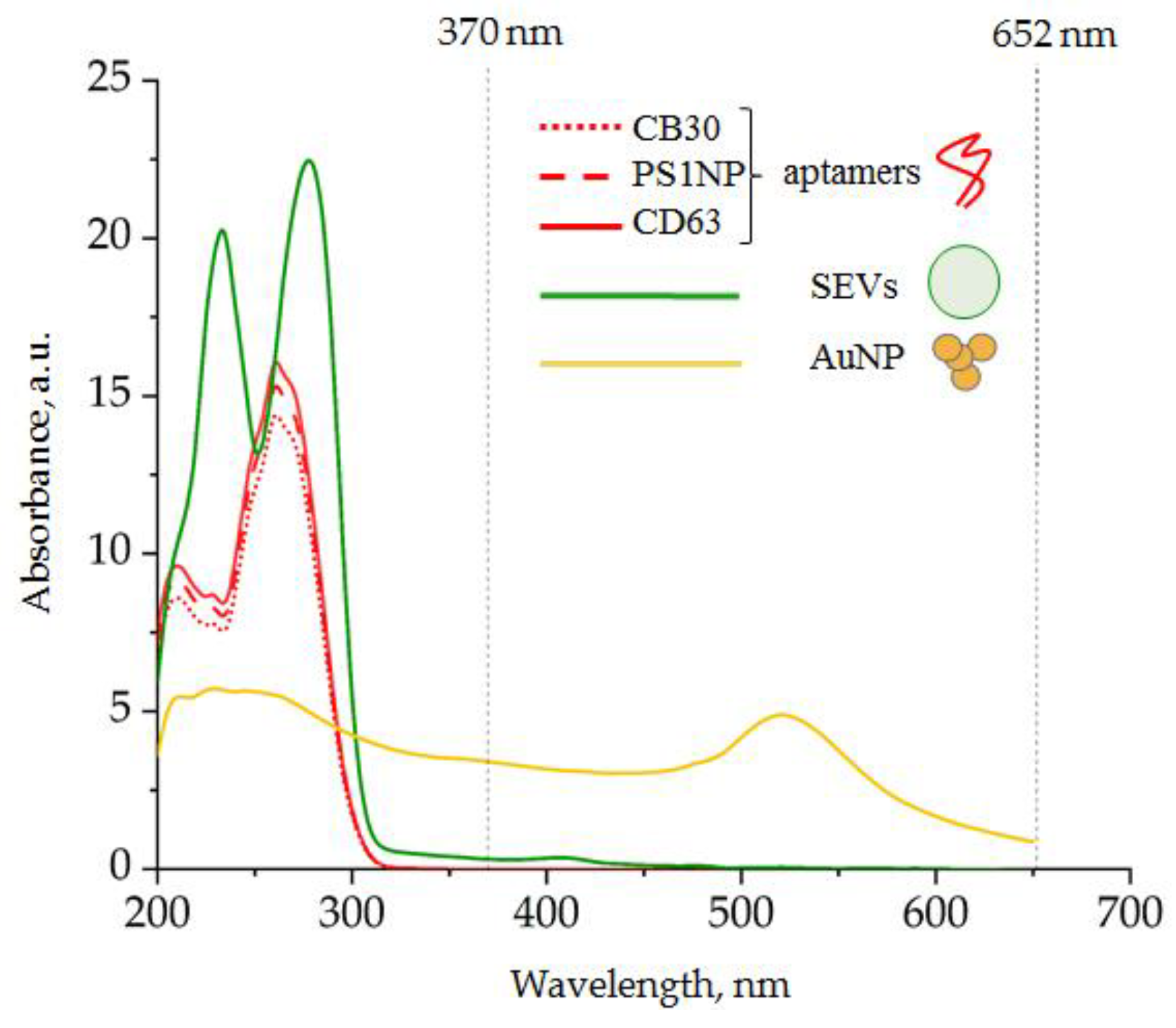
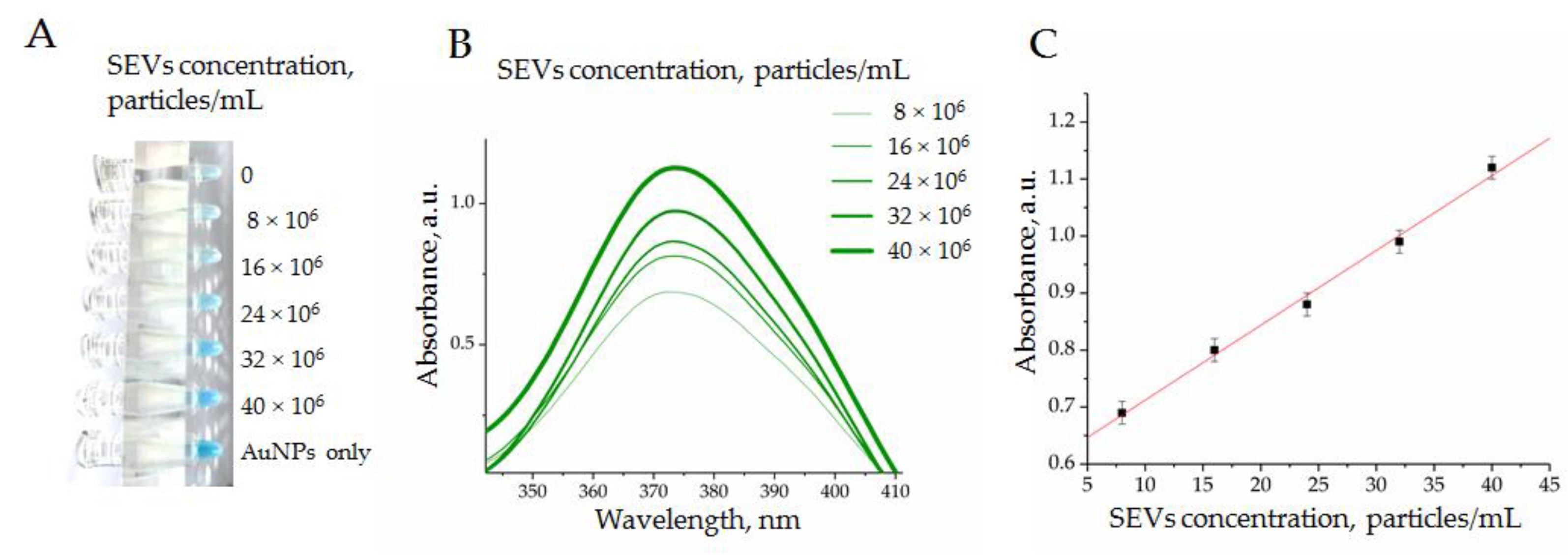
3.1.3. Spectral Analysis of the Analytic System Components
3.2. Optimization of the Analytic System Conditions
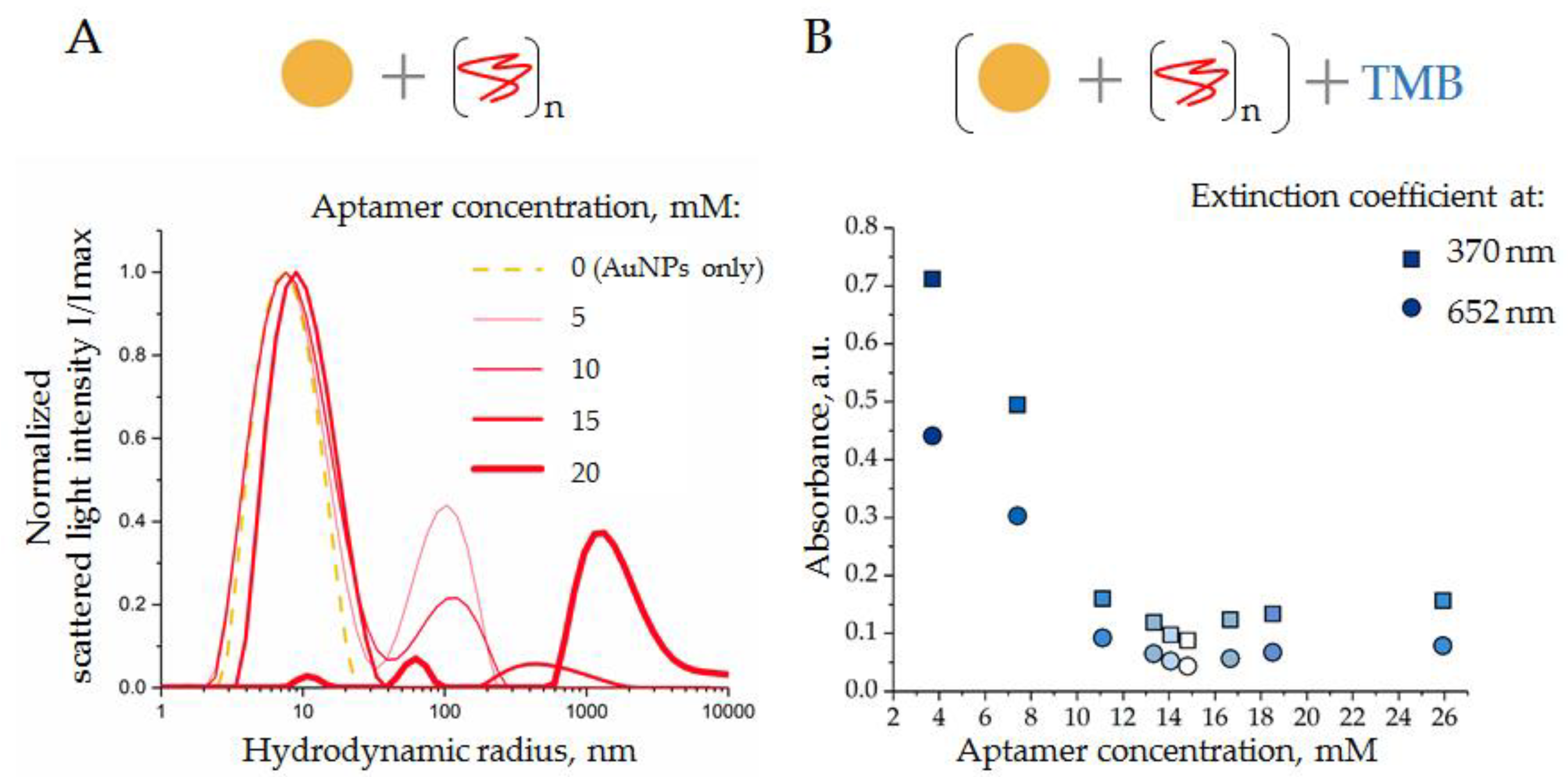
3.2.1. Interaction of AuNPs and DNA Aptamers
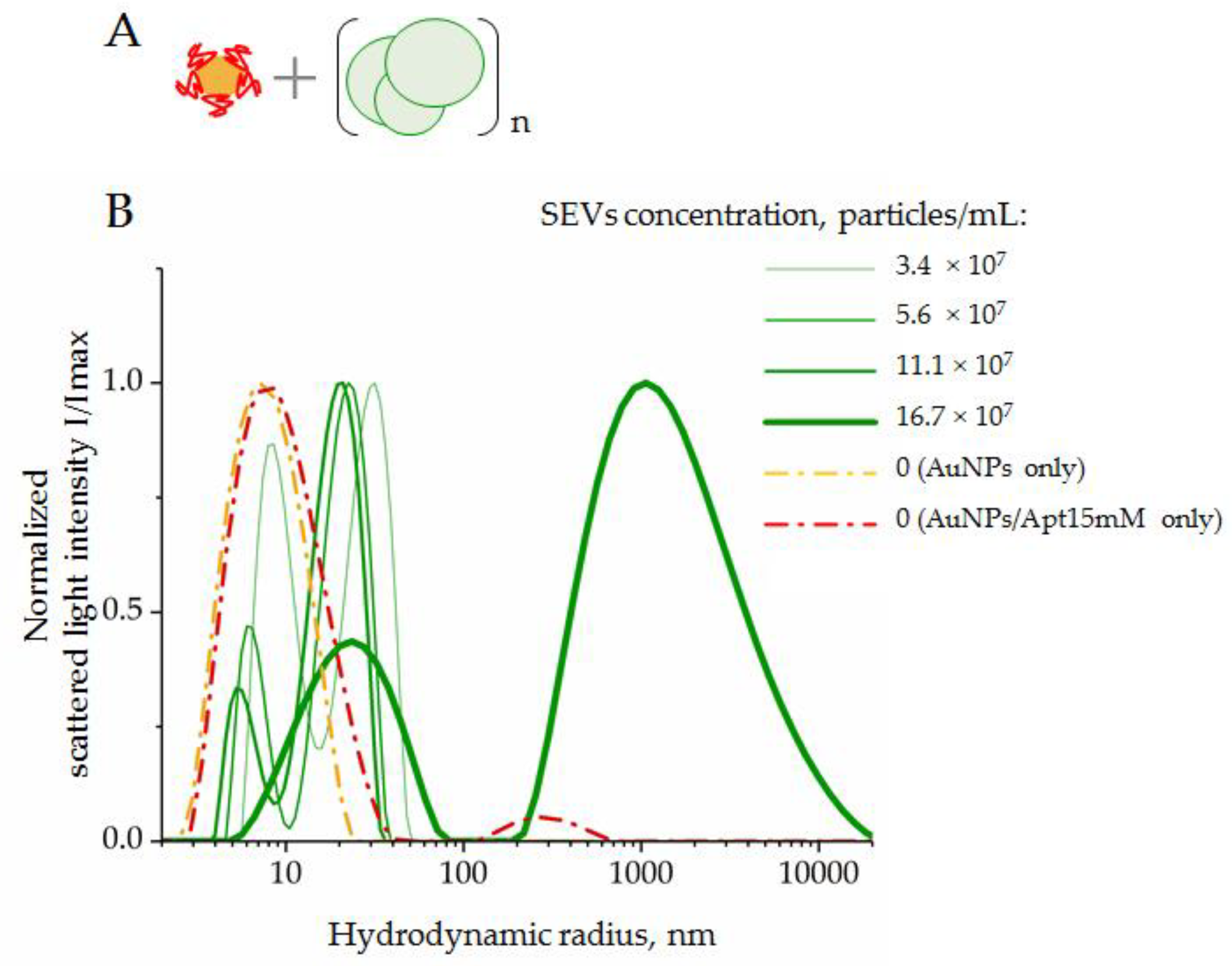
3.2.2. Interaction of SEVs with AuNP–Aptamer Complexes
3.3. Evaluation of the Clinical Performance of AuNP Aptasensor
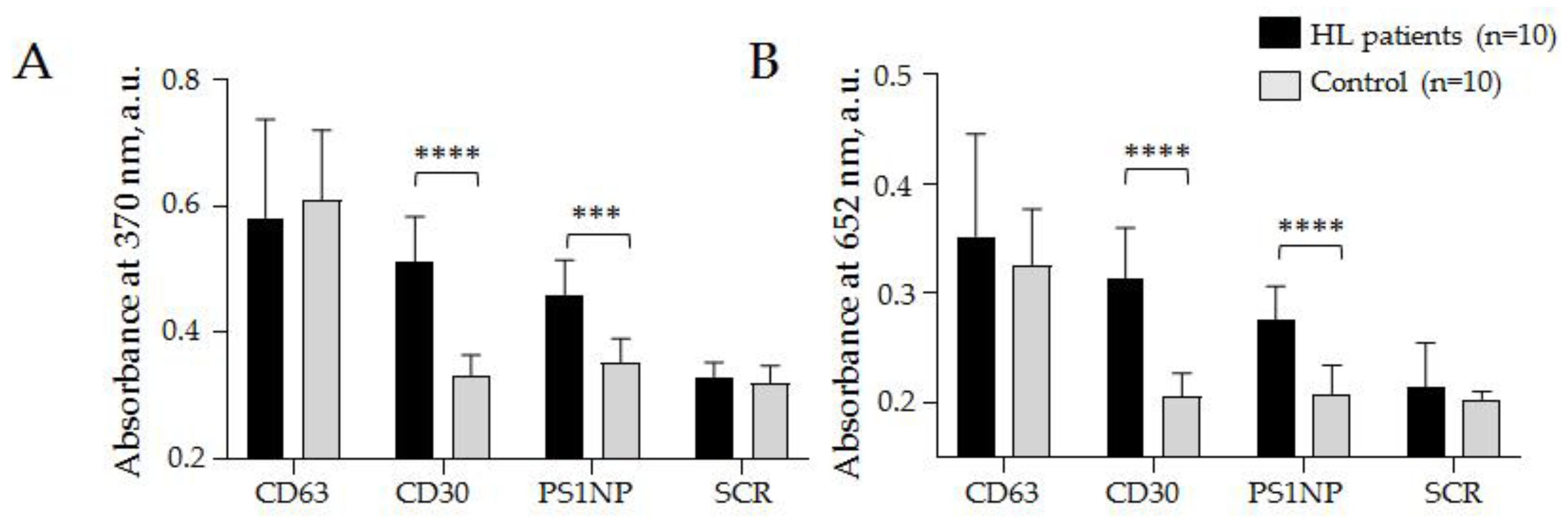
3.3.1. Quantification of CD30 + SEVs in the Plasma of cHL Patients and Healthy Donors
3.3.2. Comparison of PET-CT Data and CD30 + SEVs Relative Concentration
3.3.3. Quantification of CD30 + SEVs in the Plasma of HL Patients in the Course of Therapy
4. Discussion
4.1. The Reasons and Challenges of NP-based Sensor Development
4.2. Perspectives of AuNP Aptasensor Application for HL Management
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rossi, D.; Spina, V.; Bruscaggin, A.; Gaidano, G. Liquid biopsy in lymphoma. Haematologica 2019, 104, 648–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camus, V.; Viennot, M.; Lequesne, J.; Viailly, P.-J.; Elodie Bohers; Bessi, L.; Marcq, B.; Etancelin, P.; Dubois, S.; Picquenot, J.