Melting Curve Analysis of Aptachains: Adenosine Detection with Internal Calibration
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Oligonucleotides
2.2. Oligonucleotides Sequence Design
2.3. UV-Vis Measurements
3. Results and Discussion
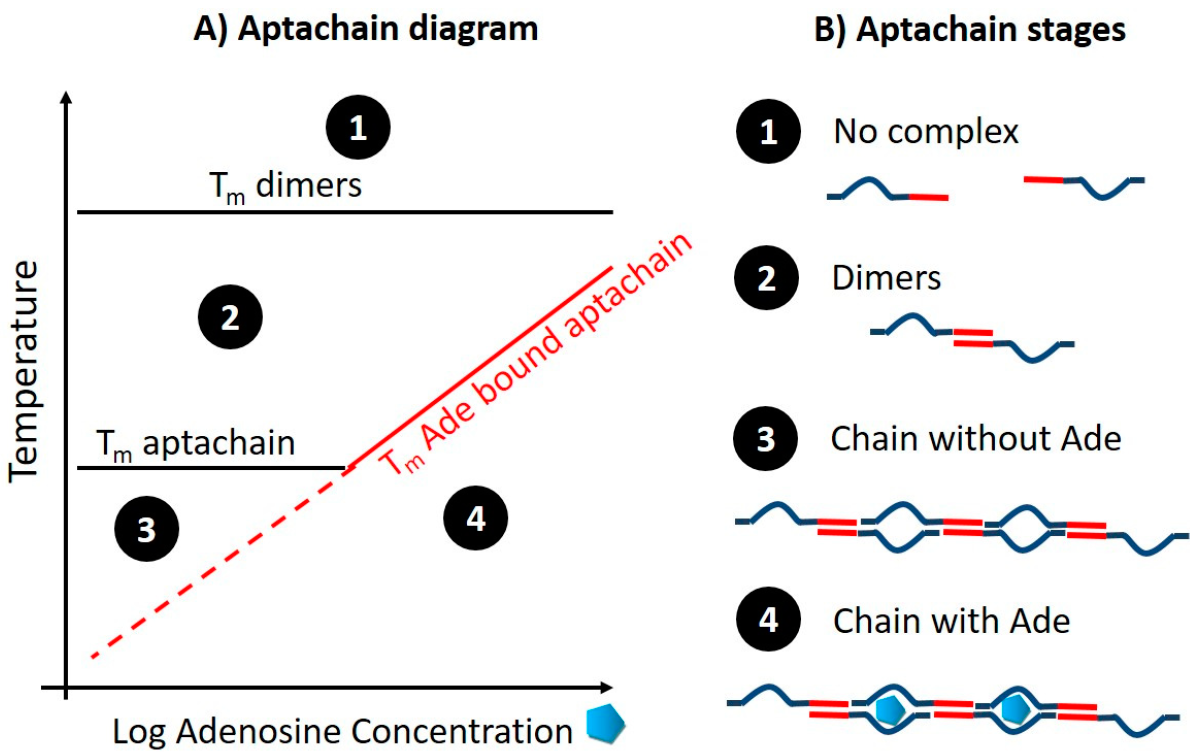
3.1. Aptachain Formation and Melting Curve Analysis through Temperature Scans
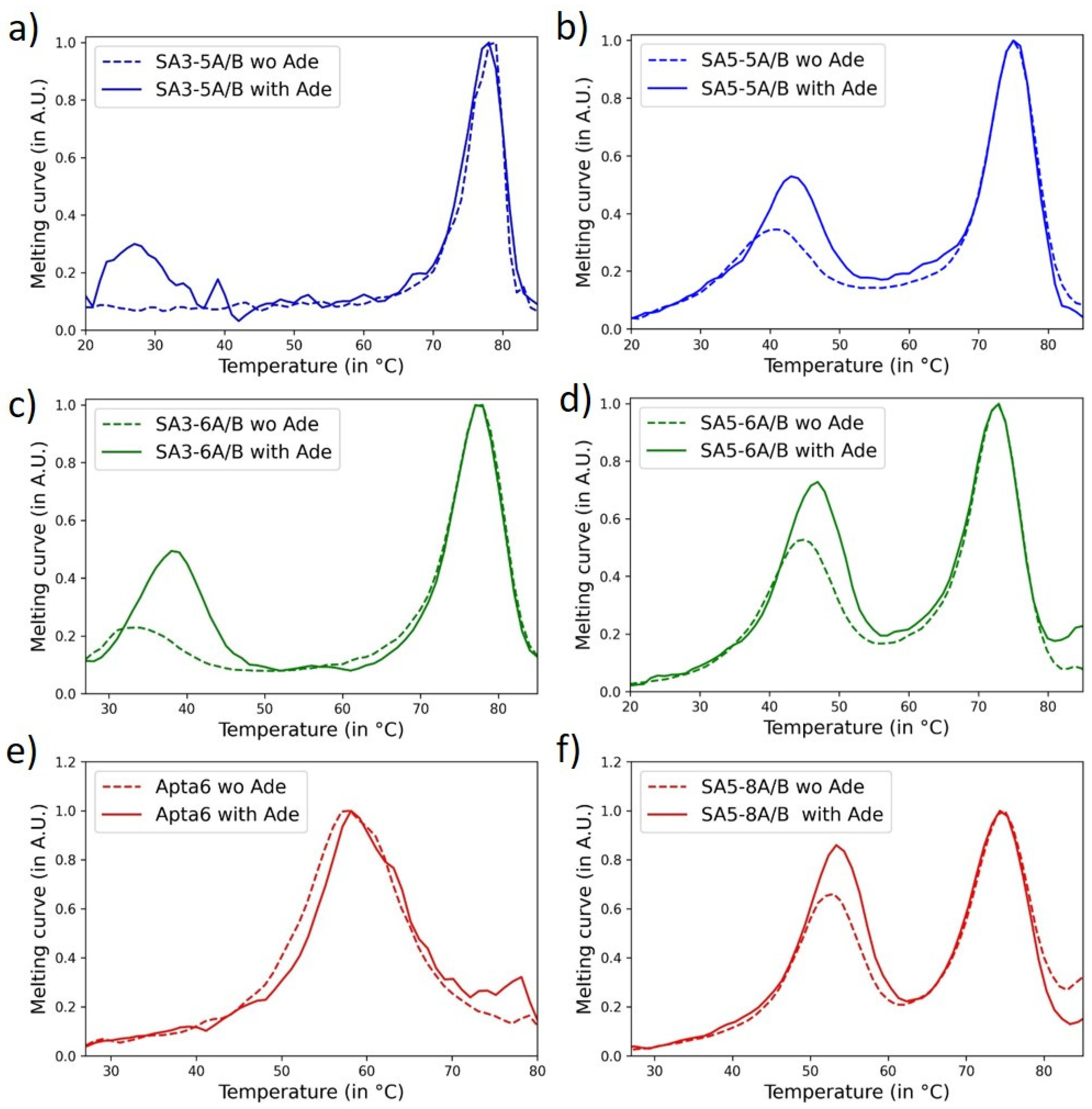
3.2. Impact of Split-Aptamer Design on Aptachain Formation
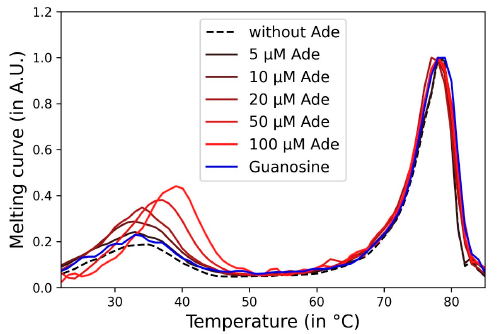
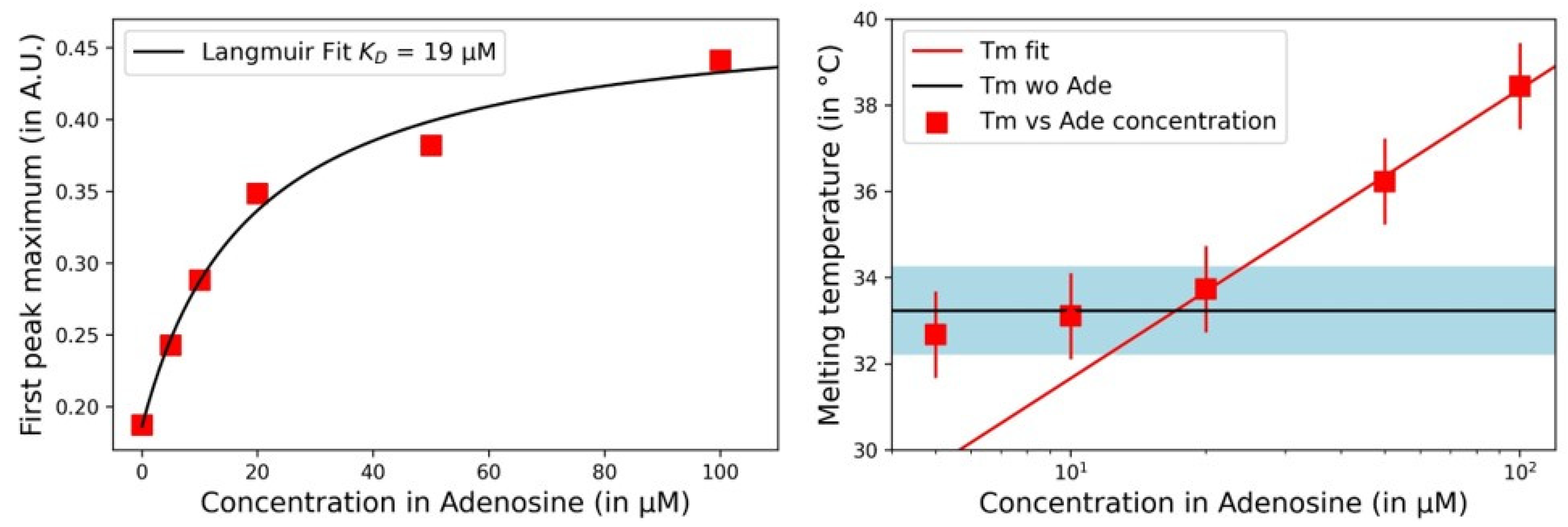
3.3. Impact of Adenosine Concentration on Aptachain Formation
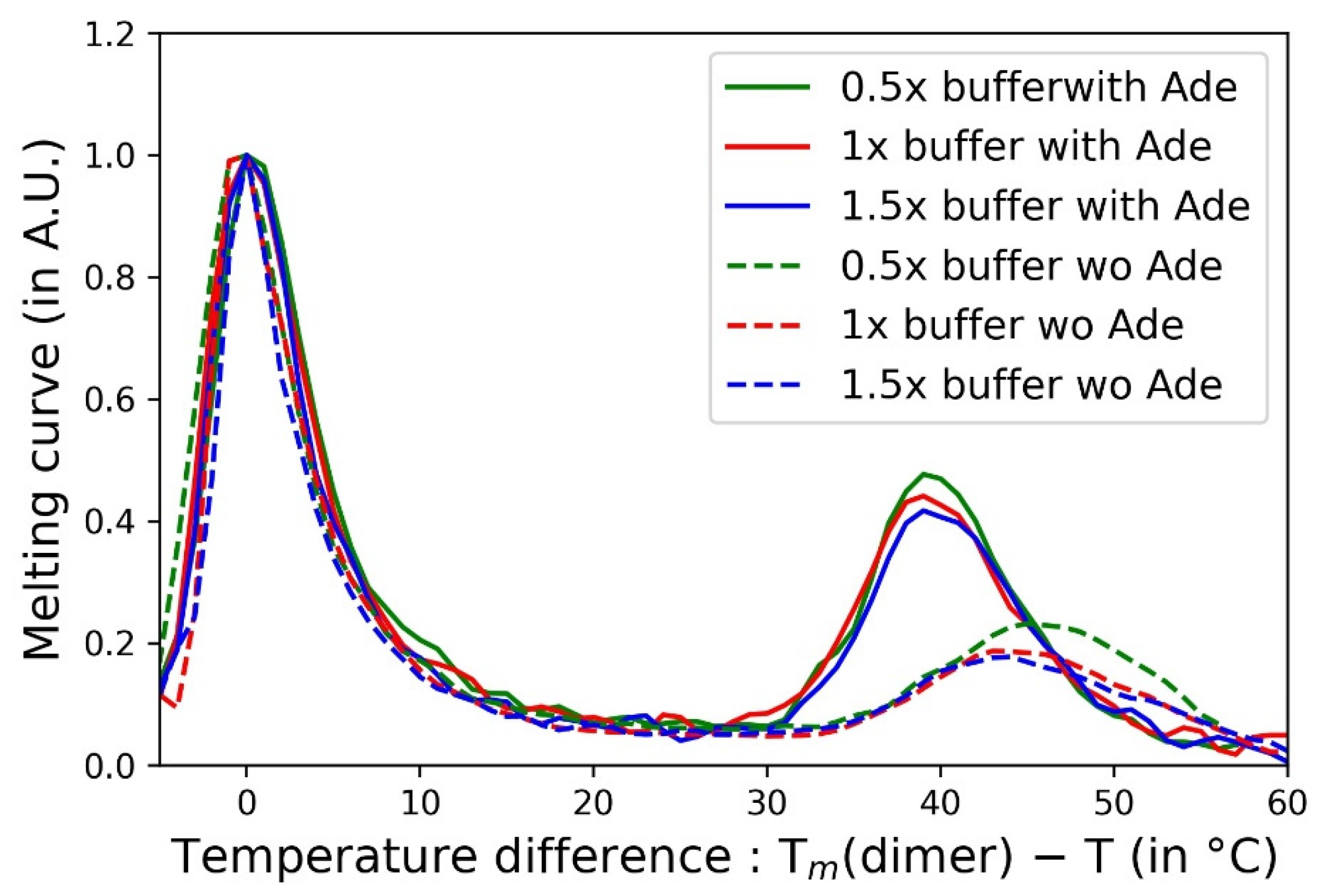
3.4. Influence of the Salt Concentration
3.5. Influence of Oligonucleotide Concentrations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schwarzenbach, R.P.; Escher, B.I.; Fenner, K.; Hofstetter, T.B.; Johnson, C.A.; von Gunten, U.; Wehrli, B. The Challenge of Micropollutants in Aquatic Systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Benotti, M.J.; Trenholm, R.A.; Vanderford, B.J.; Holady, J.C.; Stanford, B.D.; Snyder, S.A. Pharmaceuticals and Endocrine Disrupting Compounds in U.S. Drinking Water. Environ. Sci. Technol. 2009, 43, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yan, X.; Zhao, Y.; Li, L.; Wang, S. Highly Sensitive and Specific Detection of Small Molecules Using Advanced Aptasensors Based on Split Aptamers: A Review. Trac. Trends Anal. Chem. 2020, 133, 116069. [Google Scholar] [CrossRef]
- Prante, M.; Segal, E.; Scheper, T.; Bahnemann, J.; Walter, J. Aptasensors for Point-of-Care Detection of Small Molecules. Biosensors 2020, 10, 108. [Google Scholar] [CrossRef]
- Ziółkowski, R.; Jarczewska, M.; Górski, Ł.; Malinowska, E. From Small Molecules toward Whole Cells Detection: Application of Electrochemical Aptasensors in Modern Medical Diagnostics. Sensors 2021, 21, 724. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-Based Biosensors. Trac. Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Cho, E.J.; Lee, J.-W.; Ellington, A.D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef]