-M.; et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: A prospective study. Haematologica 2020, 106, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagpal, P.; Descalzi-Montoya, D.B.; Lodhi, N. The circuitry of the tumor microenvironment in adult and pediatric Hodgkin lymphoma: Cellular composition, cytokine profile, EBV, and exosomes. Cancer Rep. 2021, 4, e1311. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qin, Y.; Wan, C.; Sun, Y.; Meng, J.; Huang, J.; Hu, Y.; Jin, H.; Yang, K. Small Extracellular Vesicles: A Novel Avenue for Cancer Management. Front. Oncol. 2021, 11, 638357. [Google Scholar] [CrossRef]
- Repetto, O.; Lovisa, F.; Elia, C.; Enderle, D.; Romanato, F.; Buffardi, S.; Sala, A.; Pillon, M.; Steffan, A.; Burnelli, R.; et al. Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence. Diagnostics 2021, 11, 917. [Google Scholar] [CrossRef]
- Röthlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Wen, J.; Yuan, B.; Zu, Y. Biostable ssDNA Aptamers Specific for Hodgkin Lymphoma. Sensors 2013, 13, 14543–14557. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Li, L.; Wang, Z.; Irfan, M.; Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef]
- Chen, J.; Xu, Y.; Lu, Y.; Xing, W. Isolation and Visible Detection of Tumor-Derived Exosomes from Plasma. Anal. Chem. 2018, 90, 14207–14215. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-M.; Liu, J.-W.; Adkins, G.B.; Shen, W.; Trinh, M.P.; Duan, L.-Y.; Jiang, J.-H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, C.; Luo, J.; Tian, Y.; Zhou, N. Direct electrochemical detection of kanamycin based on peroxidase-like activity of gold nanoparticles. Anal. Chim. Acta 2016, 936, 75–82. [Google Scholar] [CrossRef]
- Tsekhmistrenko, S.I.; Bityutskyy, V.S.; Tsekhmistrenko, O.S.; Polishchuk, V.M.; Polishchuk, S.A.; Ponomarenko, N.V.; Melnychenko, Y.O.; Spivak, M.Y. Enzyme-like activity of nanomaterials. Regul. Mech. Biosyst. 2018, 9, 469–476. [Google Scholar] [CrossRef]
- Hirsch, B.; Hummel, M.; Bentink, S.; Fouladi, F.; Spang, R.; Zollinger, R.; Stein, H.; Dürkop, H. CD30-Induced Signaling Is Absent in Hodgkin’s Cells but Present in Anaplastic Large Cell Lymphoma Cells. Am. J. Pathol. 2008, 172, 510–520. [Google Scholar] [CrossRef] [Green Version]
- Dörsam, B.; Bösl, T.; Reiners, K.S.; Barnert, S.; Schubert, R.; Shatnyeva, O.; Zigrino, P.; Engert, A.; Hansen, H.P.; von Strandmann, E.P. Hodgkin Lymphoma-Derived Extracellular Vesicles Change the Secretome of Fibroblasts Toward a CAF Phenotype. Front. Immunol. 2018, 9, 1358. [Google Scholar] [CrossRef] [PubMed]