- Chen, A.; Yang, S. Replacing Antibodies with Aptamers in Lateral Flow Immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T.-H. Aptamers as a Replacement for Antibodies in Enzyme-Linked Immunosorbent Assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef]
- Seok Kim, Y.; Ahmad Raston, N.H.; Bock Gu, M. Aptamer-Based Nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [CrossRef]
- Dhiman, A.; Kalra, P.; Bansal, V.; Bruno, J.G.; Sharma, T.K. Aptamer-Based Point-of-Care Diagnostic Platforms. Sens. Actuators B Chem. 2017, 246, 535–553. [Google Scholar] [CrossRef]
- Huizenga, D.E.; Szostak, J.W. A DNA Aptamer That Binds Adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; de Prada, P.; Landry, D.W. Fluorescent Sensors Based on Aptamer Self-Assembly. J. Am. Chem. Soc. 2000, 122, 11547–11548. [Google Scholar] [CrossRef]
- Baker, B.R.; Lai, R.Y.; Wood, M.S.; Doctor, E.H.; Heeger, A.J.; Plaxco, K.W. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. J. Am. Chem. Soc. 2006, 128, 3138–3139. [Google Scholar] [CrossRef]
- McKeague, M.; DeRosa, M.C. Challenges and Opportunities for Small Molecule Aptamer Development. Available online: https://www.hindawi.com/journals/jna/2012/748913/ (accessed on 3 February 2021).
- Alkhamis, O.; Canoura, J.; Yu, H.; Liu, Y.; Xiao, Y. Innovative Engineering and Sensing Strategies for Aptamer-Based Small-Molecule Detection. Trac. Trends Anal. Chem. 2019, 121, 115699. [Google Scholar] [CrossRef]
- McKeague, M.; De Girolamo, A.; Valenzano, S.; Pascale, M.; Ruscito, A.; Velu, R.; Frost, N.R.; Hill, K.; Smith, M.; McConnell, E.M.; et al. Comprehensive Analytical Comparison of Strategies Used for Small Molecule Aptamer Evaluation. Anal. Chem. 2015, 87, 8608–8612. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Adenosine-Dependent Assembly of Aptazyme-Functionalized Gold Nanoparticles and Its Application as a Colorimetric Biosensor. Anal. Chem. 2004, 76, 1627–1632. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Fast Colorimetric Sensing of Adenosine and Cocaine Based on a General Sensor Design Involving Aptamers and Nanoparticles. Angew. Chem. 2006, 118, 96–100. [Google Scholar] [CrossRef]