- Barmin, R.; Rudakovskaya, P.; Gusliakova, O.; Sindeeva, O.; Prikhozhdenko, E.; Maksimova, E.; Obukhova, E.; Chernyshev, V.; Khlebtsov, B.; Solovev, A.; et al. Air-Filled Bubbles Stabilized by Gold Nanoparticle/Photodynamic Dye Hybrid Structures for Theranostics. Nanomaterials 2021, 11, 415. [Google Scholar] [CrossRef] [PubMed]
- Slyusarenko, M.; Nikiforova, N.; Sidina, E.; Nazarova, I.; Egorov, V.; Garmay, Y.; Merdalimova, A.; Yevlampieva, N.; Gorin, D.; Malek, A. Formation and Evaluation of a Two-Phase Polymer System in Human Plasma as a Method for Extracellular Nanovesicle Isolation. Polymers 2021, 13, 458. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.-S.; Patel, T.; Revzin, A. Development of an aptasensor for electrochemical detection of exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Parekh, P.; Kamble, S.; Zhao, N.; Zeng, Z.; Portier, B.P.; Zu, Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials 2013, 34, 8909–8917. [Google Scholar] [CrossRef] [Green Version]
- Berne, B.J.; Pecora, R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics; Dover Publications: New York, NY, USA, 2000; ISBN 0486320243. [Google Scholar]
- Tsvetkov, V.N.; Tsvetkov, N. V Electrical birefringence in solutions of rigid-chain polymers. Russ. Chem. Rev. 1993, 62, 851–876. [Google Scholar] [CrossRef]