- Chen, S.-J.; Huang, Y.-F.; Huang, C.-C.; Lee, K.-H.; Lin, Z.-H.; Chang, H.-T. Colorimetric Determination of Urinary Adenosine Using Aptamer-Modified Gold Nanoparticles. Biosens. Bioelectron. 2008, 23, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, J.; Cao, X.; Wang, L.; Li, D.; Song, S.; Ye, B.; Fan, C. Adenosine Detection by Using Gold Nanoparticles and Designed Aptamer Sequences. Analyst 2009, 134, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Feng, F.; Zhao, L.; Chen, Z.; Wang, H.; Duan, Y. A Turn-on Fluorescent Aptasensor for Adenosine Detection Based on Split Aptamers and Graphene Oxide. Analyst 2014, 139, 1843. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, J.; Tan, Z.; Wang, H. Electrochemical Impedance Spectroscopy Aptasensor for Ultrasensitive Detection of Adenosine with Dual Backfillers. Biosens. Bioelectron. 2014, 60, 218–223. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J. Aptamer-Based Strategies for Recognizing Adenine, Adenosine, ATP and Related Compounds. Analyst 2020, 145, 6753–6768. [Google Scholar] [CrossRef]
- Melaine, F.; Roupioz, Y.; Buhot, A. Gold Nanoparticles Surface Plasmon Resonance Enhanced Signal for the Detection of Small Molecules on Split-Aptamer Microarrays (Small Molecules Detection from Split-Aptamers). Microarrays 2015, 4, 41–52. [Google Scholar] [CrossRef]
- Melaine, F.; Coilhac, C.; Roupioz, Y.; Buhot, A. A Nanoparticle-Based Thermo-Dynamic Aptasensor for Small Molecule Detection. Nanoscale 2016, 8, 16947–16954. [Google Scholar] [CrossRef]
- Melaine, F.; Roupioz, Y.; Buhot, A. Small Molecule SPR Imaging Detection from Split Aptamer Microarrays. Procedia Technol. 2017, 27, 6–7. [Google Scholar] [CrossRef]
- Lopez, A.; Liu, J. Nanomaterial and Aptamer-Based Sensing: Target Binding versus Target Adsorption Illustrated by the Detection of Adenosine and ATP on Metal Oxides and Graphene Oxide. Anal. Chem. 2021, 93, 3018–3025. [Google Scholar] [CrossRef]
- Holden, M.J.; Haynes, R.J.; Rabb, S.A.; Satija, N.; Yang, K.; Blasic, J.R. Factors Affecting Quantification of Total DNA by UV Spectroscopy and PicoGreen Fluorescence. J. Agric. Food Chem. 2009, 57, 7221–7226. [Google Scholar] [CrossRef]
- Fuchs, J.; Dell’Atti, D.; Buhot, A.; Calemczuk, R.; Mascini, M.; Livache, T. Effects of Formamide on the Thermal Stability of DNA Duplexes on Biochips. Anal. Biochem. 2010, 397, 132–134. [Google Scholar] [CrossRef]
- Bhat, S.; Curach, N.; Mostyn, T.; Bains, G.S.; Griffiths, K.R.; Emslie, K.R. Comparison of Methods for Accurate Quantification of DNA Mass Concentration with Traceability to the International System of Units. Anal. Chem. 2010, 82, 7185–7192. [Google Scholar] [CrossRef]
- Bi, S.; Yue, S.; Zhang, S. Hybridization Chain Reaction: A Versatile Molecular Tool for Biosensing, Bioimaging, and Biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Beck, V.A.; Pierce, N.A. Next-Generation in Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano 2014, 8, 4284–4294. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. Triggered Amplification by Hybridization Chain Reaction. Proc. Natl. Acad. Sci. USA 2004, 101, 15275–15278. [Google Scholar] [CrossRef]
- Evanko, D. Hybridization Chain Reaction. Nat. Methods 2004, 1, 186. [Google Scholar] [CrossRef]
- Figg, C.A.; Winegar, P.H.; Hayes, O.G.; Mirkin, C.A. Controlling the DNA Hybridization Chain Reaction. J. Am. Chem. Soc. 2020, 142, 8596–8601. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Sun, R.; Huang, Z.; Luo, Z.; Zhou, C.; Wu, M.; Duan, Y.; Li, Y. The Recent Development of Hybridization Chain Reaction Strategies in Biosensors. ACS Sens. 2020, 5, 2977–3000. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhou, R.; Sun, R.; Zhang, X.; Cheng, Z.; Chen, C.; Zhu, Q. Nonlinear Hybridization Chain Reaction-Based Functional DNA Nanostructure Assembly for Biosensing, Bioimaging Applications. Biosens. Bioelectron. 2021, 173, 112814. [Google Scholar] [CrossRef]
- Lu, C.; Saint-Pierre, C.; Gasparutto, D.; Roupioz, Y.; Peyrin, E.; Buhot, A. Linear Chain Formation of Split-Aptamer Dimers on Surfaces Triggered by Adenosine. Langmuir 2017, 33, 12785–12792. [Google Scholar] [CrossRef]