- Camacho, M.R.; Etchebehere, E.; Tardelli, N.; Delamain, M.T.; Vercosa, A.F.A.; Takahashi, M.E.S.; Brunetto, S.Q.; Metze, I.G.H.L.; Souza, C.A.; Cerci, J.J.; et al. Validation of a Multifocal Segmentation Method for Measuring Metabolic Tumor Volume in Hodgkin Lymphoma. J. Nucl. Med. Technol. 2020, 48, 30–35. [Google Scholar] [CrossRef]
- Gallamini, A.; Barrington, S.F.; Biggi, A.; Chauvie, S.; Kostakoglu, L.; Gregianin, M.; Meignan, M.; Mikhaeel, G.N.; Loft, A.; Zaucha, J.M.; et al. The predictive role of interim positron emission tomography for Hodgkin lymphoma treatment outcome is confirmed using the interpretation criteria of the Deauville five-point scale. Haematologica 2014, 99, 1107–1113. [Google Scholar] [CrossRef]
- Cheson, B.D.; Fisher, R.I.; Barrington, S.F.; Cavalli, F.; Schwartz, L.H.; Zucca, E.; Lister, T.A. Recommendations for Initial Evaluation, Staging, and Response Assessment of Hodgkin and Non-Hodgkin Lymphoma: The Lugano Classification. J. Clin. Oncol. 2014, 32, 3059–3067. [Google Scholar] [CrossRef]
- Ghosh, S.; Jaiswal, A. Peroxidase-Like Activity of Metal Nanoparticles for Biomedical Applications. In Nanobiomaterial Engineering; Springer: Singapore, 2020; pp. 109–126. [Google Scholar] [CrossRef]
- Barnett, C.E. Some Applications of Wave-length Turbidimetry in the Infrared. J. Phys. Chem. 1942, 46, 69–75. [Google Scholar] [CrossRef]
- Connors, J.M.; Cozen, W.; Steidl, C.; Carbone, A.; Hoppe, R.T.; Flechtner, H.-H.; Bartlett, N.L. Hodgkin lymphoma. Nat. Rev. Dis. Primers 2020, 6, 61. [Google Scholar] [CrossRef]
- Demina, E.A.; Tumyan, G.S.; Moiseeva, T.N.; Mikhailova, N.B.; Myakova, N.V.; Rumyantsev, A.G.; Maschan, A.A.; Kaplanov, K.D.; Shmakov, R.G.; Falaleeva, N.A.; et al. Hodgkin’s lymphoma. J. Mod. Oncol. 2020, 22, 6–33. [Google Scholar] [CrossRef]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Wang, H.; Ye, B.-C. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst 2020, 145, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Kowal, J.; Théry, C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160479. [Google Scholar] [CrossRef]
- Allelein, S.; Medina-Perez, P.; Lopes, A.L.H.; Rau, S.; Hause, G.; Kölsch, A.; Kuhlmeier, D. Potential and challenges of specifically isolating extracellular vesicles from heterogeneous populations. Sci. Rep. 2021, 11, 11585. [Google Scholar] [CrossRef]
- Zabegina, L.; Nazarova, I.; Nikiforova, N.; Slyusarenko, M.; Sidina, E.; Knyazeva, M.; Tsyrlina, E.; Novikov, S.; Reva, S.; Malek, A. A New Approach for Prostate Cancer Diagnosis by miRNA Profiling of Prostate-Derived Plasma Small Extracellular Vesicles. Cells 2021, 10, 2372. [Google Scholar] [CrossRef] [PubMed]