- Wang, J.; Cheng, W.; Meng, F.; Yang, M.; Pan, Y.; Miao, P. Hand-in-Hand RNA Nanowire-Based Aptasensor for the Detection of Theophylline. Biosens. Bioelectron. 2018, 101, 153–158. [Google Scholar] [CrossRef]
- Azéma, L.; Bonnet-Salomon, S.; Endo, M.; Takeuchi, Y.; Durand, G.; Emura, T.; Hidaka, K.; Dausse, E.; Sugiyama, H.; Toulmé, J.-J. Triggering Nucleic Acid Nanostructure Assembly by Conditional Kissing Interactions. Nucleic Acids Res. 2018, 46, 1052–1058. [Google Scholar] [CrossRef]
- Neves, M.A.D.; Slavkovic, S.; Reinstein, O.; Shoara, A.A.; Johnson, P.E. A Proof of Concept Application of Aptachain: Ligand-Induced Self-Assembly of a DNA Aptamer. RSC Adv. 2019, 9, 1690–1695. [Google Scholar] [CrossRef]
- Taton, T.A.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L. The DNA-Mediated Formation of Supramolecular Mono- and Multilayered Nanoparticle Structures. J. Am. Chem. Soc. 2000, 122, 6305–6306. [Google Scholar] [CrossRef]
- Storhoff, J.J.; Lazarides, A.A.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L.; Schatz, G.C. What Controls the Optical Properties of DNA-Linked Gold Nanoparticle Assemblies? J. Am. Chem. Soc. 2000, 122, 4640–4650. [Google Scholar] [CrossRef]
- Xia, F.; Zuo, X.; Yang, R.; Xiao, Y.; Kang, D.; Vallée-Bélisle, A.; Gong, X.; Yuen, J.D.; Hsu, B.B.Y.; Heeger, A.J.; et al. Colorimetric Detection of DNA, Small Molecules, Proteins, and Ions Using Unmodified Gold Nanoparticles and Conjugated Polyelectrolytes. Proc. Natl. Acad. Sci. USA 2010, 107, 10837–10841. [Google Scholar] [CrossRef]
- Tomaszewska, E.; Soliwoda, K.; Kadziola, K.; Tkacz-Szczesna, B.; Celichowski, G.; Cichomski, M.; Szmaja, W.; Grobelny, J. Detection Limits of DLS and UV-Vis Spectroscopy in Characterization of Polydisperse Nanoparticles Colloids. J. Nanomater. 2013, 2013, 60. [Google Scholar] [CrossRef]
- Bishop, G.R.; Ren, J.; Polander, B.C.; Jeanfreau, B.D.; Trent, J.O.; Chaires, J.B. Energetic Basis of Molecular Recognition in a DNA Aptamer. Biophys. Chem. 2007, 126, 165–175. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Non-Base Pairing DNA Provides a New Dimension for Controlling Aptamer-Linked Nanoparticles and Sensors. J. Am. Chem. Soc. 2007, 129, 8634–8643. [Google Scholar] [CrossRef]