- Martín-Gracia, B.; Martín-Barreiro, A.; Cuestas-Ayllón, C.; Grazú, V.; Line, A.; Llorente, A.; de la Fuente, J.M.; Moros, M. Nanoparticle-based biosensors for detection of extracellular vesicles in liquid biopsies. J. Mater. Chem. B 2020, 8, 6710–6738. [Google Scholar] [CrossRef] [PubMed]
- Illidge, T.M.; Phillips, E.H. Progress and pitfalls with the use of image-guided personalised approaches in lymphoma. Br. J. Radiol. 2021, 94, 20210609. [Google Scholar] [CrossRef] [PubMed]
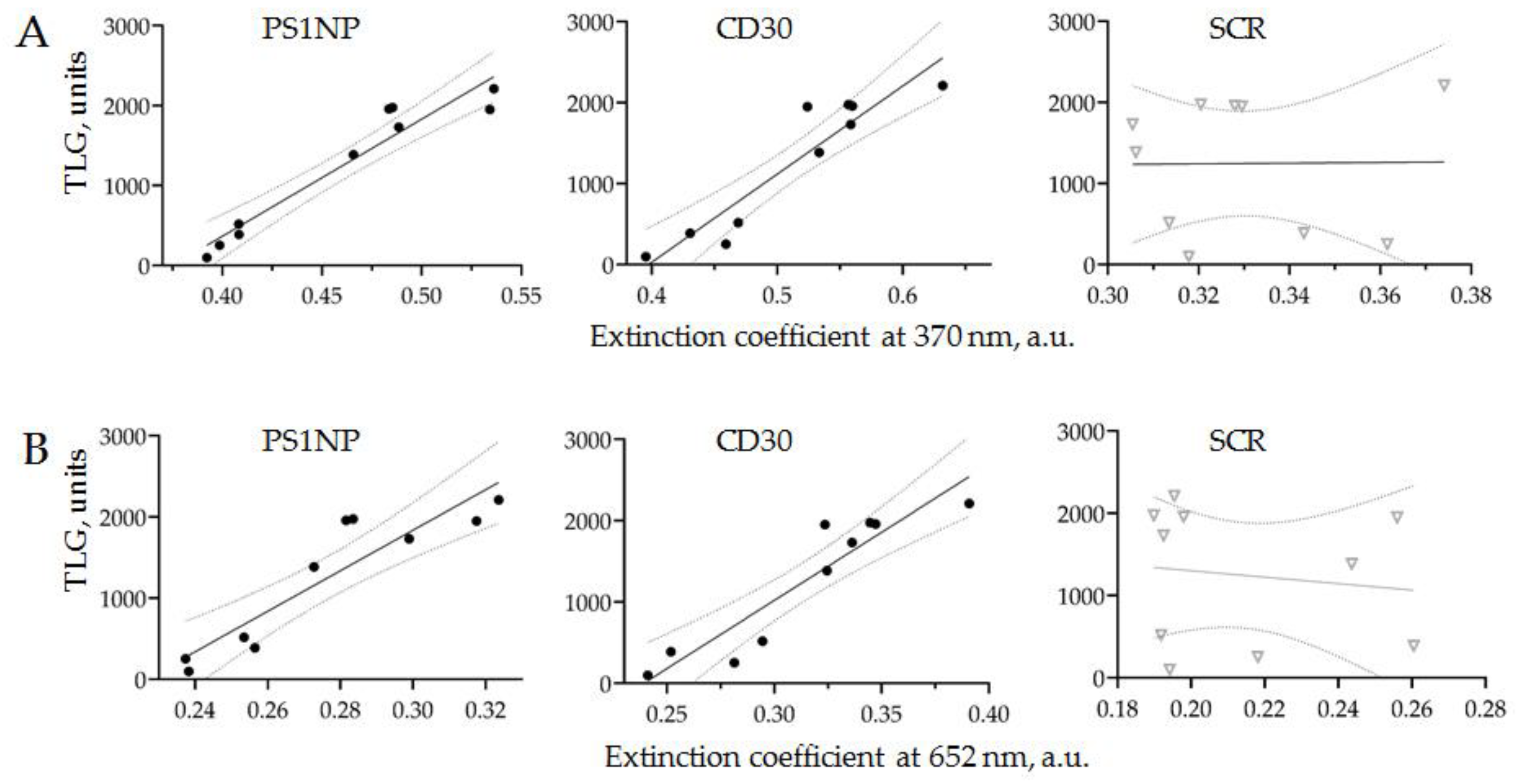

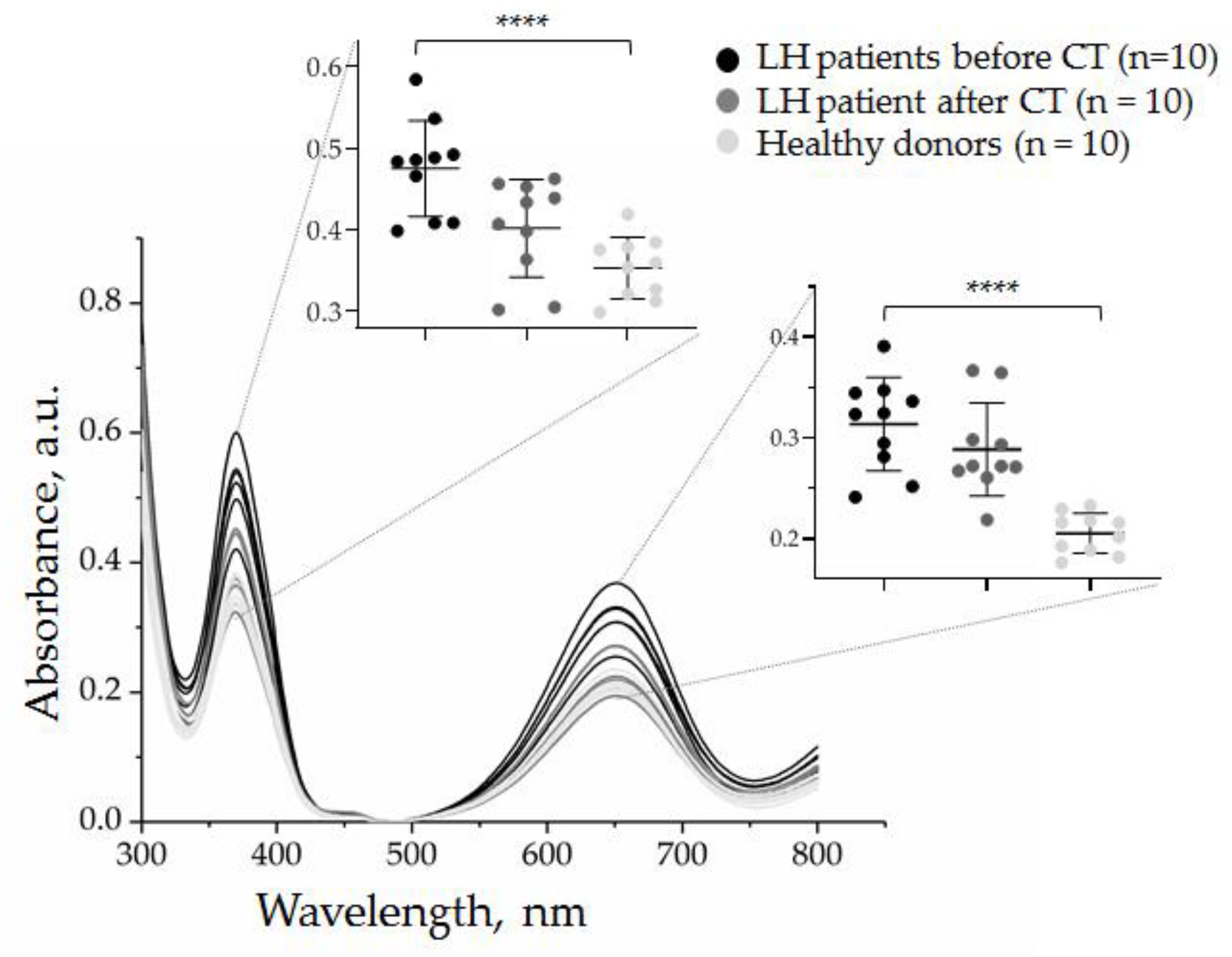
| Patient ID | Gender/Agee | Morphology * | Stage | B Symptoms | ECOG | Before Chemotherapy | After Chemotherapy | Deauville | ||
|---|---|---|---|---|---|---|---|---|---|---|
| MTV, cm3 | TLG | MTV, cm3 | TLG | |||||||
| 577848 | M/33 | MCHL | 4 | + | 1 | 563 | 1975 | 277 | 507 | 2 |
| 583352 | F/29 | MCHL | 2 | - | 0 | 72 | 254 | 0 | 0 | 2 |
| 525340 | M/30 | NSHL | 4 | + | 1 | 1033 | 2212 | 0 | 0 | 2 |
| 586446 | F/29 | NSHL | 3 | + | 2 | 310 | 1731 | 0 | 0 | 3 |
| 585071 | M/44 | NSHL | 4 | - | 1 | 251 | 1387 | 26 | 50 | 2 |
| 590335 | F/42 | NSHL | 2 | - | 0 | 91 | 520 | 0 | 0 | 2 |
| 589465 | M/54 | MCHL | 4 | + | 1 | 400 | 1959 | 0 | 0 | 3 |
| 596063 | M/27 | NSHL | 2 | - | 1 | 356 | 1951 | 0 | 0 | 2 |
| 438920 | M/29 | NSHL | 2 | - | 0 | 47 | 99 | 0 | 0 | 2 |
| 598035 | F/36 | NSHL | 4 | - | 1 | 157 | 388 | 0 | 0 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slyusarenko, M.; Shalaev, S.; Valitova, A.; Zabegina, L.; Nikiforova, N.; Nazarova, I.; Rudakovskaya, P.; Vorobiev, M.; Lezov, A.; Filatova, L.; et al. AuNP Aptasensor for Hodgkin Lymphoma Monitoring. Biosensors 2022, 12, 23. https://doi.org/10.3390/bios12010023
Slyusarenko M, Shalaev S, Valitova A, Zabegina L, Nikiforova N, Nazarova I, Rudakovskaya P, Vorobiev M, Lezov A, Filatova L, et al. AuNP Aptasensor for Hodgkin Lymphoma Monitoring. Biosensors. 2022; 12(1):23. https://doi.org/10.3390/bios12010023
Chicago/Turabian StyleSlyusarenko, Maria, Sergey Shalaev, Alina Valitova, Lidia Zabegina, Nadezhda Nikiforova, Inga Nazarova, Polina Rudakovskaya, Maxim Vorobiev, Alexey Lezov, Larisa Filatova, and et al. 2022. "AuNP Aptasensor for Hodgkin Lymphoma Monitoring" Biosensors 12, no. 1: 23. https://doi.org/10.3390/bios12010023
APA StyleSlyusarenko, M., Shalaev, S., Valitova, A., Zabegina, L., Nikiforova, N., Nazarova, I., Rudakovskaya, P., Vorobiev, M., Lezov, A., Filatova, L., Yevlampieva, N., Gorin, D., Krzhivitsky, P., & Malek, A. (2022). AuNP Aptasensor for Hodgkin Lymphoma Monitoring. Biosensors, 12(1), 23. https://doi.org/10.3390/bios12010023