- Song, K.-M.; Cho, M.; Jo, H.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold Nanoparticle-Based Colorimetric Detection of Kanamycin Using a DNA Aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef]
- Patel, M.; Dutta, A.; Huang, H. A Selective Adenosine Sensor Derived from a Triplex DNA Aptamer. Anal. Bioanal. Chem. 2011, 400, 3035–3040. [Google Scholar] [CrossRef] [PubMed]
- Slavkovic, S.; Zhu, Y.; Churcher, Z.R.; Shoara, A.A.; Johnson, A.E.; Johnson, P.E. Thermodynamic Analysis of Cooperative Ligand Binding by the ATP-Binding DNA Aptamer Indicates a Population-Shift Binding Mechanism. Sci. Rep. 2020, 10, 18944. [Google Scholar] [CrossRef] [PubMed]
- Nutiu, R.; Li, Y. Structure-Switching Signaling Aptamers. J. Am. Chem. Soc. 2003, 125, 4771–4778. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Perrier, S.; Ravelet, C.; Guieu, V.; Fize, J.; Roy, B.; Perigaud, C.; Peyrin, E. Rationally Designed Aptamer-Based Fluorescence Polarization Sensor Dedicated to the Small Target Analysis. Biosens. Bioelectron. 2010, 25, 1652–1657. [Google Scholar] [CrossRef]
- Tang, Z.; Mallikaratchy, P.; Yang, R.; Kim, Y.; Zhu, Z.; Wang, H.; Tan, W. Aptamer Switch Probe Based on Intramolecular Displacement. J. Am. Chem. Soc. 2008, 130, 11268–11269. [Google Scholar] [CrossRef]
- Zhu, Z.; Ravelet, C.; Perrier, S.; Guieu, V.; Fiore, E.; Peyrin, E. Single-Stranded DNA Binding Protein-Assisted Fluorescence Polarization Aptamer Assay for Detection of Small Molecules. Anal. Chem. 2012, 84, 7203–7211. [Google Scholar] [CrossRef]
- Halperin, A.; Buhot, A.; Zhulina, E.B. On the Hybridization Isotherms of DNA Microarrays: The Langmuir Model and Its Extensions. J. Phys. Condens. Matter 2006, 18, S463. [Google Scholar] [CrossRef]
- Fuchs, J.; Fiche, J.-B.; Buhot, A.; Calemczuk, R.; Livache, T. Salt Concentration Effects on Equilibrium Melting Curves from DNA Microarrays. Biophys. J. 2010, 99, 1886–1895. [Google Scholar] [CrossRef]
- Buhot, A.; Pingel, J.; Fiche, J.-B.; Calemczuk, R.; Livache, T. Biophysics of DNA: DNA Melting Curve Analysis with Surface Plasmon Resonance Imaging. In Introduction to Plasmonics: Advances and Applications; Szunerits, S., Boukherroub, R., Eds.; Pan Stanford Publishing: Singapore, 2015; pp. 61–88. ISBN 978-981-4613-12-5. [Google Scholar]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Chovelon, B.; Fiore, E.; Faure, P.; Peyrin, E.; Ravelet, C. A Lifetime-Sensitive Fluorescence Anisotropy Probe for DNA-Based Bioassays: The Case of SYBR Green. Biosens. Bioelectron. 2017, 90, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Ruta, J.; Perrier, S.; Ravelet, C.; Fize, J.; Peyrin, E. Noncompetitive Fluorescence Polarization Aptamer-Based Assay for Small Molecule Detection. Anal. Chem. 2009, 81, 7468–7473. [Google Scholar] [CrossRef] [PubMed]
- Perrier, S.; Guieu, V.; Chovelon, B.; Ravelet, C.; Peyrin, E. Panoply of Fluorescence Polarization/Anisotropy Signaling Mechanisms for Functional Nucleic Acid-Based Sensing Platforms. Anal. Chem. 2018, 90, 4236–4248. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.; Wang, H.; Le, X.C. Nucleic Acid Aptamers Improving Fluorescence Anisotropy and Fluorescence Polarization Assays for Small Molecules. Trac. Trends Anal. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Bai, Y.; Shu, T.; Su, L.; Zhang, X. Functional Nucleic Acid-Based Fluorescence Polarization/Anisotropy Biosensors for Detection of Biomarkers. Anal. Bioanal. Chem. 2020, 412, 6655–6665. [Google Scholar] [CrossRef]
- Fiche, J.B.; Buhot, A.; Calemczuk, R.; Livache, T. Temperature Effects on DNA Chip Experiments from Surface Plasmon Resonance Imaging: Isotherms and Melting Curves. Biophys. J. 2007, 92, 935–946. [Google Scholar] [CrossRef]
- Fiche, J.B.; Fuchs, J.; Buhot, A.; Calemczuk, R.; Livache, T. Point Mutation Detection by Surface Plasmon Resonance Imaging Coupled with a Temperature Scan Method in a Model System. Anal. Chem. 2008, 80, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Pingel, J.; Buhot, A.; Calemczuk, R.; Livache, T. Temperature Scans/Cycles for the Detection of Low Abundant DNA Point Mutations on Microarrays. Biosens. Bioelectron. 2012, 31, 554–557. [Google Scholar] [CrossRef]
- Jin, R.; Wu, G.; Li, Z.; Mirkin, C.A.; Schatz, G.C. What Controls the Melting Properties of DNA-Linked Gold Nanoparticle Assemblies? J. Am. Chem. Soc. 2003, 125, 1643–1654. [Google Scholar] [CrossRef]
- Park, S.Y.; Gibbs-Davis, J.M.; Nguyen, S.T.; Schatz, G.C. Sharp Melting in DNA-Linked Nanostructure Systems: Thermodynamic Models of DNA-Linked Polymers. J. Phys. Chem. B 2007, 111, 8785–8791. [Google Scholar] [CrossRef]
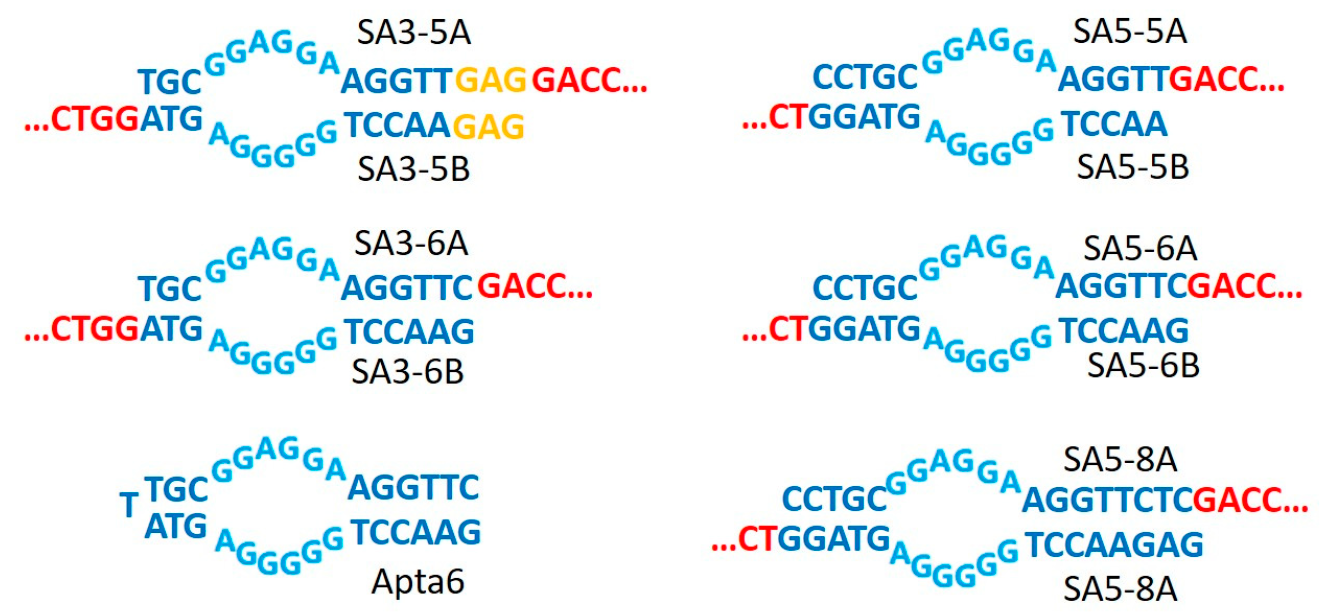

| Name | Oligonucleotide Sequences (from 5′ to 3′) |
|---|---|
| SA3-5A | TGCGGAGGAAGGTTGAGGACCATCGTGCGGGTAGGTAGACC |
| SA3-5B | GAGAACCTGGGGGAGTAGGTCTACCTACCCGCACGATGGTC |
| SA5-5A | CCTGCGGAGGAAGGTTGACCATCGTGCGGGTAGGTAGA |
| SA5-5B | AACCTGGGGGAGTAGGTCTACCTACCCGCACGATGGTC |
| SA3-6A | TGCGGAGGAAGGTTCGACCATCGTGCGGGTAGGTAGACC |
| SA3-6B | GAACCTGGGGGAGTAGGTCTACCTACCCGCACGATGGTC |
| SA5-6A | CCTGCGGAGGAAGGTTCGACCATCGTGCGGGTAGGTAGA |
| SA5-6B | GAACCTGGGGGAGTAGGTCTACCTACCCGCACGATGGTC |
| SA5-8A | CCTGCGGAGGAAGGTTCTCGACCATCGTGCGGGTAGGTAGA |
| SA5-8B | GAGAACCTGGGGGAGTAGGTCTACCTACCCGCACGATGGTC |
| Apta6 | GAACCTGGGGGAGTATTGCGGAGGAAGGTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Saint-Pierre, C.; Gasparutto, D.; Roupioz, Y.; Ravelet, C.; Peyrin, E.; Buhot, A. Melting Curve Analysis of Aptachains: Adenosine Detection with Internal Calibration. Biosensors 2021, 11, 112. https://doi.org/10.3390/bios11040112
Lu C, Saint-Pierre C, Gasparutto D, Roupioz Y, Ravelet C, Peyrin E, Buhot A. Melting Curve Analysis of Aptachains: Adenosine Detection with Internal Calibration. Biosensors. 2021; 11(4):112. https://doi.org/10.3390/bios11040112
Chicago/Turabian StyleLu, Chenze, Christine Saint-Pierre, Didier Gasparutto, Yoann Roupioz, Corinne Ravelet, Eric Peyrin, and Arnaud Buhot. 2021. "Melting Curve Analysis of Aptachains: Adenosine Detection with Internal Calibration" Biosensors 11, no. 4: 112. https://doi.org/10.3390/bios11040112
APA StyleLu, C., Saint-Pierre, C., Gasparutto, D., Roupioz, Y., Ravelet, C., Peyrin, E., & Buhot, A. (2021). Melting Curve Analysis of Aptachains: Adenosine Detection with Internal Calibration. Biosensors, 11(4), 112. https://doi.org/10.3390/bios11040112







